Abstract
Diffusion magnetic resonance imaging (dMRI) is a cutting-edge imaging method that provides a macro-scale in vivo map of the white matter pathways in the brain. The measurement of brain microstructure and the enhancement of tractography rely heavily on dMRI tissue segmentation. Anatomical MRI technique (e.g., T1- and T2-weighted imaging) is the most widely used method for segmentation in dMRI. In comparison to anatomical MRI, dMRI suffers from higher image distortions, lower image quality, and making inter-modality registration more difficult. The dMRI tractography study of brain connectivity has become a major part of the neuroimaging landscape in recent years. In this research, we provide a high-level overview of the methods used to segment several brain tissues types, including grey and white matter and cerebrospinal fluid, to enable quantitative studies of structural connectivity in the brain in health and illness. In the first part of our review, we discuss the three main phases in the quantitative analysis of tractography, which are correction, segmentation, and quantification. Methodological possibilities are described for each phase, along with their popularity and potential benefits and drawbacks. After that, we will look at research that used quantitative tractography approaches to examine the white and grey matter of the brain, with an emphasis on neurodevelopment, ageing, neurological illnesses, mental disorders, and neurosurgery as possible applications. Even though there have been substantial advancements in methodological technology and the spectrum of applications, there is still no consensus regarding the "optimal" approach in the quantitative analysis of tractography. As a result, researchers should tread carefully when interpreting the findings of quantitative analysis of tractography.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Image segmentation is significant for a variety of applications, such as image comprehension, feature extraction, processing, and interpretation. Tissue classification, tumour detection and volume estimation, blood cell outlining, surgical planning, atlas matching, and image registration are just a few of the many medical applications discovered in the scientific study of the human body [1]. Detecting pathology, monitoring illness development, or contrasting normal and abnormal subjects may all be accomplished with the use of mathematical feature extraction, modelling, and measurement approaches applied to images [2].
The abbreviations used throughout this review work can be found in Table 1.
To perform a range of dMRI visualisation and quantification tasks, including tractography and brain surface reconstruction, it is crucial to segment several types of brain tissues, such as WM, GM, and CSF. To better comprehend the structure, volume, integrity, and developmental changes in brain tissues and the blood–brain barrier (BBB), anatomical or structural MRI is often utilised [5, 6]. Examples of these types of images are T1- or T2w-weighted images, respectively.
Neuroscientists rely heavily on dMRI to learn about the structure and function of the brain's connections. dMRI is an imaging method that does not require any kind of invasive procedure, as it uses the movement of water molecules in tissue to provide contrast in the final image. It is difficult to apply anatomical MRI-based segmentation to dMRI because of low image resolution and distortions caused by echo-planar imaging (EPI) (which can significantly shorten the MRI imaging times) [7]. Acquiring more information with a dMRI can solve these issues. To facilitate registration between anatomical MRI and dMRI data, the Center for Biomedical Image Computing and Analytics (CBICA) collected high-resolution dMRI data utilising alternative phase encoding [8] to remove EPI distortions.
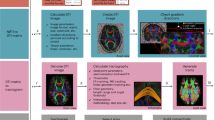
Although in many clinical scenarios when scan time is restricted, CBICA acquisition is not always accessible and is not currently viable. Therefore, techniques for segmentation which work well with dMRI data are needed. When high-quality anatomical data are missing from MRI protocols or when segmentation based on anatomical MRI is difficult to register to the dMRI space, such approaches may be helpful. MD measures the microstructural integrity, and FA is an inverse measure of the membrane density. These are often used as rotation-invariant characteristics in dMRI-based brain tissue segmentation algorithms, which are based on DTI models that give non-invasive information about the microstructures of WM in the central nervous system (CNS). Brain MRI segmentation is a fundamental step in neurology that has a wide range of applications, including quantitative analysis, operational planning, and functional imaging [1, 2]. However, medical image segmentation is a difficult task, even though MRI can accurately describe brain structures, due to poor spatial resolution and low contrast, as well as uncertain boundaries, inhomogeneity and other acquisition artefacts such as noise and object shape variability. This is compounded by a lack of anatomical models that can fully capture the possible deformations in each structure [10,11,12]. Implementing a few easy procedures as suggested in [13,14,15] can address these issues. Tumours are difficult to segment because of their diverse appearance, which includes their potential to shift in terms of size, shape, and recurrence [12]. Despite these problems, significant progress has been made in instrumentation and computer technology, resulting in major success in this field (Fig. 1).
a T1w image, b white matter (WM), c grey matter (GM), and d cerebrospinal fluid (CSF). Figure adapted from Ref. [14]
It is difficult to keep track of all the different terminology used in dMRI quantitative tractography analysis because there are so many different methodologies that have been offered over the past 2 decades. Table 2 provides a list of terms and their definitions that we will use throughout this work. In addition to the article, we also provide a graphical representation of many essential concepts that will be used in this paper (Fig. 2).
A graphic illustration of tractography is shown in this diagram. (a) An example of DWI data, is often known as dMRI. (b) Tractography was used to generate an individual streamline. (c) An example of a whole-brain tractography is made up entirely of white matter streamlines. (d) A corpus callosum is a group of streamlines that reflect an anatomical fiber tract. FA, a microstructural measure of anisotropy of water diffusion, is used to color the lines. When looking at a stream, a low FA can be noticed near the cortex and a high FA can be seen in between the two (in the deep white matter). (e) White matter tractography from the entire brain is used to create an example of a brain structural connection matrix. White matter connections between two brains' gray matter ROIs are represented by a matrix, where each row and column represent a gray matter ROI (see c for an example of brain gray matter parcellation). The value in an element of the matrix denotes the strength of this connection (quantified as the number of streamlines in this case). Figure adapted from figure 1 [53].
Quantitative tractography can be used to analyse the brain's structural connectivity in health and disease, (see Sects. 4, 5). The primary purpose of these analysis is to determine quantitative measures of connectivity (or microstructure) of a particular pathway (or pathways). Quantitative tractography analyses are divided into two primary types or styles: tract-specific studies and connectome-based analysis (this categorization is helpful but inaccurate, as some approaches incorporate aspects of both analytic styles). These studies are often hypothesis-driven and focus on specific anatomical fibre pathways [13,14,15,16]. The tract-specific analysis is becoming more popular, particularly for studying local white matter regions in health and disease person. Data-driven studies of the brain's structural connectivity are known as connectome-based analysis [17,18,19]. Tractography over the complete white matter is required for this type of analysis, which tries to understand the patterns of whole-brain connections.
Recent advances in deep learning have greatly improved the results of image segmentation [34]. Initially, algorithms like support vector machines (SVM) and non-negative matrix factorization (NMF) [35] were viewed as classic machine-learning approaches. Deep learning algorithms have recently been used in studies to increase segmentation performance. The convolutional neural network (CNN) is now the most widely used deep learning model, which is meant to automatically and adaptively learn spatial hierarchies (details) of an image [32] from low- to high-level patterns [39]. While CNN has been used for brain segmentation in multi-modal segmentation, which incorporates dMRI, its tissue segmentation performance has shown great promise [40]. However, the accuracy of cross-modality registration, which is essential to produce the multi-modal input utilised for segmentation, is still a concern for this type of application. It is impossible to develop only one standard method that suits all imaging applications.
