Abstract
Low-grade fibromyxoid sarcoma (LGFMS) is a rare soft-tissue sarcoma genetically characterized by the presence of the FUS-CREB3L2 gene fusion. While LGFMS exhibits indolent features during its early stages, the rates of recurrence, metastasis, and death from the disease are high. Presently, the role of FUS-CREB3L2 gene fusions in the unique features of LGFMS is not clear, and there is no modality to improve the clinical outcomes of patients with LGFMS; thus, extensive studies on LGFMS are required. Patient-derived cancer cell lines are critical tools for cancer research. However, no cell line has been established for LGFMS. Here, we aimed to develop a novel cell line for LGFMS and successfully established it using surgically resected tumor tissues. The cells, named NCC-LGFMS1-C1, possessed the same fusion genes as their original tumor and visible copy number variations. The cells had a fibroblastic appearance, formed spheroids when they were seeded in a low-attachment dish, and exhibited constant growth and invasion. Additionally, we demonstrated the feasibility of high-throughput drug screening using these cells. In conclusion, the NCC-LGFMS1-C1 cell line is a useful tool for studying LGFMS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Low-grade fibromyxoid sarcoma (LGFMS) is an indolent and malignant soft tissue sarcoma. It is histologically characterized by a fibroblastic appearance with alternating collagenous hypocellular and cellular myxoid nodules. The presence of deceptively bland spindle cells with whirling growth patterns and arcades of small blood vessels is also a characteristic of LGFMS. Although rare among sarcomas, because of its histological propensity to mimic other soft tissue sarcomas, the incidence of LGFMS is unclear. More than 90% of LGFMS cases have t (7;16) (q33; p11) or supernumerary ring chromosomes, both of which result in FUS-CREB3L2 gene fusions [1,2,3,4]. Other rare cases of LGFMS include FUS-CREB3L1 or EWSR1-CREB3L1 gene fusion [1, 5, 6]. The chimeric proteins of these fusion genes function as aberrant transcription factors, affecting the expression of target genes such as CD24 and MUC4 [4]. LGFMS has a unique clinical course, since the disease initially presents with indolent features. In the first year after excision of the primary tumor, the rates of recurrence and metastasis are 10% and 5%, respectively [3, 7,8,9,10,11]. However, a long follow-up study showed that the rates of recurrence, metastasis, and death from disease were 64%, 45%, and 42%, respectively [10]. Thus, novel therapies based on the understanding of the nature of LGFMS that can then inhibit disease progression are required to improve long-term prognosis.
Patient-derived cancer cells, which recapitulate the genotype and phenotype of tumor cells in original tumors, have been a vital resource for accelerating the understanding of the mechanisms of the etiology and progression of diseases. As tumor tissues have various heterogeneities in and among patients with cancers, different types of cancer models, such as cell lines and xenografts, have been developed in the last several decades. Among the various cancer models, cell lines have a unique advantage; they permit the study of the functional significance of genetic mutations and allow the cost-effective screening of the antitumor activity of novel oncology drugs. For instance, the integration of drug response and genetic data using a collection of cell lines has recently led to the discovery of predictive biomarkers for various types of malignancies [12,13,14,15,16,17,18,19,20]. However, due to the scarcity of patients with LGFMS, no cell line has been established according to Cellosaurus [21]. These observations indicate that cell lines are required to develop novel therapies for LGFMS.
Here, we report a novel LGFMS cell line, NCC-LGFMS1-C1. The cell line was established from surgically resected tumor tissue from a patient with LGFMS. To demonstrate the utility of this cell line, we characterized proliferation, spheroid formation, and invasion capabilities. Additionally, we demonstrated the practicality of use of this cell line in drug screening. To the best of our knowledge, this is the first report on an LGFMS cell line. Our cell line is anticipated to be useful for further studies on LGFMS.
