Abstract
In brain-related diseases, including Brain Tumours and Alzheimer’s, accurate and timely diagnosis is crucial for effective medical intervention. Current state-of-the-art (SOTA) approaches in medical imaging predominantly focus on diagnosing a single brain disease at a time. However, recent research has uncovered intricate connections between various brain diseases, realizing that treating one condition may lead to the development of others. Consequently, there is a growing need for accurate diagnostic systems addressing multiple brain-related diseases. Designing separate models for different diseases, however, can impose substantial overhead. To tackle this challenge, our paper introduces \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\), an innovative neural network architecture explicitly tailored for classifying brain images. The primary objective is to propose a single, robust framework capable of diagnosing a spectrum of brain-related diseases. The paper comprehensively validates \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\)’s efficacy, specifically in diagnosing Brain tumours and Alzheimer’s disease. Remarkably, the proposed model workflow surpasses current SOTA methods, demonstrating a substantial enhancement in accuracy and precision. Furthermore, it maintains a balanced performance across different classes in the Brain tumour and Alzheimer’s dataset, emphasizing the versatility of our architecture for precise disease diagnosis. \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) undergoes an ablation study to optimize its choice of the optimal optimizer, and a data growth analysis verifies its performance on small datasets, simulating real-life scenarios where data progressively increase over time. Thus, this paper signifies a significant stride toward a unified solution for diagnosing diverse brain-related diseases.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Brain imaging data, including Magnetic Resonance Imaging (MRI), are extensively utilized to investigate brain function (Du et al. 2018; Shukla et al. 2023b). The approach’s premise is that scrutinizing neuro-imaging data and detecting anomalies allow one to unravel the brain’s workings. The insights gained from neuro-imaging data can then be harnessed to enhance diagnosis and treatment. Despite the efficacy of AI solutions in addressing challenges and developing computer-assisted systems that support clinicians and expedite diagnostics, clinicians still bear the primary responsibility for meticulously examining, analyzing, and documenting any disorder in patients. The main reason for this is the current architectures’ need for more robustness and adaptability.
Numerous efforts have aimed to enhance existing architectures, promoting their application in the healthcare domain, where the capacity for critical and accurate decision-making holds paramount importance (Ozkaya and Sagiroglu 2023; Kabiraj et al. 2022; Roy et al. 2022). Specifically focusing on the brain, a multitude of diseases exist, encompassing conditions like ischemic stroke, prion diseases, Alzheimer’s, Parkinson’s, as well as various postural hazards and sclerosis (Kumar et al. 2014; Taree et al. 2020; Du et al. 2018). While encountering such diseases daily is relatively uncommon, addressing these challenges remains imperative. Furthermore, certain diseases within this spectrum are interconnected, as seen in the case of Alzheimer’s Disease (\({\mathcal{A}\mathcal{D}}\)) and Brain tumour (\({\mathcal{B}\mathcal{T}}\)), (Speidell et al. 2019; Ingeno 2019; Staff 2018). Biologically, \({\mathcal{A}\mathcal{D}}\) and \({\mathcal{B}\mathcal{T}}\) exhibit distinct cellular behaviours. Alzheimer’s is characterized by heightened cell death, whereas brain tumours are marked by uncontrolled cell proliferation akin to cancer (Lehrer 2018). Researchers made intriguing discoveries in a recent study involving a mouse model (Escarcega et al. 2022). The study uncovered that mice undergoing cancer and chemotherapy drugs experienced accelerated brain ageing. Specifically, it was found that individuals who had undergone chemotherapy had a higher prevalence of Alzheimer’s disease, a common form of dementia, than those who had never been exposed to chemotherapy. Moreover, in another independent study, it was noted that patients surviving cancerFootnote 1 were experiencing treatment-related side effects such as cognitive impairment, which subsequently increased the risk of Alzheimer’s disease (Kao et al. 2023). In summary, cancer treatment inhibits neurogenesis and increases oxidative damage, DNA damage, and inflammatory responses in the brain. Clinicians often find it hectic to diagnose multiple diseases simultaneously, rendering treatment tedious and time-consuming. Recent literature has introduced several machine learning models to aid clinicians and, in turn, enhance the accuracy of medical systems (Gupta et al. 2020; Ranga et al. 2020, 2022; Gupta et al. 2021). However, the current \({\mathcal {SOTA}}\) architecture is presently constrained in its ability to effectively identify multiple interconnected abnormalities (Kujur et al. 2022). Much ongoing research is directed toward classifying different diseases individually (Yildirim and Cinar 2020; Loddo et al. 2022; Salçin 2019).
To address these challenges to a certain extent, this paper introduces a robust average-weighted ensemble-driven architecture (named \(\mathcal {B}\)rain\({\mathcal {MRIN}}\)et or simply \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\)) for classifying brain diseases using Magnetic Resonance Imaging (MRI) images. Thus, the key contributions are as follows:
1.1 Key contributions
-
The paper introduces a novel neural architecture for brain MRI image classification, named \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\), which leverages an innovative average-weighted ensemble approach.
-
Experimental analysis validates the effectiveness of the \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) architecture in diagnosing multiple brain diseases, including Brain tumour (\({\mathcal{B}\mathcal{T}}\)) and Alzheimer’s (\({\mathcal{A}\mathcal{D}}\)). For this purpose, a comprehensive workflow for implementing the \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) architecture is proposed.
-
The proposed workflow demonstrates superior performance compared to current \({\mathcal {SOTA}}\) techniques in four-way multi-class single disease classification. The approach achieves a notable improvement of \(3\%\) in accuracy and \(2\%\) higher precision compared to all existing \({\mathcal {SOTA}}\) methods. Moreover, the \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) architecture attains the highest F1-score for both disease diagnoses within a single framework, showcasing its remarkable efficacy.
-
Additional study, involving a class-wise analysis for both diseases, reveals a well-balanced performance across various classes. Also, the conducted ablation and growth study supports the choice of regularizers and shows the efficacy of the proposed \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\).
2 Related work
This section examines the current \({\mathcal {SOTA}}\) methods in the field of disease classification concerning brain MRIs. The literature review is structured into three main subsections: first, focusing on \({\mathcal{B}\mathcal{T}}\) classification; second, \({\mathcal{A}\mathcal{D}}\) disease classification; and finally, a subsection dedicated to research on the simultaneous diagnosis of multiple diseases.