Key contributions
An overview of the quantitative investigation of segmentation and brain structural connectivity using tractography is provided in this paper. This review is designed to help researchers studying white matter segmentation, developers of quantitative analysis methods, and clinicians evaluate tractography results.
The highlights of this study are as follows:
-
1.
Presented an exhaustive study of improving the quality of brain tissue segmentation from diffusion MR images.
-
2.
The study assists clinicians in making appropriate diagnoses and subsequent treatment decisions.
-
3.
The performance and feasibility of modern systems have been demonstrated by quantitative analysis using various metrics.
-
4.
In addition, it provides readers with novel future research directions for the segmentation of brain tissue.
The rest of the paper is planned as follows: we begin with an overview of the computational processes that were used to analyse diffusion MRI data for brain fibre tractography (“An overview of the computational processes that were used to analyse diffusion MRI data for brain tissue segmentation”), followed by a review of qualitative brain tissue segmentation analysis, techniques and approaches (“Methods for improving the quality of Brain Tissue Segmentation”, “Implementing quantitative analysis: quantification approaches for tractography”), and finally a review of research that employs applications of quantitative tractography analysis to study the brain in health and disease (“Applications of quantitative tractography analysis”).
An overview of the computational processes that were used to analyse diffusion MRI data for brain tissue segmentation
In brain tissue segmentation, tractography can refer to any computational approach that calculates the anatomical trajectories of white matter fibre routes using dMRI data. Water molecule diffusion in living tissues can be measured in vivo using DWI and DTI [40]. For example, a particular tissue structure may preferentially restrict the molecular motion of water molecules, resulting in anisotropic diffusion, which is evaluated by DWI and DTI [41, 42]. To produce a 3-D field of diffusion tensors, the observed diffusion can be treated as an anisotropic Gaussian, using each voxel's diffusion tensor as a parameter [43]. It is possible to extract diffusion anisotropy from diffusion tensor measurements by applying mathematical procedures and recalculating the underlying eigenvalues.
Although this study does not include an in-depth introduction to tractography methods, we recommend that the reader study the following review papers [46, 49, 50, 52, 53] that are specific to tractography algorithms. Tractography research can be carried out using a variety of software tools, including the following: ANIMA [45], BrainSUITE [46], Camino [47], COMMIT [48], Diffusion toolkit [49], Dipy, DMIPY [50], DSI studio [51], ExploreDTI, FiberNavigator [52], FSL [53], MITK [54], MRtrix3 [55], PANDA [56], SlicerDMRI [57], TractSeg [58], and Tracula [59]. Using tractography data in any subsequent quantitative analysis is risky because of the known sensitivity of tractography to the underlying fibre-tracking techniques.
Table 3 includes important websites with free software programmes for anatomical MRI and DWI data preparation, conversion, inspection, tissue segmentation, and visualisation. An easy-to-use GUI for all OS systems, MRIcro can convert 2D (DICOM http://medical.nema.org/) to 3D (e.g., Analyze http://mayoresearch.mayo.edu/mayo/research/robb_lab/analyze.cfm) or 4D neuroimaging informatics technology initiative (NIFTI http://nifti.nimh.nih.gov/). It is possible to prepare DWI and batch processes [61] for huge data sets using command-line tools like AFNI, FreeSurfer, FSL and SPM.
A wide range of diffusion models [62, 63] and tractography methods [64, 65, 66] have been used to produce tractography results. Tractography results, on the other hand, can be affected by algorithmic parameters (such as seeding and stopping thresholds) [5]. Despite numerous studies comparing various tractography algorithms [44], no single study has been able to agree on “the best method”.
Methods for improving the quality of brain tissue segmentation
In this section, the state-of-the-art approaches and current breakthroughs in improving the quality of Brain Tissue Segmentation have been discussed.
Gegenbauer method of image reconstruction
The Gegenbauer image reconstruction method proposed by [44] is capable of reconstructing images with exponential precision, including the edges and structures in the image, without blurring any features, hence eliminating a common issue of image filtering (i.e., blurring of edges). In every high-resolution reconstruction, the first step is to figure out where the edges are. The location of the edges determines the areas in which the images can be reconstructed. The minimising technique introduced in [90] is combined with the edge-detecting method [88, 89]. The methods for edge detection and Gegenbauer reconstruction are discussed briefly in [91].
Threshold-based method
Thresholding is a popular segmentation approach that compares the intensity values of the target objects to one or more thresholds. Intensity thresholding is another term for this technique. The threshold values might be both global and local. There are two types of thresholding: (1) fixed thresholding and (2) adaptive thresholding (see Fig. 3).
In thresholding-based techniques, T2-weighted MRI are used. Pixels that are above the threshold level are assigned to a group, whereas pixels that fall below the threshold are classed as background. However, there are several artefacts present in the MRI image of the object of interest. When it comes to the object of interest, fixed thresholding-based approaches incorporate criteria such as entropy or between-class variation. A single threshold value can distinguish an image histogram with a bimodal pattern from the background. It assigns intensity values above and below the threshold to one and zero, respectively. Images are segmented by the global threshold T supplied in I (x, y), where pixels with a value of 1 indicate an object and pixels with a value of 0 indicate a background. Such a technique relies heavily on statistical fluctuations to achieve a high level of segmentation accuracy. Selecting a threshold in an increasingly complex landscape becomes more difficult as the number of areas increases. In terms of brain tissue segmentation, it should be observed that more than two tissues are required (i.e., WM, GM, and CSF).
Mixture models
The intensity levels of different substructures and tissues in brain MR images vary significantly. Images are characterised using statistical mixture models. The maximum-likelihood (ML) or maximum posterior probability (MAP) criterion is used to assess the probability distribution of intensity in an image. Gaussian Mixture Model (GMM) is a well-known statistical model in neuroscience that is frequently employed. A Gaussian distribution is used to estimate the intensity of pixels (or voxels) in a given region. The GMM parameters are then estimated using the expectation maximisation (EM) technique, which maximises the likelihood of the observed image.
Methods for feature extraction and classification
Brain tissue segmentation relies heavily on feature extraction and classification methods. T2-weighted MR images are used in this procedure. The fundamental objective of this strategy is to identify the most effective and discriminating elements in the MRI brain image. After that, classification is carried out based on the distinguishing characteristics. DWT, Gabor filter, and statistical approaches such as grey level co-occurrence matrix and grey level run length matrix [12] have been published in the literature. Due to distortions like noise and IIH, feature extraction from MRI is still difficult. In addition, feature extraction methods are limited by excessive dimensionality. There are several techniques for dealing with dimensionality, including PCA (principal component analysis), LDA (linear discriminant analysis), and so on. They can identify a small range of features that are important for correct classification.