Materials and methods
Patient history
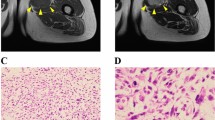
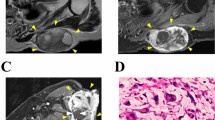
The patient was a 45-year-old woman with LGFMS. The patient visited the previous hospital with the chief complaint of a mass on her epigastrium. Computed tomography detected a mass in the rectus abdominis muscle (Fig. 1a). Thus, the patient was referred to the National Cancer Center Hospital (Tokyo, Japan) for further treatment. Magnetic resonance imaging confirmed a soft tissue tumor in the rectus abdominis muscle (Fig. 1b, c). The tumor was diagnosed as LGFMS upon needle biopsy. Because there was no evidence of distant metastasis, a wide resection was performed. Pathologically, the tumor consisted of swirling fascicles of mildly atypical long spindle cells within the fibromyxoid stroma (Fig. 1d) and was diffusely positive for MUC4 based on immunohistochemistry (Fig. 1e). Fluorescence in situ hybridization (FISH) analysis confirmed FUS rearrangement (Fig. 1f). A part of the resected tumor at the time of surgery was used to establish the cell line described in this study. The ethical committee of the National Cancer Center approved the use of clinical materials for this study, and written informed consent was obtained from the patient donor.
Clinical and pathological data. A Computed tomography showing a mass in the rectus abdominis muscle. Magnetic resonance imaging showing a mass with (B) iso intensity in T1-weighted image and (C) moderate high and low intensity in T2-weighted image. Yellow arrows indicate the tumor. D H&E staining showing swirling fascicles of mildly atypical long spindle cells within fibromyxoid stroma. E MUC4 immunohistochemistry was diffusely positive. F FISH assay using a FUS break-apart probe set displayed split green and orange or isolated green signals in the great majority of cells, indicating FUS rearrangement (orange arrows)
Donor tumor analyses
Histological analysis
Histological examination was performed on 4 μm-thick sections obtained from a representative paraffin-embedded tumor sample. Sections were deparaffinized and stained with hematoxylin and eosin (HE).
Immunohistochemistry
Immunohistochemistry was performed using deparaffinized tumor samples. The sections were exposed to 3% hydrogen peroxide for 15 min to block endogenous peroxidase activity and then washed in deionized water for 2–3 min. The preparations were subjected to heat-induced epitope retrieval. A primary antibody against MUC4 was used (clone 8G7; 1:400; Santa Cruz Biotechnology, Dallas, TX, USA). The slides were incubated for 1 h at room temperature and subsequently labeled with peroxidase (EnVision system, Dako, Santa Clara, CA, USA).
FISH
FISH analysis was performed on 4 µm-thick sections of the tumor sample. Break-apart probes (Vysis FUS Dual Color Break Apart Probe; Abbott Molecular, Abbott Park, IL, USA) were used to detect FUS genes. Images of the samples were collected for FISH using the Metafer Slide Scanning Platform (MetaSystems, Altlussheim, Germany).
Cell culture procedure
Surgically resected tumor tissue was used for cell line establishment, as previously described [22]. The cells were maintained in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (Gibco, Grand Island, NY, USA) supplemented with 5% heat-inactivated fetal bovine serum (Gibco), 100 μg/mL penicillin, and 100 μg/mL streptomycin (Nacalai Tesque, Kyoto, Japan), 0.4 µg/mL hydrocortisone (Sigma-Aldrich, St. Louis, MO, USA), 5 ng/mL EGF (Sigma-Aldrich), 10 ng/mL bFGF (Sigma-Aldrich), 5 µg/mL insulin (Sigma-Aldrich), and 10 µM Y-27632 (Selleck Chemicals, Houston, TX, USA: Rock inhibitor) at 37 °C in a humidified atmosphere with 5% CO2. The cells were maintained for more than 1 year under tissue culture conditions and were passaged more than 30 times.
Authentication and quality control of the cell line
We examined short tandem repeats (STRs) in 10 loci using the GenePrint 10 system (Promega, Madison, WI, USA) according to our previous report [22]. In brief, genomic DNA was extracted from the original tumor tissue or tissue-cultured cells using AllPrep DNA/RNA Mini kits (Qiagen, Hilden, Germany) and quantified using a NanoDrop 8000 instrument (Thermo Fisher Scientific, Waltham, MA, USA). We amplified the STRs of the extracted DNA by PCR and analyzed them using a 3500xL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). We examined the STR data using GeneMapper software (Applied Biosystems) and compared the obtained STR profiles with those of previously reported cell lines using the Cellosaurus function [21]. We amplified the DNA sequence unique to mycoplasma using the e-Myco Mycoplasma PCR Detection Kit (Intron Biotechnology, Gyeonggi-do, Korea) and observed the results with gel electrophoresis.