2.1 Brain tumour classification
This subsection delves into the literature closely related to the conducted study and the proposed network on \({\mathcal{B}\mathcal{T}}\) classification using MRI. A study by Aamir et al. (2022) aims to automate the detection of brain tumours using MRI scans, mitigating time-consuming and error-prone techniques. The approach involves pre-processing MRI images, extracting features using pre-trained models, and combining these features through the partial least square method for identification. Srinivas et al. (2022) conduct a comprehensive study of pre-trained models for classification from MRI images. The models utilized include VGG-16 (Simonyan and Zisserman 2015), ResNet-50 (He et al. 2016), and Inception-V3 (Szegedy et al. 2017), with VGG-16 outperforming others for \({\mathcal{B}\mathcal{T}}\) localization. Mehnatkesh et al. (2023) propose an optimization-based deep convolutional ResNet model combined with a novel evolutionary algorithm to optimize the architecture and hyperparameters of deep ResNet (He et al. 2016). Asif et al. (2023a) propose a novel deep stacked ensemble model named BMRI-NET, comprising DenseNet-201 (Huang et al. 2017), ResNet-152v2, and InceptionResNetv2. Özkaraca et al. (2023) propose a modular deep learning model that retains the advantages of known transfer learning methods on DenseNet, VGG-16, and basic Convolutional Neural Network (CNN) for MRI classification, using tenfold cross-validation for testing. Saurav et al. (2023) presents a novel lightweight attention-guided CNN architecture that uses channel-attention blocks focused on regions relevant to MRI classification. Asif et al. (2023b) employ pre-trained models (Xception (Fran et al. 2017), DenseNet-121, InceptionResNetv2, DenseNet-201, and ResNet-152v2), modifying the final architectures with deep, dense blocks and softmax layers to enhance multi-class classification accuracy. Abd El-Wahab et al. (2023) propose an architecture with 13 layers, including trainable convolutional layers, average pooling, fully connected layers, and a softmax layer. The training involves five iterations and fivefold cross-validation for retraining, achieving \(98.63\%\) average accuracy with transfer learning and \(98.86\%\) using retrained fivefold cross-validation to classify 3 classes. Zulfiqar et al. (2023) presents a transfer learning-based approach for multi-class classification via fine-tuning pre-trained EfficientNets (Tan and Le 2019). The work employs different variants of modified EfficientNets under various experimental settings, incorporating Grad-CAM visualization on MRI sequences. Thanki and Kaddoura (2022) apply a hybrid learning technique to binary classification, proposing a learning approach and comprehensively comparing existing supervised learning approaches. Yazdan et al. (2022) presents a dual-fold solution for \({\mathcal{B}\mathcal{T}}\) classification through MRI: a multi-scale CNN for robust classification and minimizing Rician Noise impact to improve model performance. The study focuses on four classes with the aim of outperforming the existing \({\mathcal {SOTA}}\) methods.
2.2 Alzheimer’s disease classification
This subsection delves into the literature closely related to the conducted study and the proposed network for Alzheimer’s classification using MRI. Shukla et al. (2023a) presents various \({\mathcal {SOTA}}\) on CNN models built from scratch for binary and multi-class classification comparison. They achieve \(94\%\) accuracy in multi-class and \(99\%\) in binary classification. Nancy Noella and Priyadarshini (2023) explore diverse machine learning classifiers, including bagged ensemble, Iterative Dichotomiser 3 (ID3), Naive Bayes, and Multi-class Support Vector Machine (SVM), for Alzheimer’s classification. Balasundaram et al. (2023) propose an approach that utilizes a reduced version of datasets to achieve accurate predictions, enabling faster training. Their method involves image segmentation to isolate the hippocampus region from brain MRI images. They compare models trained on segmented and unsegmented images. Salehi et al. (2023) introduce a Long Short-Term Memory (LSTM) network that leverages the temporal memory characteristics of LSTMs. The network efficiently studies patterns using stratified shuffled split cross-validation inherent in MRI scans, achieving an accuracy of \(98.67\%\) in binary classification. Ghazal et al. (2022) adopt a transfer learning approach for multi-class classification using AlexNet (Krizhevsky et al. 2012), achieving an overall accuracy of \(91.70\%\). Shanmugam et al. (2022) propose an approach focused on the early detection of multistage cognitive impairment by targeting cognitive functions and memory loss. They use transfer learning primarily with pre-trained networks, such as GoogleNet (Ballas et al. 2015), AlexNet, and ResNet-18. Marwa et al. (2023) presents an analysis pipeline that involves a lightweight CNN architecture and 2-dimensional (2d) T1-weighted MRI. The pipeline not only proposes a fast and accurate diagnosis module but also suggests both global and local classifications for \({\mathcal{A}\mathcal{D}}\) . Samhan et al. (2022) introduces a cost-effective CNN network achieving \(100\%\) accuracy on train data for \({\mathcal{A}\mathcal{D}}\) classification which is an overfit model.
2.3 Interconnected and multiple disease classification
This subsection delves into the literature closely related to the conducted study and the proposed network for both \({\mathcal{B}\mathcal{T}}\) and \({\mathcal{A}\mathcal{D}}\) classification using MRI (Majd et al. 2019; Roe et al. 2010; Sánchez-Valle et al. 2017).
Kujur et al. (2022) propose a stratified k-fold cross-validation method utilizing CNNs trained from scratch, ResNet-50, InceptionV3, and Xception, to detect Alzheimer’s and brain tumours simultaneously. Acquarelli et al. (2022) assess the value of CNNs in diagnosing Brain tumours and Alzheimer’s disease, addressing challenges related to limited case numbers in datasets and resulting in interpretability in terms of relevant regions. Chandaran et al. (2022) reviews pre-trained CNNs for classifying multiple diseases, including \({\mathcal{A}\mathcal{D}}\), \({\mathcal{B}\mathcal{T}}\), Hemorrhage, Parkinson’s, and Stroke, using transfer learning. Namachivayam and Puviarasan (2023) develop a computerized brain disease detection system focusing on Alzheimer’s, tumour, and Parkinson’s diseases. They employ a transfer learning approach with InceptionV3 and VGG-19 models for efficient disease detection. Arabahmadi et al. (2022) conducts a comprehensive review of existing deep learning methods applied to MRI data to classify multiple diseases. Ismail et al. (2022) propose a multimodal image fusion technique to combine MRI neuro-images with modular sets of images. They employ a CNN with three classifiers–softmax, SVM, and random forest–to forecast and classify Alzheimer’s brain multimodal progression and Mild Cognitive Impairment (MCI) disease through high-dimensional magnetic resonance characteristics.
3 Material and methodology
This section presents a thorough outline of the datasets used in this research, the pre-processing techniques applied, and the architecture proposed for classifying multiple diseases in brain MRI. The upcoming section will assess the efficacy of the proposed architecture in identifying both \({\mathcal{B}\mathcal{T}}\) and \({\mathcal{A}\mathcal{D}}\).