Parcellation/division of the whole-brain tractogram segmentation
A whole-brain tractogram, which depicts the white matter of the entire brain, is used to enable quantitative analysis of all potential white matter connections in the entire brain (Fig. 4).
a Whole-brain tractogram has been calculated using tractography on DWI data, b anatomical tracts have been recovered from the tractogram. c Finally, a structural connection matrix has been constructed by performing whole-brain tractography segmentation across all pairs of FreeSurfer cortical areas. Figure adapted from [51]
Fiber clustering and cortical parcellation methods are the two most commonly used approaches [31]. A connection matrix can be constructed using the cortical-parcellation-based approaches, which can then be analysed using graph theory techniques (as discussed in Sect. 5.6) [42, 56].
However, despite their relative lack of use, fibre clustering tractography parcellation methods continue to be utilised in applications such as disease categorization and between-population statistical analysis. As a side note, the terminology "fibre clustering" has been widely used in the scientific literature, even though "streamline clustering" is a more accurate term. Cortical parcellation-based techniques focus on grey matter, not the brain's white matter. They focus on the structural connection between various grey matter ROIs when parcelling tractography according to a cortical (and sometimes a subcortical) grey matter parcellation [17, 21, 27, 31, 49, 56, 67, 68]. Tissue segmentation is done by obtaining the streamlines that connect two ROIs. Therefore, the choice of a cortical parcellation scheme has a major impact on the final tractography segmentation. T1 or T2 weighted MRI is used to compute cortical parcellation in the majority of approaches. Freesurfer Desikan–Killiany cortex parcellation is the most commonly used cortical parcellation; however, several other cortical parcellation methods have been widely employed [18, 39, 46].
In addition, a functional cortical parcellation calculated using functional MRI data has been used in a variety of cortical-parcellation-based approaches. It is also possible to look at the cortical surface as a “parcellation”, i.e., locate the lines that connect the vertices of the cortex. However, as of now, there is no consensus on which brain parcellation technique is the most effective.
Implementing quantitative analysis: quantification approaches for tractography
The goal of tractography quantification is to retrieve quantitative measurements that may be used to examine the structural connectivity of white matter pathways in the brain. This section introduces quantitative measurements that can be computed from tractography (“White matter tractography”). In “The domain of analysis: measurement of white matter fibre pathways in the brain”, we will discuss how to extract these measurements from each white matter fibre pathway and how to execute filtering techniques to limit the risk of bias in the retrieved measures (“Quantitative analysis of data using filtering techniques”). We explain how the retrieved metrics can be employed for tract-specific investigation of anatomical white matter tracts (“Diffusion tensor imaging-based quantitative tractography (qtDTI)”). Finally, we explain Statistical or machine-learning approaches for fibre tract-tissue analysis (“Statistical or machine learning approaches for fibre tract-tissue analysis”).
White matter tractography
White matter tractography techniques estimate the connection patterns between distinct brain areas using local estimations of fibre orientation at each voxel. There have been numerous WMT algorithms to date. These algorithms begin by selecting a starting point or seed, which can be either a voxel or an exact place that can be characterised by Cartesian coordinates in the brain space. There are three types of WMT algorithms: deterministic, stochastic, and global. Each seed point is assigned a unique trajectory by the deterministic algorithms. This path connects two distinct areas of the brain in the final analysis. In probabilistic algorithms, propagation fronts or Monte Carlo approaches [36,37,38,39,40,41,42,43] are used to produce multiple alternative seed pathways. Weights are used in these algorithms to link the seed to a particular cluster of voxels or regions of the brain. Global optimization methods [44,45,46,47] use smoothness and signal fit to find the best path between two brain regions.
The domain of analysis: measurement of white matter fibre pathways in the brain
After the selection of a quantitative measure of interest (“White matter tractography”) and a brain tissue segmentation approach (“Methods for improving the quality of brain tissue segmentation”), there are a variety of methods for extracting the measure inside a single white matter fibre pathway. This is necessary to conduct tract-specific analysis (“Diffusion tensor imaging-based quantitative tractography (qtDTI)”) and to generate a connectivity matrix (“Statistical or machine learning approaches for fibre tract-tissue analysis”). A scalar value can be used to summarise data, or data collected along the fibre path can be used. Scalar values are the most often used method for extracting quantitative information from fibre pathways. For tract-specific analysis, this is a common method to connectome-based analysis, which is necessary. While some quantitative metrics (e.g., NOS or tract volume) yield a single scalar value per pathway, others (e.g., sampled values of a quantitative metric) involve the calculation of some statistic to obtain such a scalar. The most commonly utilised microstructural measure is the mean of the fibre path. Additionally, other summary statistics such as the median, maximum, and lowest were used [50, 53]. According to how microstructural metrics are computed along fibre paths, a summary statistic can be generated in a variety of methods. In a microstructure image, it can be calculated within a binary mask that defines a fibre pathway (e.g., the mask means FA) [4, 19, 44, 47]. Additionally, it can be estimated using the average of each point’s microstructural measurements [28, 32, 39, 53, 69]. We are still debating which summary statistic is the most appropriate for data collected along a fibre pathway. When comparing outlier-resistant statistics such as microstructure parameter distributions along a streamline to mean distributions, researchers have found that studies have indicated that the median statistic is more robust than the mean statistic [50, 53]. According to one study, the maximum and minimum values in machine learning-based disease classification are more discriminative than the mean value [59].
Quantitative analysis of data using filtering techniques
The filtering methods outlined in “Mixture models” can also be utilised to produce "microstructure-informed" tractography, which aims to derive quantitative estimates of white matter fibre routes and eliminate potential bias in connection estimation. These approaches assign a specific microstructural property to each reconstructed path, assuming that the microstructure attributes corresponding to a single streamline remain constant. This is based on the fact that the microstructure properties of the magnetic resonance signal are sensitive to a feasible resolution, and so we can assume that on average, the microstructure qualities of a single axon are not affected by a group of axons following the same trajectory. A whole-brain tractogram is converted to pixel-wise fibre density using SIFT [15], SIFT2 [16], which offers a value for each streamline that may be used (for example) to determine the connectome edge weights. Deconvolution of the dMRI signal measured on the streamlines is similar in LiFE [33], COMMIT [15, 16, 38], COMMIT 2 [37], and COMMIT2tree [31] utilising traditional multicompartment models [32] assign a single contribution, or "weight," to each of them. We have seen better results than with the usual NOS when using these methods to explore the characteristics of both healthy and diseased brains. Complementing tractography with biophysical models of tissue microstructure such as the COMMIT, COMMIT2 and COMMIT2tree allow one to access more quantitative and biologically informative features of individual bundles, such as average axon diameter, myelin content and bundle-specific T2 [25].