Genetic analysis
DNA amplification and sequencing
We extracted DNA and total RNA from the excised tissue or cultured cells using the AllPrep DNA/RNA Mini Kit (Qiagen). Complementary DNA was generated by reverse transcription (RT) reaction with Superscript III reverse transcriptase e (Invitrogen, Carlsbad, CA, USA). We amplified the FUS-CREB3L2 fusion transcript with the FUS forward primer FUS-F (5′-GTAAAACGACGGCCAGAGCCAGTCCACGGACACTTCAGGC-3′) and CREB3L2 reverse primer CREB3L2-R (5'-CAGGAAACAGCTATGACGAATTTTCTTCAGGGCCTTCTCC-3′), using Platinum® Taq DNA Polymerase High Fidelity (Thermo Fisher Scientific). We further amplified the PCR products with the same forward and reverse primers for the junction to use for Sanger sequence analysis. Direct sequencing was performed using the BigDye v3.1 Cycle Sequencing Kit (Applied Biosystems) on the Applied Biosystems 3130xL by GENEWIZ (GENEWIZ, South Plainfeld, NJ, USA). We matched the obtained sequence data against the FUS (NM_004960.4) and CREB3L2 (NM_194071.4) sequences using the Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Single-nucleotide polymorphism (SNP) array and analysis
SNP array genotyping was performed to identify copy number alterations using an Infinium OmniExpressExome-8 v. 1.4 BeadChip (Illumina, San Diego, CA) according to the manufacturer’s instructions. In brief, we extracted genomic DNA from tumor cells and normal white blood cells and amplified it for the reaction with the array slides in an iScan system (Illumina). We calculated log R ratios and B allele frequencies using Genome Studio 2011.1 with cnvPartition v3.2.0 (Illumina) and KaryoStudio Data Analysis Software v. 1.0 (Illumina). We used human genome data, the hg19 version, for annotation mapping. We smoothed the whole-genome (except for chromosomes X and Y) log10 ratio (tumor/reference) value and detected abnormal copy number regions using the circular binary segmentation algorithm [23, 24] with the R version 4.0.3 (R Foundation for Statistical Computing, http://www.R-project.org) and DNAcopy package version 1.64.0 (Bioconductor, https://bioconductor.org/) [25]. Amplifications were defined as regions with copy number > 3. We defined deletions as regions with copy number < 1 in the tumor cells. Among identified genes with copy number alterations, we searched for “cancer-related genes” using “Cancer Gene Census” in the Catalog of Somatic Mutations in Cancer (COSMIC) database (GRCh 37 v91) [25].
Evaluation of cell physiology
Spheroid formation assay
We performed spheroid formation assays, as previously described [22]. In brief, we seeded 1 × 105 cells/well into 96-well plates (96-well Clear Round Bottom Ultra Low Attachment Microplate; Corning, Inc., Corning, NY, USA) in the medium. The cells were maintained in a humidified atmosphere of 5% CO2 at 37 °C for 3 days. We confirmed spheroid formation using microscopic observations. The spheroids were embedded in iPGell (Genostaff, Tokyo, Japan) according to the manufacturer’s instructions and fixed with 10% formalin neutral buffer solution. We generated four-micrometer-thick paraffin sections and stained them with HE for microscopic observations.
Cell proliferation assay
The cells were seeded at a density of 2.5 × 104 cells/well in 24-well culture plates on day 0. The number of cells was counted at multiple time points. Doubling time was calculated based on the growth curve.