3.1 Datasets
This investigation primarily employed two datasets:
-
Brain tumour MRI dataset (\({\mathcal {BTD}}\)) (Nickparvar 2021): encompasses 7023 MRI images of human brains categorized into four distinct classes: glioma, meningioma, no tumour, and pituitary. These images are collected from three sources primarily—Figshare (Cheng 2017), SARTAJ,Footnote 2 and Br35H (Hamada 2020).
-
Alzheimer disease dataset (\({\mathcal {ADD}}\)) (Dubey 2019): consists of four distinct categories: Mild Demented, Moderate Demented, Non Demented, and Very Mild Demented. The dataset as a whole is composed of 6400 images.
Illustrated in Figs. 1 and 2 are sample images from each class extracted from the \({\mathcal {BTD}}\) and \({\mathcal {ADD}}\) datasets, respectively. The figures illustrate the variation in dataset samples from different classes. For instance, in Alzheimer’s data, it is evident that classifying between different classes is a tedious and time-consuming process. This exerts pressure on clinicians, leading to reduced accuracy and efficiency. This observation underscores the need for automated machine learning models to assist clinicians.
3.2 Dataset pre-processing and analysis
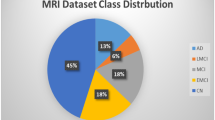
All images obtained from the specified datasets underwent consistent resizing to dimensions of 224 \(\times \) 224, followed by rescaling within the range of 0 to 1. Upon meticulous examination, it became evident that the training datasets displayed a degree of imbalance, as depicted in Fig. 3. This imbalance could potentially lead to bias, overfitting, and underfitting during training, potentially compromising performance outcomes.
As evident from Fig. 3, the \({\mathcal {BTD}}\) dataset demonstrates a relatively balanced distribution, while there is a notable imbalance within the \({\mathcal {ADD}}\) dataset. To address these concerns and rectify the imbalance, we employed the Synthetic Minority Over-sampling Technique (SMOTE) (Chawla et al. 2002) with a batch size of 7000. Following the application of the SMOTE up-scaling technique, the count of training data increased to 8000 for the \({\mathcal {BTD}}\) dataset and 12800 for the \({\mathcal {ADD}}\) dataset, mitigating the identified imbalance issues to a certain extent. Thus, the current paper proceeds to propose a novel architecture framework, \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\), to help assist clinicians in achieving precise multi-disease classification.
3.3 Proposed architecture: \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\)
\(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) represents an ensemble-based Convolutional Neural Network (CNN) designed to classify brain MRI images. This network is constructed by combining two distinct CNN architectures, denoted as M1 and M2, developed from the ground up. M1 encompasses 34 layers, while M2 is composed of 26 layers. Fusing these two CNNs through an average-weighted ensemble methodology enhances the robustness and efficacy of \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) in managing the intricate task of analyzing brain MRI images. For an overview of the \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) architecture and the data dimension at each layer during training, please refer to Fig. 4. Both the M1 and M2 architectures process rescaled brain images with a consistent dimension of \(224 \times 224 \times 3\).This format standardizes the images to a resolution of 224 pixels in height and 224 pixels in width, with three RGB channels. Essentially, each image is treated as a \(224 \times 224\) pixel image with three color channels, enabling the models to perceive the images using red, green, and blue intensities to effectively classify brain MRI images.
In M1, four convolutional blocks follow the input layer, each comprising a Maxpooling-2d layer and three consecutive 2d convolutional and batch normalization layers. The neuron counts for these blocks’ layers are 16, 32, 64, and 128. Similarly, M2 has four convolutional blocks with Maxpooling-2d layers, 2d convolutional layers, and batch normalization layers. The neuron counts within these blocks are set to 16, 32, 64, and 128.
The primary distinction between M1 and M2 lies in the convolutional layer count within each block, resulting in distinct feature extraction capabilities. The sub-architectures yield soft predictions collectively used to generate a hard prediction. \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) offers significant advantages in scenarios with limited available data, particularly when the existing data exhibit high similarity, presenting challenges in discerning unique features for precise classification.
The outputs generated by M1 and M2 form the soft predictions. Subsequently, the average-weighted layer processes these soft predictions to produce the ultimate hard prediction. Due to the distinctive characteristics of the sub-architecture models, they adapt to the same distribution in varying manners. The training process persists until each model reaches its point of convergence or until no further reduction in the loss value is achieved.
In terms of the equations used for training, let \(SP_i\) denote the soft prediction from the \(i^{th}\) model and \(w_i\) represent the weight factor associated with \(SP_i\). The generalized formula for the average-weighted ensemble, involving n models to compute the final soft prediction (Final SP), is as follows:
Here, \(w_i \ge 0\ \forall \ i\), and \(\sum _i^n w_i = 1\). In this formulation, we compute the FinalSP by taking the weighted sum of the soft predictions from all n models. The weight assigned to each model represents its contribution to the final prediction, and these weights are normalized to ensure their combined sum equals 1.
Note that \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) consists of two distinct sub-architectures (\(n=2\)), namely, M1 and M2. There is a possibility of expanding the number of sub-architectures, ensuring the robustness and applicability of the architecture. The next section tests the applicability of \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) in multi-disease classification for \({\mathcal{B}\mathcal{T}}\) and \({\mathcal{A}\mathcal{D}}\) (Speidell et al. 2019; Kao et al. 2023; Escarcega et al. 2022; Ingeno 2019; Staff 2018).
4 Proposed workflow for \({\mathcal{B}\mathcal{T}}\) and \({\mathcal{A}\mathcal{D}}\) diagnosis
Section elucidates the comprehensive workflow for employing \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) to detect both \({\mathcal{A}\mathcal{D}}\) and \({\mathcal{B}\mathcal{T}}\). Prior evidence shows that both diseases are interconnected (Kao et al. 2023; Escarcega et al. 2022; Roe et al. 2010). The workflow commences with the collection of medical records in the form of MRI scans for both ailments. Subsequently, two distinct pipelines were established, each specialized for diagnosing one of the diseases. The initial pipeline is trained using Brain tumour Data (\({\mathcal {BTD}}\)), resulting in a well-trained \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) architecture. Similarly, another pipeline is trained using the Alzheimer Disease Data (\({\mathcal {ADD}}\)) dataset.