Diffusion tensor imaging-based quantitative tractography (qtDTI)
Scalar metrics and tractography are employed in qtDTI technology to estimate bundle-specific properties that characterise the structural aspects of fibre bundles, such as average fiber bundle length (FBL), total length and average scalar metrics (AD, RD, MD, and FA). Anisotropy-weighted FBL and intracranial volume (ICV)-normalized length can also be combined to generate more sophisticated composite measurements [17]. Tractography can be halted due to microstructural changes that reduce anisotropy or cause sudden changes in fibre orientation [36)] Microstructural abnormalities and variations in bundle metrics can be used to diagnose white matter damage associated with inflammation in multiple sclerosis, which can produce these microstructural changes [42]. Tract-specific changes can be detected with the use of qtDTI technology rather than traditional DTI measurements like FA, which are more limited in their ability to detect tract-wide changes [17]. White matter structure and function in healthy and diseased populations should benefit greatly from this novel method.
Statistical or machine learning approaches for fibre tract-tissue analysis
Once a quantitative measure for individual fibre routes has been obtained, several tract-specific analytic techniques are available, including hypothesis-driven statistical analysis and data-driven machine learning. These approaches may use a summary statistic for each tract, or they may analyse data along a particular tract. For example, a hypothesis-driven approach is utilised to examine whether there are differences in the tracts of interest between groups (e.g., between health and disease, or between different subtypes of a disease). Assuming that the chosen microstructural measure has been extracted from the tracts of interest, statistical group-wise comparison methods such as the Student’s t test, ANOVA, or other more advanced statistical analysis methods are used to compute the level of group differences such as p values [18, 33, 38, 85]. Using a regression model (e.g., a generalised linear model [12] or support vector regression [36]), the correlation between the tract quantitative measurement and a behavioural or disease symptom score can be evaluated. It has been used to explore how white matter fibre pathways are impacted by clinical outcomes [42, 70, 80] and how they grow during human neurodevelopment [17, 29, 60]. If there are several quantitative measurements and/or multiple tracts evaluated, it is necessary to compensate for multiple comparisons. It is not uncommon for researchers to employ false discovery rate (FDR) and Bonferroni corrections to adjust multiple comparisons [20, 42].
Data-driven and machine learning techniques are used to perform tasks like illness categorization and prediction in another tract-specific analysis strategy. Individual fibre paths are handled as feature descriptors in machine learning analysis, and feature descriptors are used to train a model using a set of training samples that have known information (such as a support vector machine) (e.g., labels such as disease or healthy control). The model can then be used to forecast new samples after it has been properly trained. The machine-learning-based methods, on the other hand, strive to provide predictive relevance instead of only finding white matter structures with statistical differences (Table 4).
Applications of quantitative tractography analysis
Recent advances in dMRI-based tractography, which provides quantitative measures of fibre tracts, have expanded neuroanatomy methodologies and become a crucial technology for studying the brain's white matter in a variety of applications over the last 2 decades. This section focuses on various important topics, such as development, ageing, neurological illnesses, mental disorders, and neurosurgery. These findings can be divided into two categories, one for tract-specific studies and the other for analyses based on the connectome. The first category focuses on main fibre channels or localised structural connections to capture the fine-grained characteristics of brain circuits. Clustering analysis and graph theoretical analysis is used in the second category because they view the white matter as a complex system and hence use these techniques to reduce features or quantify topological properties.
Mapping of specific white matter tracts
White Matter Tracts have been widely employed to illustrate in vivo mapping of the brain's white matter pathways [6,7,8,9, 51, 52, 57, 58, 71,72,73,74,75,76]. There were initial efforts to replicate the major fibre patterns of classical anatomy in WMT investigations. WMT was able to detect the corpus callosum, superior longitudinal fasciculus, corona radiata fibres, and front-occipital fasciculus fibres. The WMT fibre reconstruction's closeness to known anatomy was the first confirmation of WMT's ability to map discrete white matter structures noninvasively. Very few anatomical structures may be located on an individual specimen through the use of invasive post-mortem anatomical techniques including fibre dissections and fibre tracing operations (as unveiling a structure of interest involves a specific sequence of dissection steps, which render unusable the specimen for future investigations). When using WMT, you can perform as many ‘in vivo’ anatomy and physiology as you like on a single dataset because it is a three-dimensional computational technique. Recent tractography research has provided new insights into the anatomy and development of brain segmentation. As a result of these findings, we now have a better grasp of the linguistic pathways [74] and the U-fibers [75] as well as other connection fibres in the brain.
White matter parcellation
WMT can be used to segment the brain volume occupied by several white matter tracts. Consequently, WMT allows for the segmentation of white matter into various regions that correspond to different structures. Using any other imaging approach, this information would be impossible to obtain. The segmented tracts can be analysed using anisotropy, and diffusivity data, or data from other imaging modalities, for morphometric analysis (volumetric, cross-sectional area, etc.) According to the constituent structures, white matter can be segmented as illustrated in Fig. 5. This approach has lately been used to study white matter integrity in a variety of brain disorders. Frontal and limbic lobe tract segmentation in patients with schizophrenia [77] and temporal epilepsy [78], frontostriato-thalamic connections in Tourette syndrome [79], and hippocampo and amygdalo fusiform pathways in autism [80] are a few instances. Anisotropy and diffusivity variations between populations were studied using ROIs derived from the segmented volumes. White matter microstructure in both normal and sick populations was analysed.
White matter tractography can mark brain voxels according to their white matter structure. Image tractogram of the projection (green), association (red), and callosal (blue) fibres are mapped into the brain space to show their position in relation to other brain regions. External capsule (ec); internal capsule (ic); posterior region of corona radiata (pcr). (b) A similar technique was implemented to identify the corpus callosum (purple), superior longitudinal fasciculus (yellow), cingulum (green), uncinate fasciculus (dark red), inferior occipitofrontal fasciculus (orange), inferior longitudinal fasciculus (brown), corticobulbar tract (light blue), corticospinal tract (white), fornix, and stria terminalis (light yellow). Several sagittal and axial slices indicate tract locations. Figure adapted from [92]
Gray matter parcellation
Primate brain connectivity maps show that different parts of the brain have different connectivity patterns [81]. Using WMT, researchers have been able to identify distinct regions of grey matter based on patterns of connection [82, 83, 84]. Clustering was used to differentiate the supplementary motor area (SMA) from the pre-SMA regions of the premotor cortex, based on unique patterns of distributed connectivity across the brain. In addition to the parietal cortex, Broca's area, and cingulate gyrus, other grey matter regions have been segmented using these techniques.