Real-time quantitative cell analysis (RTCA)
To examine the potential for invasion, we used RTCA according to the manufacturer’s instructions. Briefly, we layered Matrigel at a concentration of 9.3 mg/mL (BD Biosciences, Franklin Lakes, NJ, USA) onto the membrane in the upper chamber and seeded 4 × 104 cells onto it. MG63 osteosarcoma cells (JCRB, Osaka, Japan) were used as controls. We added the medium used to maintain the cells in the lower chamber. The tumor cells on the Matrigel-coated membrane migrated to the bottom chamber and adhered to the electronic sensors on the underside of the membrane. The attached tumor cells influenced the electrical impedance of the electronic sensors. We estimated the invasion capability of the cells based on the positive correlation between impedance and the number of cells. The impedance was monitored every 15 min for 72 h, and the results were plotted as a function of time after seeding.
Screening for the antiproliferative effects of anticancer reagents
The cells were seeded in a 384-well plate (Thermo Fisher Scientific, Fair Lawn, NJ, USA) at 5 × 103 cells/well in the medium and incubated at 37 °C in a humidified atmosphere of 5% CO2 with the Bravo automated liquid handling platform (Agilent Technologies, Santa Clara, CA, USA). On the following day, we added 214 anticancer agents, including FDA-approved drugs (Selleck Chemicals, Houston, TX, USA), at a concentration of 10 µM, using the Bravo automated liquid handler. A list of anticancer agents is presented in Supplementary Table 1. After 72 h, the survival rates were assessed using the CCK-8 reagent according to the manufacturer’s protocol. The response readout relative to the DMSO control was calculated in terms of relative growth inhibition.
We then performed dose–response experiments to validate the hits available in the pilot screening. We determined the IC50, the sample concentration required to inhibit cell growth by 50% in comparison with the growth of the cell control, from the dose–response curves. Serial dilutions of the compounds at different concentrations were dispensed in 384-well plates, using the Bravo Automated liquid handler. Cells were collected by trypsinization. We then dispensed 25 μL of single-cell suspensions (5 × 103 cells) into each well using the Bravo Automated liquid handler. The plates were incubated for 72 h at 37 °C and 5% CO2.
Cell viability was determined using the CCK-8 assay. A multimode Epoch multiplate reader (BioTek, Winooski, VT, USA) was used for signal quantification. We plotted the signal values resulting from normalized cell numbers against the drug concentrations tested in order to obtain IC50 values using GraphPad Prism 9.1.1 software. This analysis was performed in duplicate.
Results
Authentication of the cell line
We authenticated the NCC-LGFMS1-C1 cell line by analyzing the status of its STRs. The STRs of NCC-LGFMS1-C1 were compared with those of the corresponding original tumor tissue, and it was confirmed that all of the examined STRs were identical (Table 1). We checked if any cell line available in public cell banks possessed the STR patterns of our NCC-LGFMS1-C1 cells using the Cellosaurus cell line database. We also confirmed that a DNA sequence unique to Mycoplasma was absent in the tissue culture medium of the NCC-LGFMS1-C1 cells (data not shown).
Characterization of the cell line
We confirmed the presence of the FUS-CREB3L2 gene fusion in the original tumor tissue as well as in NCC-LGFMS1-C1 cells (Fig. 2). While the SNP array indicated genomic instability, there was not a considerable amount of copy number alterations in NCC-LGFMS1-C1 cells (Fig. 3).
NCC-LGFMS1-C1 cells exhibited a fibroblastic appearance under monolayer culture conditions when evaluated using phase contrast microscopy (Fig. 4a, b). NCC-LGFMS-C1 cells formed spheroids when they were seeded in a low-attachment tissue culture plate (Fig. 4c, d). In spheroids, they had a pleomorphic and atypical appearance. Based on the growth curve, NCC-LGFMS1-C1 cells showed constant proliferation (Fig. 4e), and the population doubling time during the logarithmic growth phase was determined to be approximately 149.9 h. We also found that NCC-LGFMS1-C1 had an increased invasibility compared to MG63 cells (Fig. 4f).