Once these pipelines are trained and the model weights are learned, they can be deployed for real-world testing. During testing, a single MRI scan of a patient is input into the system, enabling experts to identify both diseases simultaneously. This approach offers a vital advantage: if a patient undergoing brain tumour treatment exhibits symptoms of Alzheimer’s, the system can identify these signs during routine tumour checkups. This enables the timely initiation of Alzheimer’s treatment. Hence, the methodology accommodates the potential detection of singular or multiple diseases. This simultaneous diagnosis of interconnected diseases enhances efficiency, promotes a comprehensive understanding, and facilitates holistic treatment approaches. By diagnosing shared features, the approach leads to more effective treatments. Figure 5 shows a visual representation of the complete workflow. We now validate the efficacy of the proposed workflow against \({\mathcal {SOTA}}\) methods.
5 Experimental analysis
First, compare the proposed methodology with the \({\mathcal {SOTA}}\) in individual pipelines, i.e., approaches that address either \({\mathcal{B}\mathcal{T}}\) or \({\mathcal{A}\mathcal{D}}\) exclusively. Subsequently, compare the performance with \({\mathcal {SOTA}}\) methods designed for multi-disease classification. This encompasses studies that focus solely on \({\mathcal{B}\mathcal{T}}\) and \({\mathcal{A}\mathcal{D}}\), as well as works proposed for handling more than two diseases. While the proposed architectural framework can encompass more than two diseases, it is left as an intriguing avenue for future exploration. This study restricts the analysis to the simultaneous classification of two diseases. The complete experimental code is made available anonymously.Footnote 3 The dataset employed in this research has already been thoroughly examined in Sect. 3.1.
5.1 Implementation and reproducibility
Experimental setup: A Google Colaboratory Notebook with 24GB of RAM, running Python-3 on a T4 GPU, conducted all experiments. Additionally, \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) incorporates hyperparameters—specifically, learning rate, epochs, and batch size—set at 0.001, 100, and 32, respectively, across all pipelines. The baseline \({\mathcal {SOTA}}\) methods retained parameters as originally reported in their respective papers. The training-test split consistently followed previous literature. Furthermore, the activation function for feature extraction is ELU, softmax activation is used for multi-class classification, and the data split ratio is 75% for training, 5% for validation, and 15% for testing.
Code repository: The GitHub repository provides public access to the comprehensive source code, facilitating proper code reproducibility. The repository consists primarily of three files: visualization.py, train.py, and requirements.txt. A detailed step-by-step README outlines commands for replicating the Python environment using requirements.txt, including the names and versions of all libraries. Subsequently, it provides instructions for utilizing train.py for model training and visualization.py for reproducing experimental analyses. Additionally, for direct real-world deployment and testing, the repository includes trained model weights saved in the HDF5 binary data format (h5py), compatible with the TensorFlow framework. Individual README files and comments in each line of code offer additional support. Section 3.1 covers dataset details, and code comments explain file paths for different datasets.
5.2 Performance metrics
-
Accuracy (Acc): measures the proportion of correctly predicted instances to the total number of instances
$$\begin{aligned} \text {Acc} = \frac{\text {Number of Correct Predictions} \times 100}{\text {Total Number of Instances}}. \end{aligned}$$(2) -
Loss: is a measure of the dissimilarity between predicted values and actual values, often used during training to guide the optimization process. It is typically inversely related to accuracy; as the loss decreases, accuracy tends to improve. In other words, minimizing the loss function leads to higher accuracy. We consider the categorical cross-entropy loss function (\(\mathcal {L}(\cdot )\)), which for N classes is computed as follows:
$$\begin{aligned} \mathcal {L}(y, \hat{y}) = -\sum _{i=1}^{N} y_i \log (\hat{y}_i), \end{aligned}$$(3)where y and \(\hat{y}\) are true distributions (ground truth) of the categories and predicted distribution of the categories, often obtained from a neural network or other classifier respectively. These are typically represented as a one-hot encoded vector.
-
Precision: quantifies the accuracy of positive predictions made by a model. It measures the proportion of true positive predictions out of all instances the model predicted as positive
$$\begin{aligned} \text {Precision} = \frac{\text {TP}}{\text {TP} + \text {FP}}, \end{aligned}$$(4)where True Positives (TP) are instances correctly predicted as positive, and False Positives (FP) are instances incorrectly predicted as positive when they are negative.
-
Recall: also known as sensitivity or true positive rate, is a metric in classification that measures the ability of a model to correctly identify all relevant instances from the total number of actual positive instances
$$\begin{aligned} \text {Recall} = \frac{\text {TP}}{\text {TP} + \text {FN}}, \end{aligned}$$(5)where False Negatives (FN) are instances incorrectly predicted as negative when actually positive.
-
F1-score: is a composite metric that integrates precision and recall into a single measurement to comprehensively assess a model’s efficacy. When the evaluation calls for accounting for false positives and False negatives inside a single indicator, its relevance stands out, making it a crucial tool for evaluating overall performance. When class distributions are unbalanced, or the effects of false positives and negatives are noticeable, this significance increases. Further, the F1-score acts as a deciding factor, favoring models with higher F1-scores as the best choice when different classifiers excel in recall and precision individually. It is computed as follows:
$$\begin{aligned} \text {F1-Score} = 2 \times \frac{\text {Precision} \times \text {Recall}}{\text {Precision} + \text {Recall}}. \end{aligned}$$(6)
5.3 Training \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) pipeline
First, training the individual pipeline in the proposed workflow on \({\mathcal {ADD}}\) and \({\mathcal {BTD}}\) datasets is performed separately in line with Fig. 5. Figure 6a, b for \({\mathcal{B}\mathcal{T}}\) and \({\mathcal{A}\mathcal{D}}\) diagnosis, respectively, illustrates the train and test phase learning curves. The learning curves for model accuracies exhibit a smooth progression without any indications of overfitting. The loss is inversely related to accuracy following a similar observation and is omitted from the current version of the paper.
5.4 Comparison against single disease diagnosis
This subsection compares the performance of \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) against works individually in classifying \({\mathcal {BTD}}\) dataset. The train and test data split results are reported in Table 1. The metrics used for comparison are accuracy, precision, recall, and F1-score.
Observation: In the \({\mathcal {ADD}}\) dataset, our \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) achieves the highest levels of accuracy and precision. The recall value of \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) approaches 100 (though not best), indicating minimal occurrences of false negatives. While \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) demonstrates superior precision, and Loddo et al. (2022) exhibits greater recall, analysis turn to the F1-score to determine the optimal classifier. Notably, the average F1-score across all classes favors the \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) model, showing its efficacy. Similar observations can be made in \({\mathcal {BTD}}\) dataset, showing the proposed architecture’s robustness and adaptability to different brain-related disease diagnoses.