Development of brain fibre tractography
The development from a neural tube to a multilayer connectome requires highly organised mutations in white matter. DMRI tractography has been used to study brain development from the middle fetal stage through adulthood over the past decade [14, 46, 49, 53, 55, 60, 61]. When it comes to the early stages of development, most tractography investigations are focused on discovering the pathways that neurons use to migrate and the creation of major tracts in the brain. It is possible to see the fornix and cerebellum bundles at 13 weeks of gestation using ex vivo dMRI images on infant brain tissues, but only the corpus callosum is visible by 15 weeks [26]. Studies reveal that the basic structural pathways are already recognised by term, and the radial and tangential pathways organisation are evident by 17 weeks [42]. Traditional histology research [24, 48] supports these findings. Methodological breakthroughs like motion correction algorithms have made it possible to conduct in vivo tractography on infants’ brains, and these studies show tremendous potential for future early brain investigations. By applying quantitative dMRI measurements from the early years [24, 46, 46, 53, 61] to the adolescent stage [25, 43], we may better map the spatiotemporal maturation patterns of rebuilt tracts. These studies, despite focusing on diverse developmental ranges, consistently report large age-related increases in FA and MD in extensive fibre tracts during postnatal growth [53]. For long tracts of development, such as from infancy to childhood or from adolescence to maturity [33,34,35] or from adolescence into adulthood, studies have also characterised many typical nonlinear diffusion trajectory (piecewise, exponential and quadratic) metrics. According to one study, researchers can use multivariate analysis to estimate the growth of tracts at the subsystem level; for example, they can find out how many months it takes for a linguistic bundle to mature by looking at its maturational calendars [25].
Brain fibre tractography in neurosurgery
Tractography in vivo is essential for the planning and implementation of surgery by neurosurgeons because white matter tracts that have been displaced or otherwise impacted by the tumour can be visualised and localised. Aside from a few hitches, tractography has shown to be a useful tool for neurosurgery patients in terms of pinpointing lesions and improving their quality of life and overall survival. However, in vivo tractography provides critical spatial information about particular anatomical pathways for surgery planning, even though quantitative image-based metrics of local tracts are of limited utility in the field of neurosurgery. The trade-off between preserving function and maximising resection is critical in malignant lesion surgery. Direct electrical stimulation (DES) is the gold standard for intraoperative identification of eloquent regions [26, 27, 32, 41]. DES intensities and distances to reconstructed tracts have recently shown a high degree of connection when used with recent intraoperative MRI-based in vivo tractography applications [32]. It has also been shown that intraoperative MRI can identify fibre tracts that were not previously detected in preoperative tracings [16]. Intraoperative tractography can help patients with malignancies near the superior longitudinal fasciculus retain their ability to speak when used in conjunction with navigated transcranial magnetic stimulation (nTMS) [32, 41]. Tractography estimation may be the sole viable option for measuring the white matter tracts involved in language area surgery when awake operation is impractical or unavailable. There have been numerous studies using in vivo tractography to examine lesions in the supratentorial and infratentorial regions of the brain [31, 37, 40], examining not only the larger tracts but also the finer fibre structures [87]. The traditional surgical technique could benefit from a move away from local tumour topography to network-guided "oncological disconnection surgery" if the brain connectome idea is adopted [28, 85, 86]. Classical neurosurgery holds that cancers involving the sensorimotor, language, and visual cortices, which are all considered to be "eloquent" brain regions, should not be removed, whereas tumours in other, "non-eloquent" regions can be removed with little thought [26]. A consistent location of "eloquent" areas across patients appears erroneous, however, because of the possibility of functional compensation and the presence of individual variance [25]. For functional mapping, researchers have combined tractography atlases with intraoperative electrical stimulation to create probabilistic atlases of white matter pathways and structural hubs that define a minimal common brain connectome with low inter-individual variability and low postlesional compensation potential. It may be possible to use information from previous "structural and functional skeletons" of the brain to guide the removal of specific neural tracts during tumour surgery [28].
Limitations
Several drawbacks of qtDTI technology should be addressed. To begin, qtDTI is highly sensitive to imaging errors driven by partial-volume averaging of fibre bundle populations with varying degrees of myelination, orientation, and/or axon diameter. Partial-volume confounds can be avoided to some extent by reducing the voxel size and increasing gradient strengths and the number of orientations. These limitations likewise apply to tractography, which is predicated on the underlying assumptions of the diffusion tensor analysis. In general, DTI calculates the average water diffusion behaviour (including its anisotropy features) inside the pixel volume and makes the assumption that the water diffusion only shows one preferred orientation of motion. This is because DTI assumes that the anisotropy of the water diffusion is constant. Therefore, volume averaging may become a major source of inaccuracy when the image pixel size is raised or if disproportionally high slice widths are employed (both of which are frequent methods to improve SNR). Moreover, magnetic field inhomogeneity may occur at the brain and bone interface when the rapid echo-planar MRI technique is utilised to acquire images [18]. Furthermore, when the pixel volume has two or more fibre populations crossing at nonparallel angles, such as when fibres cross, the one-fiber population diffusion tensor model may fail. Several methods, such as q-space imaging [19] and high angular resolution diffusion imaging [20,21,22,23], may prove useful for resolving multi-population fibre structures. Finally, small changes in brain architecture might confound tractography investigations. Procedures must be developed so researchers can seed the areas they wish to examine. Atrophy presents a unique set of difficulties for research involving older populations. DTI signals could be reduced as a result of these sorts of difficulties [46]. When it comes to detecting and identifying white matter pathways, diffusion tensor imaging based tractography has a lot of promise for a number of possible applications. It is possible that this will allow for a better understanding of how a change in the brain circuitry is connected with neuropsychiatric disorder.
Discussion and conclusion
Tractography provides a high-level overview of how it can be used to quantitatively analyse the anatomical connectivity of the brain in health and disease. For quantitative analysis of tractography, we looked at the methodologies that are involved in each of the primary processing steps. The white matter of the brain has also been studied using quantitative tractography approach.
Researchers and clinicians who use tractography in their work should use the utmost caution when interpreting biological findings from quantitative results. There is no direct correlation between reconstructed streamlines and nerve fibres [23, 51], and fundamental diffusion metrics are just assumptions from local diffusion properties, which are not direct measures of tissue qualities [11]. To avoid errors in quantitative dMRI analysis and brain connection studies utilising tractography, several review papers have been published [10, 27, 31, 36].
Research is underway to improve biological specificity to the type of tissue change, by improving the information that is obtained at the acquisition level [19, 24, 28, 30, 39, 48] by proposing advanced mathematical modelling and machine learning techniques [33, 37, 49]. False-positive and false-negative tracking outcomes are common in quantitative tractography analysis [46, 79].
Tractography has made it possible to examine the connections in the brain's white matter in both healthy and diseased individuals for their lives. Overall, we conclude that, despite significant advances in methodological technology and application breadth, there is still no consensus regarding the "optimal" approach for quantitative tractography analysis, and researchers should exercise caution when interpreting results in research and clinical applications.
Brain tissue segmentation: future applications and innovations
As a novel technique, qtDTI is still in its early stages of development, and additional study is needed to determine reproducibility and reliability. Studies using histological techniques and known indicators of white-matter disease will support the method validity (e.g., WMH). For further information on how white matter integrity and cognitive development in children are linked, we can use qtDTI technology. DTI, in combination with fibre tractography, may help us learn more about the ways in which different parts of the brain are linked to one another and how these connections affect things like emotional stability and memory. This may improve our understanding of how the brain functions, but to do such work we must couple DTI with measures of function, including fMRI studies or neuropsychological testing. Inter-regional connections in the brain are particularly dense in complex cognitive areas like mood control [53]. DTI has the potential to become yet another helpful tool as we move forward with this work. It will also help us learn more about the potential role of connection abnormalities in neuropsychiatric diseases. Till now, the majority of neuropsychiatric studies utilising this method have compared people with mental illness to healthy controls to look for abnormalities. This is an essential beginning: it can support existing hypotheses of the neurological underpinnings of various diseases and help to identify regions demanding of further investigation.