Characterization of NCC-LGFMS1-C1 cells. NCC-LGFMS1-C1 cells presented with a fibroblastic appearance under 2D culture conditions (A and B). Spheroid formation of NCC-LGFMS1-C1 cells was observed when they were seeded in an ultra-low attachment plate (C and D). Growth curve of NCC-LGFMS1-C1 assessed using RTCA (E). The Y-axis indicates the cell proliferation plot of the NCC-LGFMS1-C1 cells, and the x-axis represents the day after passage. The invasion ability of NCC-LGFMS1-C1 cells was compared to that of MG63 cells acting as the control (F)
Anticancer agent response
We examined the cell viability of NCC-LGFMS1-C1 cells after treatment with 214 anticancer agents at a fixed concentration of 10 µM. The results are summarized in Supplementary Table 2. Among the 214 anticancer agents examined, we selected 24 agents that either exerted significant antiproliferative effects on NCC-LGFMS1-C1 cells or were frequently used as standard chemotherapy for sarcomas and examined their IC50 values (Table 2). The growth suppressive effects of the six agents that demonstrated the lowest IC50 values are shown in Fig. 5.
Growth curves for determining the IC50 values of the anticancer agents. Growth curves calculated for determining the IC50 values of the investigated anticancer agents (A–F). Viability of cells treated with different concentrations of anticancer agents. Each anticancer agent is labeled below the graph
Discussion
Cell lines are a fundamental bio-resource for cancer research that enables the functional study of critical genes and permits the evaluation of the antitumor effects of novel drugs. The paucity of cell lines can hinder the progress of research and development. Establishment of patient-derived cancer cell lines requires fresh tumor tissues or cells, and it is difficult to obtain such materials in rare cancers such as LGFMS. In this study, we established NCC-LGFMS1-C1, a novel cell line, derived from a patient with LGFMS. To the best of our knowledge, NCC-LGFMS1-C1 is the first LGFMS cell line.
We characterized the NCC-LGFMS1-C1 cells in terms of their genetic background, proliferation capability, spheroid formation, and invasion. FUS-CREB3L2 fusion is specific for LGFMS, except for nosologically related sclerosing epithelioid fibrosarcoma [26,27,28]. Our observations were consistent with previous reports that have stated the absence of recurrent genomic imbalances or single-nucleotide variants, except recurrent microdeletions in association with the t(7;16) chromosomal breakpoints and a gain of 7q in cases with ring chromosomes [4, 26]. The properties of FUS-CREB3L2 gene fusion in the etiology of LGFMS has not yet been established, and the clinical utility of this gene fusion is unclear. Thus, NCC-LGFMS1-C1 cell line will be a useful resource for studying the genetic background of LGFMS, including this aspect.
We also demonstrated that NCC-LGFMS1-C1 cells could be used for drug screening studies. We found that the drugs exerted remarkable growth suppression on NCC-LGFMS1-C1 cells. However, these drugs should be further validated using additional cell lines from different patients with LGFMS. In addition, the results obtained by tissue-cultured cell lines should be confirmed using other patient-derived cancer models, such as xenografts, before considering the clinical trials, because all cancer models are imperfect and capture only a portion of specific cancer types.
In a similar vein, more LGFMS cell lines should also be established, because the clinical and pathological features of LGFMS are diverse, and experimental findings obtained using a limited number of cell lines cannot provide conclusive results. Because of the extreme rarity of this disease, a multi-institutional collaboration study needs to be conducted, and the established cell lines should be shared within the research community.
We found that dasatinib and vinblastine exerted remarkable antiproliferative effects on NCC-LGFMS1-C1. Dasatinib is a tyrosine kinase inhibitor that targets BCR-ABL, KIT, the nonreceptor kinase SRC family, and ephrin receptor kinases. It is used as a first- and second-line treatment for patients with chronic myelogenous leukemia [29, 30]. The clinical utility of dasatinib as a first-line treatment for gastrointestinal stromal tumors has been explored in a single-arm phase 2 trial [31]. Brodin et al. reported that the response to dasatinib treatment could be predicted by examining the chemosensitivity of the ex vivo culture of patient-derived cells in sarcomas with translocations [32]. Our results indicate the possible clinical utility of dasatinib in LGFMS and the potential power of patient-derived cell lines. Vinblastine, a vinca alkaloid and a chemical analog of vincristine, is a microtubule dynamics inhibitor that prevents cell division during metaphase [33]. The FDA has approved the use of vinblastine as a palliative treatment for breast cancer, choriocarcinoma, Hodgkin lymphoma, non-Hodgkin sarcoma, Kaposi sarcoma, mycosis fungoides, and testicular germ cell tumors (National Cancer Institute, https://www.cancer.gov/about-cancer/treatment/drugs/vinblastinesulfate). The clinical utility of vinblastine has not been reported in soft-tissue sarcomas, and our results may warrant further investigation of vinblastine for the treatment of LGFMS.