5.5 Comparison against multiple disease diagnosis
This subsection compares the performance of \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) against works that classify multiple diseases using the same neural architecture. Table 2 reports the train and test data results. The metrics used for comparison are accuracy, precision, recall, and F1-score.
Observation: \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) attains comparable levels of accuracy and other metrics in the \({\mathcal {ADD}}\) dataset. The test performance closely aligns with the training performance, underscoring the efficacy of the approach. In contrast, real-world test performance insights in Kujur et al. (2022) make it challenging to assess its practical utility. Additionally, Namachivayam and Puviarasan (2023) focuses on a slightly different dataset that does not consider class-wise distribution within each disease category (\({\mathcal{A}\mathcal{D}}\), \({\mathcal{B}\mathcal{T}}\), Parkinson), opting to directly classify between different disease categories—a distinct line of work. Similarly, Acquarelli et al. (2022) compares the model using the Matthews Correlation Coefficient, an uncommon metric in existing literature. Consequently, this paper excludes this metric from the comparative study.
5.6 Class-wise performance analysis of \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\)
To conduct a class-wise performance analysis of \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) class reports for both datasets, reporting Precision, Recall, and F1-Score are generated and reported in Table 3. Figure 7 also visualizes the confusion matrices for the four class classifications.
Observation: It is evident from the results that there is a minimal disparity in metric values across classes in both datasets, indicating that \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) contributes to achieving balanced class-wise performance.
5.7 Ablation and data growth study of \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\)
This subsection analyzes the performance of the proposed architecture across various parameters, including dataset size and optimizers. Additionally, it further examines the algorithm’s runtime.
5.7.1 Study on different regularizers
The model architecture’s performance is initially assessed in the first study using widely popular optimizers: (Kingma and Ba 2014), AdaGrad (Duchi et al. 2011), Stochastic Gradient Descent (SGD), and RMSprop (Ruder 2016). Tuned to their widely adopted default hyperparameter values suitable for most machine learning algorithms, these values are set as default in the TensorFlow library. The default values are as follows—Adam (\(\beta _1 = 0.9, \beta _2 = 0.999, \epsilon = e^{-7}\)), AdaGrad (initial accumulator value = \(0.1, \epsilon = e^{-7}\)), SGD (momentum = 0.0), and RMSProp (\(\rho = 0.9\), momentum = 0.0, \(\epsilon =e^{-7}\)). Figure 8a, b illustrates the results for the \({\mathcal {ADD}}\) and \({\mathcal {BTD}}\) datasets, respectively, and summarizes the observations:
Observations: (a) In the \({\mathcal {ADD}}\) dataset, SGD outperforms other optimizers among train accuracies and is followed by Adam. However, in test accuracies, the performance of SGD drops slightly, while Adam achieves stable and the highest performance.
(b) In the \({\mathcal {BTD}}\) dataset, although SGD achieves the highest train accuracies, Adam exhibits comparatively stable train and test accuracies, making it an ideal choice for the tumour dataset.
Thus, the above ablation study validates using Adam as the optimizer in our experiments.
5.7.2 Data growth study on accuracy as metric
The performance of \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) across different dataset sizes is analyzed in this experiment. The model’s efficiency in real-world deployments, where data per class are initially limited and tend to increase over time, is assessed. Initially conducted by considering only \(p\%\) of the data from each class, the experiment evaluates the model’s efficiency using the accuracy metric. The study considers p values ranging from 20 to 100 with increments of 20. The plots for train and test accuracy on \({\mathcal {ADD}}\) and \({\mathcal {BTD}}\) datasets are depicted in Fig. 9a, b, respectively. Summarizing the observations:
Observations: Both datasets consistently exhibit stable train accuracies across varying dataset sizes, suggesting the architecture’s ability to extract features from input MRI images. While the test accuracies initially show slight decrements, a significant improvement is observed when \(40\%\) of data from each class are provided. This improvement brings the performance to a level comparable to many \({\mathcal {SOTA}}\) baselines reported in Table 1, demonstrating the effective performance of our model architecture, even on small datasets.
5.7.3 Data growth study on runtime
This study presents the average runtime of the proposed model architecture across different dataset sizes. The findings are depicted in Fig. 10. It is evident that in the \({\mathcal {ADD}}\) dataset, a 20% increase in data results in a corresponding 20% increase in runtime. Similarly, for the \({\mathcal {BTD}}\) dataset, the runtime increases by approximately 20–25% with every 20% increase in data size.
6 Discussion
In this section, a detailed discussion is carried out for both results in Tables 1 and 2.
6.1 Comparison against single disease diagnosis
\({\mathcal {ADD}}\) dataset: It can be observed from Table 1 that the initial approach by Acharya et al. (2019) employed shearlet transformation in conjunction with the k-nearest neighbors (KNN) method, achieving only \(94.54\%\) accuracy. Subsequently, introducing automatic feature extraction through the Deep Neural Network (DNN) AlexNet and SVM ensemble, as proposed by Mohammed et al. (2021), slightly improves accuracy while enhancing precision and recall. Efforts to address the class imbalance and growing parameters in DNN, as undertaken by Ahmed et al. (2022) using synthetic oversampling, prove ineffective and lead to deteriorated results. The classification of Alzheimer’s disease in MRI data sees improvement when Yildirim and Cinar (2020) leveraged feature selection with Convolutional Neural Networks (CNN). It was later that Bangyal et al. (2022) observed that incorporating domain ontology into CNN could serve as an alternative method to enhance accuracy and focus on precision and recall metrics. To mitigate the complexity and training time load associated with deep networks, El-Latif et al. (2023); Murugan et al. (2021) and Balasundaram et al. (2023) utilized small datasets and segmentation, respectively, for faster training. Notably, the work by Loddo et al. (2022) using the power of deep ensembles ranks as the second-best among the compared literature. In conclusion, the proposed model, \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\), surpasses the existing \({\mathcal {SOTA}}\) on all evaluation metrics.
\({\mathcal {BTD}}\) dataset: The initial approach by Sachdeva et al. (2016) involved using contour models to delineate tumour regions, followed by applying a genetic algorithm, resulting in an accuracy of \(95.23\%\). Another study by Mohsen et al. (2017) explored the use of fuzzy c-means for segmentation and applied Discrete Wavelet Transform (DWT) followed by Principal Component Analysis (PCA) for feature extraction, achieving an initial accuracy of \(93.94\%\), later improved to \(96.97\%\) Mohsen et al. (2018). Approximately a year later, Salçin (2019) adopted the faster R-CNN architecture to reduce the overhead of feature selection and harness the power of deep learning. Combining these ideas, Shafi et al. (2021) proposed an ensemble model comprising the region of interest (ROI) and collective normalization, Lloyd max quantization for feature extraction, and Support Vector Machines (SVM) as base learners, resulting in an improved accuracy of \(97.82\%\). In contrast, Dewan et al. (2023) streamlined the pre-processing feature extraction, employing pre-trained models to achieve a comparable performance of \(97\%\). On the contrary, proposed \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) achieves the highest accuracy of \(99.97\%\) without significant pre-processing overhead and excels in performance across almost all metrics.