Furthermore, measurements (for example, FBL) may be used to improve a given organism's sensitivity to injury and abnormal growth. qtDTI measures can be applied to infants and children, but further research is needed to determine their relevance, particularly with myelin water fraction mapping [8]. Experimenting with diffusion models that are more complex than the single-tensor model can provide a wealth of new information. Crossing bundle reconstruction and the quantification of characteristics such as fibre dispersion and free water contamination could benefit from these techniques. This constraint is presently being addressed through the use of methods that use more advanced models, such as limited spherical convolution, diffusion ball-and-sticks and neurite orientation dispersion and density imaging [53]. In addition, there is much to understand about the qtDTI and other imaging modalities interaction. When qtDTI and fMRI are used together, the model of brain integrity can be better understood since it provides a parallel perspective of both structural and functional integrity.
Data Availability
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Change history
23 June 2023
A Correction to this paper has been published: https://doi.org/10.1007/s13760-023-02301-2
References
Hasan A, Meziane F, Aspin R, Jalab H (2016) Segmentation of brain tumors in MRI images using three-dimensional active contour without edge. Symmetry 8(11):132
Gordillo N, Montseny E, Sobrevilla P (2013) State of the art survey on MRI brain tumor segmentation. Magn Reson Imaging 31(8):1426–1438
Bahadure N, Kumar Ray A, Pal TH (2017) Image analysis for MRI based brain tumor detection and feature extraction using biologically inspired BWT and SVM. Int J Biomed Imaging 2017:1–12
Wong KP (2005) Medical image segmentation: methods and applications in functional imaging. In: Suri JS, Wilson DL, Laxminarayan S (eds) Handbook of biomedical image analysis. Topics in biomedical engineering international book series. Springer, Boston. https://doi.org/10.1007/0-306-48606-7_3
Jeurissen B, Tournier J-D, Sijbers J (2015) Tissue-type segmentation using non-negative matrix factorization of multi-shell diffusion-weighted MRI images. In: ISMRM 23th Annual Meeting, Toronto, Ontario, Canada, volume 23, p. 349
Kumazawa S, Yoshiura T, Honda H, Toyofuku F, Higashida Y (2010) Partial volume estimation and segmentation of brain tissue based on diffusion tensor MRI. Med Phys 37(4):1482–1490
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324
CBICA Homepage, https://ipp.cbica.upenn.edu/. Last accessed 10 May 2022
Mah Y-H, Jager R, Kennard C, Husain M, Nachev P (2014) A new method for auto- mated high-dimensional lesion segmentation evaluated in vascular injury and applied to the human occipital lobe. Cortex 56:51–63
Kumar R, Rani S, Sarkar A, Talukdar FA (2017) GPU-based level set method for MRI brain tumor segmentation using modified probabilistic clustering. IGI Global, pp 1053–1078
Ilunga-Mbuyamba E, Avina-Cervantes JG, Garcia-Perez A, de Jesus-Romero-Troncoso R, Aguirre-Ramos H, Cruz-Aceves I (2017) Localized active contour model with background intensity compensation applied on automatic MR brain tumor segmentation. Neurocomputing 220:84–97
Soltaninejad M, Zhang L, Lambrou T, Yang G, Allinson N, Ye X (2017) MRI brain tumor segmentation and patient survival prediction using random forests and fully convolutional networks. In: Int. MICCAI Brain lesion Workshop. Springer. pp 204–15
Parveen SA (2015) Detection of brain tumor in MRI images, using combination of fuzzy C-means and SVM. In: 2nd Int. Conf. Signal Processing and Integrated Networks (SPIN), pp 98–102
Soltaninejad M, Zhang L, Lambrou T, Yang G, Allinson N, Ye X (2017) MRI brain tumor segmentation using random forests and fully convolutional networks. In: International MICCAI Brain lesion Workshop, pp 279–83
Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A (2018) Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats’ challenge. arXiv preprint https://arXiv.org/181102629
Steven AJ, Zhuo J, Melhem ER (2014) Diffusion kurtosis imaging: an emerging technique for evaluating the microstructural environment of the brain. Am J Roentgenol 202(1):26–33
Sun P, Wu Y, Chen G, Wu J, Shen D, Yap P-T (2019) Tissue segmentation using sparse non-negative matrix factorization of spherical mean diffusion MRI data. In: International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, pp 69–76
Tabesh A, Jensen JH, Ardekani BA, Helpern JA (2011) Estimation of tensors and tensor-derived measures in diffusional kurtosis imaging. Magn R83eson Med 65(3):823–836
Tong Q, He H, Gong T, Li C, Liang P, Qian T, Sun Y, Ding Q, Li K, Zhong J (2020) Multicenter dataset of multi-shell diffusion MRI in healthy traveling adults with identical settings. Sci Data 7(1):1–7
Tournier J-D, Calamante F, Connelly A (2007) Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage 35(4):1459–1472
Veraart J, Fieremans E, Jelescu IO, Knoll F, Novikov DS (2016) Gibbs ringing in diffusion MRI. Magn Reson Med 76(1):301–314
Wasserthal J, Neher P, Maier-Hein KH (2018) Tractseg-fast and accurate white matter tract segmentation. Neuroimage 183:239–253
Wen Y, He L, von Deneen KM, Lu Y (2013) Brain tissue classification based on DTI using an improved fuzzy C-means algorithm with spatial constraints. Magn Reson Imaging 31(9):1623–1630
Wu M, Chang L-C, Walker L, Lemaitre H, Barnett AS, Marenco S, Pierpaoli C (2008) Comparison of EPI distortion correction methods in diffusion tensor mri using a novel framework. Medical image computing and computer-assisted intervention. Springer, pp 321–329
Yap P-T, Zhang Y, Shen D (2015) Brain tissue segmentation based on diffusion MRI using L0 sparse-group representation classification. Medical image computing and computer-assisted intervention. Springer, pp 132–139
Zhang F, Cho KIK, Tang Y, Zhang T, Kelly S, Di Biase M, Xu L, Li H, Matcheri K, Whitfield-Gabrieli S et al (2020) MK-curve improves sensitivity to identify white matter alterations in clinical high risk for psychosis. Neuroimage 226:117564
Zhang F, Ning L, O’Donnell LJ, Pasternak O (2019) MK-curve—characterizing the relation between mean kurtosis and alterations in the diffusion MRI signal. Neuroimage 196:68–80