We conclude that NCC-LGFMS1-C1 will be a useful patient-derived cancer cell line for investigating the molecular background that underlies tumor characteristics and developing novel therapeutic strategies for LGFMS. Because of the complexity of the disease, additional cell lines should be established for further research on LGFMS.
References
Mertens F, Fletcher CD, Antonescu CR, et al. Clinicopathologic and molecular genetic characterization of low-grade fibromyxoid sarcoma, and cloning of a novel FUS/CREB3L1 fusion gene. Lab Invest. 2005;85:408–15.
Matsuyama A, Hisaoka M, Shimajiri S, et al. Molecular detection of FUS-CREB3L2 fusion transcripts in low-grade fibromyxoid sarcoma using formalin-fixed, paraffin-embedded tissue specimens. Am J Surg Pathol. 2006;30:1077–84.
Guillou L, Benhattar J, Gengler C, et al. Translocation-positive low-grade fibromyxoid sarcoma: clinicopathologic and molecular analysis of a series expanding the morphologic spectrum and suggesting potential relationship to sclerosing epithelioid fibrosarcoma: a study from the French Sarcoma Group. Am J Surg Pathol. 2007;31:1387–402.
Möller E, Hornick JL, Magnusson L, Veerla S, Domanski HA, Mertens F. FUS-CREB3L2/L1-positive sarcomas show a specific gene expression profile with upregulation of CD24 and FOXL1. Clin Cancer Res. 2011;17:2646–56.
Lau PP, Lui PC, Lau GT, Yau DT, Cheung ET, Chan JK. EWSR1-CREB3L1 gene fusion: a novel alternative molecular aberration of low-grade fibromyxoid sarcoma. Am J Surg Pathol. 2013;37:734–8.
Rubinstein JC, Visa A, Zhang L, Antonescu CR, Christison-Lagay ER, Morotti R. Primary low-grade fibromyxoid sarcoma of the kidney in a child with the alternative EWSR1-CREB3L1 gene fusion. Pediatric Dev Pathol. 2014;17:321–6.
Billings SD, Giblen G, Fanburg-Smith JC. Superficial low-grade fibromyxoid sarcoma (Evans tumor): a clinicopathologic analysis of 19 cases with a unique observation in the pediatric population. Am J Surg Pathol. 2005;29:204–10.
Evans HL. Low-grade fibromyxoid sarcoma. A report of 12 cases. Am J Surg Pathol. 1993; 17:595–600.
Evans HL. Low-grade fibromyxoid sarcoma. A report of two metastasizing neoplasms having a deceptively benign appearance. Am J Clin Pathol. 1987; 88:615–9.
Evans HL. Low-grade fibromyxoid sarcoma: a clinicopathologic study of 33 cases with long-term follow-up. Am J Surg Pathol. 2011;35:1450–62.
Goodlad JR, Mentzel T, Fletcher CD. Low grade fibromyxoid sarcoma: clinicopathological analysis of eleven new cases in support of a distinct entity. Histopathology. 1995;26:229–37.
Barretina J, Caponigro G, Stransky N, et al. The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5.
Basu A, Bodycombe NE, Cheah JH, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–61.
Seashore-Ludlow B, Rees MG, Cheah JH, et al. Harnessing Connectivity in a Large-Scale Small-Molecule Sensitivity Dataset. Cancer Discov. 2015;5:1210–23.
Rees MG, Seashore-Ludlow B, Cheah JH, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol. 2016;12:109–16.