6.2 Comparison against multiple disease diagnosis
The approach by Kujur et al. (2022) incorporates a CNN model after addressing class imbalance. Performance evaluation involves standard architectures like ResNet50, InceptionV3, and Xception, resulting in the best performance of \(98.51\%\) and \(84.26\%\) on \({\mathcal {ADD}}\) and \({\mathcal {BTD}}\), respectively.
In contrast, the proposed model adopts a novel CNN architecture with two separate parallel pipes to enhance feature learning. This innovative approach yields superior metric performance, with the highest accuracy reaching \(97.89\%\) and \(99.97\%\) on \({\mathcal {ADD}}\) and \({\mathcal {BTD}}\) dataset, respectively.
7 Conclusion and future directions
This paper introduces a novel architecture designed to classify brain images. The primary objective is to offer a unified and robust solution for diagnosing brain-related diseases. The current study substantiates the effectiveness of \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) in the context of Brain tumour and Alzheimer’s disease diagnosis. The proposed model workflow surpasses current \({\mathcal {SOTA}}\) methods while demonstrating balanced performance across different classes within each disease. The model undergoes an ablation study to validate the selection of Adam as the optimizer and assess its effectiveness in terms of both accuracy and runtime across varying dataset sizes. One minor limitation of the paper could be the absence of real-world testing, which was omitted due to cost constraints, as the authors had no access to such resources. Another drawback is not leveraging the recent advancements in the potential capabilities of human-in-the-loop medical systems. An intriguing avenue for future research involves extending the application of this architecture to the simultaneous diagnosis of conditions, such as Parkinson’s disease and other closely related or interrelated brain disorders. Also, considering segmentation state-of-art methods as pre-processing before feeding into deep networks can also be a potential direction to improvise metrics Pal et al. (2022); Gangopadhyay et al. (2022); Roy et al. (2017b, 2017a); Roy and Shoghi (2019); Roy et al. (2017b).
Furthermore, an intriguing avenue for exploration involves assessing the performance of the \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) workflow trained on Alzheimer’s and tumour data against a singular \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) architecture trained on MRI images that encompass four distinct classes: ‘No Disease,’ ‘Tumour,’ ‘Alzheimer,’ and ‘Tumour and Alzheimer.’ Currently, the scope of this study is constrained due to the unavailability of an MRI dataset featuring patients recovering from tumours with mild symptoms of Alzheimer’s. Despite this limitation, the direction shows promise, as a comparable architecture to \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\) achieves a train and test accuracy of \(98\%\) and \(97\%\) on ‘No Disease,’ ‘Tumour,’ and ‘Alzheimer’ classes. However, its applicability is restricted when diagnosing both diseases simultaneously, as the ‘Tumour’ class only includes patients with tumours and no signs of Alzheimer’s, and the same applies to the ‘Alzheimer" class. Additionally, theoretical analysis of the workflow can be a possible research direction.
Data availability
All datasets used in the experiments are publicly available and cited.
Code availability
The code has been made publicly available at https://github.com/sg-research08/Brain-Analysis.
Notes
thanks to advancements in medical science.
References
Aamir M, Rahman Z, Dayo ZA, Abro WA, Uddin MI, Khan I, Imran AS, Ali Z, Ishfaq M, Guan Y et al (2022) A deep learning approach for brain tumour classification using mri images. Comput Electric Eng 101:108105
Abd El-Wahab BS, Nasr ME, Khamis S, Ashour AS (2023) Btc-fcnn: fast convolution neural network for multi-class brain tumour classification. Health Inform Sci Syst 11(1):3
Acharya UR, Fernandes SL, WeiKoh JE, Ciaccio EJ, Fabell MKM, Tanik UJ, Rajinikanth V, Yeong CH (2019) Automated detection of Alzheimer’s disease using brain mri images-a study with various feature extraction techniques. J Med Syst 43:1–14
Acquarelli J, van Laarhoven T, Postma GJ, Jansen JJ, Rijpma A, van Asten S, Heerschap A, Buydens LM, Marchiori E (2022) Convolutional neural networks to predict brain tumour grades and Alzheimer’s disease with mr spectroscopic imaging data. PLoS ONE 17(8):e0268881
Ahmed G, Er MJ, Fareed MMS, Zikria S, Mahmood S, He J, Asad M, Jilani SF, Aslam M (2022) Dad-net: Classification of Alzheimer’s disease using Adasyn oversampling technique and optimized neural network. Molecules 27(20):7085
Arabahmadi M, Farahbakhsh R, Rezazadeh J (2022) Deep learning for smart healthcare–a survey on brain tumour detection from medical imaging. Sensors 22(5):1960
Asif S, Zhao M, Chen X, Zhu Y (2023a) Bmri-net: A deep stacked ensemble model for multi-class brain tumour classification from mri images. Interdiscipl Sci Comput Life Sci 15:1–16
Asif S, Zhao M, Tang F, Zhu Y (2023b) An enhanced deep learning method for multi-class brain tumour classification using deep transfer learning. Multimed Tools Appl 82:1–28
Balasundaram A, Srinivasan S, Prasad A, Malik J, Kumar A (2023) Hippocampus segmentation-based Alzheimer’s disease diagnosis and classification of mri images. Arab J Sci Eng 48:1–17
Ballas N, Yao L, Pal C, Courville A (2015) Delving deeper into convolutional networks for learning video representations. arXiv preprint arXiv:1511.06432
Bangyal WH, Rehman NU, Nawaz A, Nisar K, Ibrahim AAA, Shakir R, Rawat DB (2022) Constructing domain ontology for Alzheimer disease using deep learning based approach. Electronics 11(12):1890
Chandaran SR, Muthusamy G, Sevalaiappan LR, Senthilkumaran N (2022) Deep learning-based transfer learning model in diagnosis of diseases with brain magnetic resonance imaging. Acta Polytech Hung 19(5):127–147
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) Smote: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357
Cheng J (2017) 4. brain tumour dataset. 10.6084/m9.figshare.1512427.v5