Zhang F, Noh T, Juvekar P, Frisken SF, Rigolo L, Norton I, Kapur T, Pujol S, Wells W, Yarmarkovich A, Kindlmann G, Wassermann D, San-Jose-Estepar R, Rathi Y, Kikinis R, Johnson HJ, Westin C-F, Pieper S, Golby AJ, O’Donnell LJ (2020) SlicerDMRI: diffusion MRI and tractography research software for brain cancer surgery planning and visualization. JCO Clin Cancer Inform. 4:299–309
Sakkalis V (2011) Review of advanced techniques for the estimation of brain connectivity measured with EEG/MEG. Comput Biol Med 41:1110–1117
Gatys LA, Ecker AS, Bethge M (2016) Image style transfer using convolutional neural networks. In: IEEE Conference on Computer Vision and Pattern Recognition, pp 2414 – 2423
Sarubbo S, Tate M, De Benedictis A, Merler S, Moritz-Gasser S, Herbet G, Duau H (2020) Mapping critical cortical hubs and white matter pathways by direct electrical stimulation: an original functional atlas of the human brain. Neuroimage 205:116237
Sarwar T, Seguin C, Ramamohanarao K, Zalesky A (2020) Towards deep learning for connectome mapping: a block decomposition framework. Neuroimage 212:116654
Sbardella E, Tona F, Petsas N, Pantano P (2013) Dti measurements in multiple sclerosis: evaluation of brain damage and clinical implications. Mult Scler Int 2013(2013):671730
Schaefer A, Kong R, Gordon EM, Laumann TO, Zuo XN, Holmes AJ, Eickho SB, Yeo BT (2018) Local-global parcellation of the human cerebral cortex from intrinsic functional connectivity MRI. Cereb Cortex 28:3095–3114
Devi CN, Chandrasekharan A, Sundararaman VK, Alex ZC (2015) Neonatal brain MRI segmentation: a review. Comput Biol Med 64:163–178
Schurr R, Filo S, Mezer AA (2019) Tractography delineation of the vertical occipital fasciculus using quantitative t1 mapping. Neuroimage 202:116121
Seguin C, van den Heuvel MP, Zalesky A (2018) Navigation of brain networks. Proc Natl Acad Sci U S A 115:6297–6302
Seguin C, Razi A, Zalesky A (2019) Inferring neural signalling directionality from undirected structural connectomes. Nat Commun 10:4289
Seguin C, Tian Y, Zalesky A (2020) Network communication models improve the behavioral and functional predictive utility of the human structural connectome. Netw Neurosci 4:980–1006
Sepasian N, ten Thije Boonkkamp J, Ter Haar Romeny B, Vilanova Bartroli A (2012) Multivalued geodesic ray-tracing for computing brain connections using diffusion tensor imaging. SIAM J Imag Sci 5:483–504
Shahab S, Stefanik L, Foussias G, Lai MC, Anderson KK, Voineskos AN (2018) Sex and diffusion tensor imaging of white matter in schizophrenia: a systematic review plus meta-analysis of the corpus callosum. Schizophr Bull 44:203–221
Pecheva D, Yushkevich P, Batalle D, Hughes E, Aljabar P, Wurie J, Hajnal JV, Edwards AD, Alexander DC, Counsell SJ et al (2017) A tract-specific approach to assessing white matter in preterm infants. Neuroimage 157:675–694
Miller DJ, Duka T, Stimpson CD, Schapiro SJ, Baze WB, McArthur MJ, Fobbs AJ, Sousa AM, Sestan N, Wildman DE et al (2012) Prolonged myelination in human neocortical evolution. Proc Natl Acad Sci 109:16480–16485
Gottlieb D, Shu CW, Solomonoff A, Vandeven H (1992) On the Gibbs phenomenon I: recovering exponential accuracy from the Fourier partial sum of a nonperiodic analytic function. J Comp Appl Math 43:81–98
Misic B, Betzel RF, Nematzadeh A, Goni J, Gria A, Hagmann P, Flammini A, Ahn YY, Sporns O (2015) Cooperative and competitive spreading dynamics on the human connectome. Neuron 86:1518–1529
Mito R, Raelt D, Dhollander T, Vaughan DN, Tournier JD, Salvado O, Brodtmann A, Rowe CC, Villemagne VL, Connelly A (2018) Fibre-specific white matter reductions in Alzheimer’s disease and mild cognitive impairment. Brain 141:888–902
Cui LB, Wei Y, Xi YB, Gria A, De Lange SC, Kahn RS, Yin H, Van den Heuvel MP (2019) Connectome-based patterns of first episode medication-naive patients with schizophrenia. Schizophr Bull 45:1291–1299
Cui Z, Zhong S, Xu P, Gong G, He Y (2013) Panda: a pipeline toolbox for analyzing brain diffusion images. Front Hum Neurosci 7:42
Daducci A, Dal-Palu A, Descoteaux M, Thiran JP (2016) Microstructure informed tractography: pitfalls and open challenges. Front Neurosci 10:247
Daducci A, Dal Palu A, Lemkaddem A, Thiran JP (2013) A convex optimization framework for global tractography. In: 2013 IEEE 10th International Symposium on Biomedical Imaging, IEEE. pp 524–527
Daducci A, Dal Palu A, Lemkaddem A, Thiran JP (2014) COMMIT: convex optimization modeling for microstructure informed tractography. IEEE Trans Med Imaging 34:246–257
Damatac CG, Chauvin RJ, Zwiers MP, van Rooij D, Akkermans SE, Naaijen J, Hoekstra PJ, Hartman CA, Oosterlaan J, Franke B et al (2020) White matter microstructure in attention deficit/hyperactivity disorder: a systematic tractography study in 654 individuals. Biol Psychiatry Cogn Neurosci Neuroimaging 7:979–988
Zhanga F, Daduccib A, Yong H, Schiavib S, Seguing C, Smithi R, Yeh C-H, Zhao T, O’Donnell LJ (2021) Quantitative mapping of the brain’s structural connectivity using diffusion MRI tractography: a review. arXiv, pp 1–29
Davis SW, Dennis NA, Buchler NG, White LE, Madden DJ, Cabeza R (2009) Assessing the effects of age on long white matter tracts using diffusion tensor tractography. Neuroimage 46:530–541
De Witte NA, Mueller SC (2017) White matter integrity in brain networks relevant to anxiety and depression: evidence from the human connectome project dataset. Brain Imaging Behav 11:1604–1615
Zhang W, Olivi A, Hertig SJ, Van Zijl P, Mori S (2008) Automated fiber tracking of human brain white matter using diffusion tensor imaging. Neuroimage 42:771–777
Zhang Y, Zhang J, Oishi K, Faria AV, Jiang H, Li X, Akhter K, Rosa-Neto P, Pike GB, Evans A et al (2010) Atlas-guided tract reconstruction for automated and comprehensive examination of the white matter anatomy. Neuroimage 52:1289–1301
Zhang Z, Descoteaux M, Zhang J, Girard G, Chamberland M, Dunson D, Srivastava A, Zhu H (2018) Mapping population-based structural connectomes. Neuroimage 172:130–145
Zhao T, Cao M, Niu H, Zuo XN, Evans A, He Y, Dong Q, Shu N (2015) Age-related changes in the topological organization of the white matter structural connectome across the human lifespan. Hum Brain Mapp 36:3777–3792