Haverty PM, Lin E, Tan J, et al. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature. 2016;533:333–7.
Iorio F, Knijnenburg TA, Vis DJ, et al. A Landscape of Pharmacogenomic Interactions in Cancer. Cell. 2016;166:740–54.
Behan FM, Iorio F, Picco G, et al. Prioritization of cancer therapeutic targets using CRISPR-Cas9 screens. Nature. 2019;568:511–6.
Townsend EC, Murakami MA, Christodoulou A, et al. The public repository of xenografts enables discovery and randomized phase II-like trials in mice. Cancer Cell. 2016;29:574–86.
Bairoch A. The Cellosaurus, a cell-Line knowledge resource. J Biomol Tech. 2018;29:25–38.
Yoshimatsu Y, Noguchi R, Tsuchiya R, et al. Establishment and characterization of NCC-CDS2-C1: a novel patient-derived cell line of CIC-DUX4 sarcoma. Hum Cell. 2020;33:427–36.
Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72.
Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–63.
Tate JG, Bamford S, Jubb HC, et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47:D941–7.
Arbajian E, Puls F, Antonescu CR, et al. In-depth genetic analysis of sclerosing epithelioid fibrosarcoma reveals recurrent genomic alterations and potential treatment targets. Clin Cancer Res. 2017;23:7426–34.
Prieto-Granada C, Zhang L, Chen HW, et al. A genetic dichotomy between pure sclerosing epithelioid fibrosarcoma (SEF) and hybrid SEF/low-grade fibromyxoid sarcoma: a pathologic and molecular study of 18 cases. Genes Chromosomes Cancer. 2015;54:28–38.
Dewaele B, Libbrecht L, Levy G, et al. A novel EWS-CREB3L3 gene fusion in a mesenteric sclerosing epithelioid fibrosarcoma. Genes Chromosomes Cancer. 2017;56:695–9.
Jabbour E, Kantarjian HM, Saglio G, et al. Early response with dasatinib or imatinib in chronic myeloid leukemia: 3-year follow-up from a randomized phase 3 trial (DASISION). Blood. 2014;123:494–500.
Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401.
Montemurro M, Cioffi A, Dômont J, et al. Long-term outcome of dasatinib first-line treatment in gastrointestinal stromal tumor: a multicenter, 2-stage phase 2 trial (Swiss Group for Clinical Cancer Research 56/07). Cancer. 2018;124:1449–54.
Brodin BA, Wennerberg K, Lidbrink E, et al. Drug sensitivity testing on patient-derived sarcoma cells predicts patient response to treatment and identifies c-Sarc inhibitors as active drugs for translocation sarcomas. Br J Cancer. 2019;120:435–43.
Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–65.
Acknowledgements
We thank Drs. F. Nakatani, E. Kobayashi, T. Komatsubara (Department of Musculoskeletal Oncology), and Drs. C. Sato, T. Shibayama, H. Tanaka (Department of Diagnostic Pathology), National Cancer Center Hospital, for sampling tumor tissue specimens from surgically resected materials. We also appreciate the technical assistance provided by Ms. Y. Kuwata, R. Sasaki and T. Akiyama (Division of Rare Cancer Research). We appreciate the technical support provided by Ms. Y. Shiotani, Mr. N. Uchiya, and Dr. T. Imai (Central Animal Division, National Cancer Center Research Institute). We would also like to thank Editage (www.editage.jp) for their help with English language editing and their constructive comments on the manuscript. This research was technically assisted by the Fundamental Innovative Oncology Core in the National Cancer Center.
Funding
This research was supported by the Japan Agency for Medical Research and Development (grant number 20ck0106537h0001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics approval
The ethical committee of the National Cancer Center approved the use of clinical materials for this study (approval number 2004–050).
Informed consent
Written informed consent for publication was provided by the patient.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yoshimatsu, Y., Noguchi, R., Sin, Y. et al. Establishment and characterization of NCC-LGFMS1-C1: a novel patient-derived cell line of low-grade fibromyxoid sarcoma. Human Cell 34, 1919–1928 (2021). https://doi.org/10.1007/s13577-021-00612-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00612-1