Dewan JH, Thepade SD, Deshmukh P, Deshmukh S, Katpale P, Gandole K (2023) Comparative analysis of deep learning models for brain tumour detection using transfer learning. In: 2023 4th International Conference for Emerging Technology (INCET), pp 1–7. IEEE
Du Y, Fu Z, Calhoun VD (2018) Classification and prediction of brain disorders using functional connectivity: promising but challenging. Front Neurosci 12:525
Dubey S (2019) Alzheimer’s dataset ( 4 class of images)
Duchi J, Hazan E, Singer Y (2011) Adaptive subgradient methods for online learning and stochastic optimization. J Mach Learn Res 12(7):2121–2159
El-Latif AAA, Chelloug SA, Alabdulhafith M, Hammad M (2023) Accurate detection of Alzheimer’s disease using lightweight deep learning model on mri data. Diagnostics 13(7):1216
Escarcega RD, Patil AA, Manchon JFM, Urayama A, Dabaghian YA, Morales R, McCullough LD, Tsvetkov AS (2022) Chemotherapy as a risk factor for Alzheimer’s disease. Alzheimer’s Dement 18:e067196
Fran C et al (2017) Deep learning with depth wise separable convolutions. In: IEEE Conference on computer vision and pattern recognition (CVPR)
Gangopadhyay T, Halder S, Dasgupta P, Chatterjee K, Ganguly D, Sarkar S, Roy S (2022) Mtse u-net: an architecture for segmentation, and prediction of fetal brain and gestational age from mri of brain. Netw Model Anal Health Inform Bioinform 11(1):50
Ghazal TM, Abbas S, Munir S, Khan M, Ahmad M, Issa GF, Zahra SB, Khan MA, Hasan MK (2022) Alzheimer disease detection empowered with transfer learning. Comput Mater Continua 70(3):5005–5019
Gupta S, Meena J, Gupta O (2020) Neural network based epileptic eeg detection and classification. ADCAIJ Adv Distrib Comput Artif Intell J 9(2):23–32
Gupta S, Ranga V, Agrawal P (2021) Epilnet: a novel approach to iot based epileptic seizure prediction and diagnosis system using artificial intelligence. ADCAIJ Adv Distrib Comput Artif Intell J 10(4):435
Hadjouni M, Elmannai H, Saad A, Altahe A, Elaraby A (2023) A novel deep learning approach for brain tumours classification using mri images. Traitement du Signal 40(3):108105
Hamada A (2020) Br35h: brain tumour detection 2020
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on computer vision and pattern recognition, pp. 770–778
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE Conference on computer vision and pattern recognition, pp 4700–4708
Ingeno L (2019) Hormone therapy for prostate cancer may raise risk of Alzheimer’s, dementia
Ismail WN, F. Rajeena PP, Ali MA (2022) Multforad: Multimodal mri neuroimaging for Alzheimer’s disease detection based on a 3d convolution model. Electronics 11(23):3893
Kabiraj A, Meena T, Reddy PB, Roy S (2022) Detection and classification of lung disease using deep learning architecture from x-ray images. In: International Symposium on visual computing, pp 444–455. Springer
Kao YS, Yeh CC, Chen YF (2023) The relationship between cancer and dementia: an updated review. Cancers 15(3):640
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems 25
Kujur A, Raza Z, Khan AA, Wechtaisong C (2022) Data complexity based evaluation of the model dependence of brain mri images for classification of brain tumour and alzheimer’s disease. IEEE Access 10:112117–112133. https://doi.org/10.1109/ACCESS.2022.3216393
Kumar V, Abbas AK, Fausto N, Aster JC (2014) Robbins and Cotran pathologic basis of disease, professional edition e-book. Elsevier health sciences
Lehrer S (2018) Glioma and Alzheimer’s disease. J Alzheimer’s Dise Rep 2(1):213–218
Loddo A, Buttau S, Di Ruberto C (2022) Deep learning based pipelines for Alzheimer’s disease diagnosis: a comparative study and a novel deep-ensemble method. Comput Biol Med 141:105032
Majd S, Power J, Majd Z (2019) Alzheimer’s disease and cancer: when two monsters cannot be together. Front Neurosci 13:155
Marwa EG, Moustafa HED, Khalifa F, Khater H, AbdElhalim E (2023) An mri-based deep learning approach for accurate detection of Alzheimer’s disease. Alex Eng J 63:211–221
Mehnatkesh H, Jalali SMJ, Khosravi A, Nahavandi S (2023) An intelligent driven deep residual learning framework for brain tumour classification using mri images. Expert Syst Appl 213:119087
Mohammed BA, Senan EM, Rassem TH, Makbol NM, Alanazi AA, Al-Mekhlafi ZG, Almurayziq TS, Ghaleb FA (2021) Multi-method analysis of medical records and mri images for early diagnosis of dementia and Alzheimer’s disease based on deep learning and hybrid methods. Electronics 10(22):2860
Mohsen H, El-Dahshan E, El-Horbaty E, Salem A (2017) Brain tumour type classification based on support vector machine in magnetic resonance images. Annals Of “Dunarea De Jos” University Of Galati, Mathematics, Physics, Theoretical mechanics, Fascicle II, Year IX (XL) 1
Mohsen H, El-Dahshan ESA, El-Horbaty ESM, Salem ABM (2018) Classification using deep learning neural networks for brain tumours. Future Comput Inform J 3(1):68–71
Murugan S, Venkatesan C, Sumithra M, Gao XZ, Elakkiya B, Akila M, Manoharan S (2021) Demnet: a deep learning model for early diagnosis of Alzheimer diseases and dementia from mr images. Ieee Access 9:90319–90329
Namachivayam A, Puviarasan N (2023) Computerized brain disease classification using transfer learning. Int J Intell Syst Appl Eng 11(7s):536–544
Nancy Noella R, Priyadarshini J (2023) Machine learning algorithms for the diagnosis of Alzheimer and Parkinson disease. J Med Eng Technol 47(1):35–43
Nickparvar M (2021) Brain tumour mri dataset
Özkaraca O, Bağrıaçık Oİ, Gürüler H, Khan F, Hussain J, Khan J, Laila Ue (2023) Multiple brain tumour classification with dense cnn architecture using brain mri images. Life 13(2):349
Ozkaya C, Sagiroglu S (2023) Glioma grade classification using cnns and segmentation with an adaptive approach using histogram features in brain mris. IEEE Access 11:52275–52287. https://doi.org/10.1109/ACCESS.2023.3273532
Pal D, Reddy PB, Roy S (2022) Attention uw-net: a fully connected model for automatic segmentation and annotation of chest x-ray. Comput Biol Med 150:106083
Ranga V, Gupta S, Meena J, Agrawal P (2020) Automated human mind reading using eeg signals for seizure detection. J Med Eng Technol 44(5):237–246