Zhao T, Mishra V, Jeon T, Ouyang M, Peng Q, Chalak L, Wisnowski JL, Heyne R, Rollins N, Shu N et al (2019) Structural network maturation of the preterm human brain. Neuroimage 185:699–710
Zhao T, Sheng C, Bi Q, Niu W, Shu N, Han Y (2017) Age-related differences in the topological efficiency of the brain structural connectome in amnestic mild cognitive impairment. Neurobiol Aging 59:144–155
Zhao T, Xu Y, He Y (2019) Graph theoretical modeling of baby brain networks. Neuroimage 185:711–727
Zhao X, Tian L, Yan J, Yue W, Yan H, Zhang D (2017) Abnormal rich-club organization associated with compromised cognitive function in patients with schizophrenia and their unaffected parents. Neurosci Bull 33:445–454
Ziyan U, Sabuncu MR, Grimson WEL, Westin CF (2009) Consistency clustering: a robust algorithm for group-wise registration, segmentation and automatic atlas construction in diffusion MRI. Int J Comput Vision 85:279–290
Zollei L, Jaimes C, Saliba E, Grant PE, Yendiki A (2019) Tracts constrained by underlying infant anatomy (traculina): an automated probabilistic tractography tool with anatomical priors for use in the newborn brain. Neuroimage 199:1–17
Yeh CH, Smith RE, Dhollander T, Connelly A (2017) Mesh-based anatomically-constrained tractography for effective tracking termination and structural connectome construction. In: Proceedings of the ISMRM, no. 0058
Yeh CH, Smith RE, Liang X, Calamante F, Connelly A (2016) Correction for diffusion MRI fibre tracking biases: the consequences for structural connectomic metrics. Neuroimage 142:150–162
Yeh FC, Badre D, Verstynen T (2016) Connectometry: a statistical approach harnessing the analytical potential of the local connectome. Neuroimage 125:162–171
Yeh FC, Panesar S, Fernandes D, Meola A, Yoshino M, Fernandez-Miranda JC, Vettel JM, Verstynen T (2018) Population-averaged atlas of the macroscale human structural connectome and its network topology. Neuroimage 178:57–68
Urban G, Bendszus M, Hamprecht F, Kleesiek J (2014) Multimodal brain tumor segmentation using deep convolutional neural networks. In: MICCAI BraTS (Brain Tumour Segmentation) Challenge. Proceedings, Winning Contribution, pp 31–35
Vijayakumar C, Gharpure DC (2011) Development of image-processing software for automatic segmentation of brain tumors in MRI images. J Med Phys/Assoc Med Phys India 36(3):147
Wang SH, Phillips P, Sui Y, Liu B, Yang M, Cheng H (2018) Classification of Alzheimer’s disease based on eight-layer convolutional neural network with leaky rectified linear unit and max pooling. J Med Syst 42(5):85
Rondina JM, Ferreira LK, de Souza Duran FL, Kubo R, Ono CR, Leite CC et al (2018) Selecting the most relevant brain regions to discriminate Alzheimer’s disease patients from healthy controls using multiple kernel learning: a comparison across functional and structural imaging modalities and atlases. Neuroimage Clin 17:628–641
Saha P, Udupa J (2001) Optimum image thresholding via class uncertainty and region homogeneity. IEEE Trans Pattern Anal Mach Intell 12(7):689–706
Salman Y (2009) Modified technique for volumetric brain tumour measurements. J Biomed Sci Eng 2:16–19
Salman Y, Badawi A, Assal M, Alian S (2005) New automatic technique for tracking brain tumor response. In: International conference on biological and medical physics, pp 1–4
Sanchez A, Mammone N, Morabito FC, Marino S, Adeli H (2019) A novel methodology for automated differential diagnosis of mild cognitive impairment and the Alzheimer’s disease using EEG signals. J Neurosci Methods 322:88–95
Oxtoby NP, Garbarino S, Firth NC et al (2017) Data-driven sequence of changes to anatomical brain connectivity in sporadic Alzheimer’s disease. Front Neurol 8:580
Betzel RF, Bassett DS (2018) Specificity and robustness of long-distance connections in weighted, interareal connectomes. Proc Natl Acad Sci U S A 115:E4880–E4889
Raffelt D, Sadeghian F, Connor H. Connelly A (2015) Decreased apparent fiber density in the optic pathways correlates with glaucoma disease severity. In: Proc ISMRM, p 2213
Vaughan DN, Raffelt D, Curwood E et al (2017) Tract-specific atrophy in focal epilepsy: disease, genetics, or seizures? Ann Neurol 81:240–250
Mito R, Raffelt D, Dhollander T et al (2018) Fibre-specific white matter reductions in Alzheimer’s disease and mild cognitive impairment. Brain 141:888–902
Liang X, Yeh C-H, Connelly A, Calamante F (2019) Robust identification of rich-club organization in weighted and dense structural connectomes. Brain Topogr 32:1–16
Xing X-X, Zuo X-N (2018) The anatomy of reliability: a must read for future human brain mapping. Sci Bull 63:1606–1607
Zuo X-N, Xu T, Milham MP (2019) Harnessing reliability for neuroscience research. Nat Hum Behav 3:768–771
Tzourio-Mazoyer N, Landeau B, Papathanassiou D et al (2002) Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 15:273–289
Glasser MF, Coalson TS, Robinson EC et al (2016) A multi-modal parcellation of human cerebral cortex. Nature 536:171–178
Gelb A, Tadmor E (1999) Detection of edges in spectral data. Appl Comput Harmon Anal 7:101–135
Gelb A, Tadmor E (2000) Detection of edges in spectral data II: nonlinear enhancement. SIAM J Numer Anal 38:1389–1408
Archibald R, Gelb A (2002) A method to reduce the Gibbs ringing artifact in MRI scans while keeping tissue boundary integrity. IEEE Trans Med Imaging 21:305–319
Archibald R, Chen K, Gelb A, Renautc R (2003) Improving tissue segmentation of human brain MRI through preprocessing by the Gegenbauer reconstruction method. Neuroimage 20:489–502
Wakana S, Jiang H, Nagae-Poetscher LM, van Zijl PC, Mori S (2004) Fiber tract-based atlas of human white matter anatomy. Radiology 230:77–87
Author information
Authors and Affiliations
Contributions
PRK: conceptualization, methodology, and writing original draft preparation. RKJ: visualization, writing—review, and supervision. AK: validation and investigation. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest in publishing the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, P.R., Jha, R.K. & Katti, A. Brain tissue segmentation in neurosurgery: a systematic analysis for quantitative tractography approaches. Acta Neurol Belg 124, 1–15 (2024). https://doi.org/10.1007/s13760-023-02170-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13760-023-02170-9