Ranga V, Gupta S, Agrawal P, Meena J (2022) Pathological analysis of blood cells using deep learning techniques. Recent Adv Comput Scie Commun (Formerly: Recent Patents on Computer Science) 15(3):397–403
Roe CM, Fitzpatrick A, Xiong C, Sieh W, Kuller L, Miller J, Williams M, Kopan R, Behrens MI, Morris J (2010) Cancer linked to Alzheimer disease but not vascular dementia. Neurology 74(2):106–112
Roy S, Shoghi KI (2019) Computer-aided tumour segmentation from t2-weighted mr images of patient-derived tumour xenografts. In: Image Analysis and Recognition: 16th International Conference, ICIAR 2019, Waterloo, ON, Canada, August 27–29, 2019, Proceedings, Part II 16, pp. 159–171. Springer
Roy S, Bhattacharyya D, Bandyopadhyay SK, Kim TH (2017a) An effective method for computerized prediction and segmentation of multiple sclerosis lesions in brain mri. Comput Methods Programs Biomed 140:307–320
Roy S, Bhattacharyya D, Bandyopadhyay SK, Kim TH (2017b) An iterative implementation of level set for precise segmentation of brain tissues and abnormality detection from mr images. IETE J Res 63(6):769–783
Roy S, Meena T, Lim SJ (2022) Demystifying supervised learning in healthcare 4.0: a new reality of transforming diagnostic medicine. Diagnostics 12(10):2549
Ruder S (2016) An overview of gradient descent optimization algorithms. arXiv preprint arXiv:1609.04747
Sachdeva J, Kumar V, Gupta I, Khandelwal N, Ahuja CK (2016) A package-sfercb-"segmentation, feature extraction, reduction and classification analysis by both svm and ann for brain tumours". Appl Soft Comput 47:151–167
Salçin K et al (2019) Detection and classification of brain tumours from mri images using faster r-cnn. Tehnički glasnik 13(4):337–342
Salehi W, Baglat P, Gupta G, Khan SB, Almusharraf A, Alqahtani A, Kumar A (2023) An approach to binary classification of Alzheimer’s disease using lstm. Bioengineering 10(8):950
Samhan LF, Alfarra AH, Abu-Naser SS (2022) Classification of Alzheimer’s disease using convolutional neural networks. Int J Acad Inform Syst Res (IJAISR) 6(3):18–23
Sánchez-Valle J, Tejero H, Ibáñez K, Portero JL, Krallinger M, Al-Shahrour F, Tabarés-Seisdedos R, Baudot A, Valencia A (2017) A molecular hypothesis to explain direct and inverse co-morbidities between Alzheimer’s disease, glioblastoma and lung cancer. Sci Rep 7(1):1–12
Saurav S, Sharma A, Saini R, Singh S (2023) An attention-guided convolutional neural network for automated classification of brain tumour from mri. Neural Comput Appl 35(3):2541–2560
Shafi A, Rahman MB, Anwar T, Halder RS, Kays HE (2021) Classification of brain tumours and auto-immune disease using ensemble learning. Inform Med Unlock 24:100608
Shanmugam JV, Duraisamy B, Simon BC, Bhaskaran P (2022) Alzheimer’s disease classification using pre-trained deep networks. Biomed Signal Process Control 71:103217
Shukla A, Tiwari R, Tiwari S (2023) Alz-convnets for classification of Alzheimer disease using transfer learning approach. SN Computer Science 4(4):404
Shukla A, Tiwari R, Tiwari S (2023) Review on Alzheimer disease detection methods: automatic pipelines and machine learning techniques. Sci 5(1):13
Simonyan K, Zisserman A (2015) Very deep convolutional networks for large-scale image recognition. In: 3rd International Conference on Learning Representations (ICLR 2015). Computational and Biological Learning Society
Speidell AP, Demby T, Lee Y, Rodriguez O, Albanese C, Mandelblatt J, Rebeck GW (2019) Development of a human apoe knock-in mouse model for study of cognitive function after cancer chemotherapy. Neurotox Res 35:291–303
Srinivas C, NP, KS, Zakariah M, Alothaibi YA, Shaukat K, Partibane B, Awal H et al (2022) Deep transfer learning approaches in performance analysis of brain tumour classification using mri images. J Healthc Eng 2022
Staff N (2018) Gene tied to Alzheimer’s may be associated with cancer-related cognitive problems
Szegedy C, Ioffe S, Vanhoucke V, Alemi A (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Proceedings of the AAAI Conference on artificial intelligence, Volume 31
Tan M, Le Q (2019) Efficientnet: rethinking model scaling for convolutional neural networks. In: International Conference on machine learning, pp 6105–6114. PMLR
Taree A, Eslami V, Emamzadehfard S (2020) Approach to brain magnetic resonance imaging for non-radiologists. J Neurol Res 10(5):173–176
Thanki R, Kaddoura S (2022) Dual learning model for multiclass brain tumour classification. In: International Conference on dependability and complex systems, pp 350–360. Springer
Yazdan SA, Ahmad R, Iqbal N, Rizwan A, Khan AN, Kim DH (2022) An efficient multi-scale convolutional neural network based multi-class brain mri classification for samd. Tomography 8(4):1905–1927
Yildirim M, Cinar A (2020) Classification of alzheimer’s disease mri images with cnn based hybrid method. Ingénierie des Systèmes d Inf 25(4):413–418
Zulfiqar F, Bajwa UI, Mehmood Y (2023) Multi-class classification of brain tumour types from mr images using efficientnets. Biomed Signal Process Control 84:104777
Funding
No funding.
Author information
Authors and Affiliations
Contributions
All authors have contributed equally. Shivam and Sudip were more involved in technical areas of deep learning. Deepti was more involved in helping us with medical concerns and knowledge.
Corresponding author
Ethics declarations
Conflict of interest
No potential competing interest was reported by the authors.
Ethics approval
Not applicable.
Consent for publication
The paper is the authors’ own original work, which has not been previously published elsewhere. The paper is not currently being considered for publication elsewhere. The paper reflects the author’s own research and analysis truthfully and completely. The paper properly credits the meaningful contributions of co-authors and co-researchers.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ghosh, S., Deepti & Gupta, S. \(\mathcal {B}\text {rain}{\mathcal{M}\mathcal{N}}\text {et}\): a unified neural network architecture for brain image classification. Netw Model Anal Health Inform Bioinforma 13, 11 (2024). https://doi.org/10.1007/s13721-024-00443-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-024-00443-8