Abstract
Background
Lung adenocarcinoma (LUAD) is the most common histological type of lung cancer with lower survival rates. Recent advancements in targeted therapies and immunotherapies targeting immune checkpoints have achieved remarkable success, there is still a large percentage of LUAD that lacks available therapeutic options. Due to tumor heterogeneity, the diagnosis and treatment of LUAD are challenging. Exploring the biology of LUAD and identifying new biomarker and therapeutic targets options are essential.
Method
We performed single-cell RNA sequencing (scRNA-seq) of 6 paired primary and adjacent LUAD tissues, and integrative omics analysis of the scRNA-seq, bulk RNA-seq and whole-exome sequencing data revealed molecular subtype characteristics. Our experimental results confirm that CDC25C gene can serve as a potential marker for poor prognosis in LUAD.
Results
We investigated aberrant gene expression in diverse cell types in LUAD via the scRNA-seq data. Moreover, multi-omics clustering revealed four subgroups defined by transcriptional profile and molecular subtype 4 (MS4) with poor survival probability, and immune cell infiltration signatures revealed that MS4 tended to be the immunosuppressive subtype. Our study revealed that the CDC25C gene can be a distinct prognostic biomarker that indicates immune infiltration levels and response to immunotherapy in LUAD patients. Our experimental results concluded that CDC25C expression affects lung cancer cell invasion and migration, might play a key role in regulating Epithelial-Mesenchymal Transition (EMT) pathways.
Conclusions
Our multi-omics result revealed a comprehensive set of molecular attributes associated with prognosis-related genes in LUAD at the cellular and tissue level. Identification of a subtype of immunosuppressive TME and prognostic signature for LUAD. We identified the cell cycle regulation gene CDC25C affects lung cancer cell invasion and migration, which can be used as a potential biomarker for LUAD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Lung cancer continues to be the foremost cause of global cancer-related mortality and constitutes 22.7% of malignant tumors [1, 2]. Lung adenocarcinoma (LUAD) is the most common histological subtype of non-small cell lung cancer (NSCLC) and accounts for approximately 40% of lung malignancies [3, 4]. Recent advancements in targeted therapies and immunotherapies targeting immune checkpoints have achieved remarkable success. Despite this progress, there is still a large percentage of LUAD that lacks available therapeutic options. This may be due to the lack of known genetic mutations or the difficulty in targeting oncogenic mutations. Accordingly, the challenge of effectively stratifying patients for treatment with appropriate therapeutic approaches and finding new molecular targets remain a crucial priority.
Varied evidence confirms that the process of tumor progression is closely related to the tumor microenvironment (TME), gene expression profiles and epigenetic modifications [5,6,7,8]. The scRNA-seq technologies have ushered in breakthroughs understanding TME of lung cancer [9, 10]. Comparing the bulk RNA-seq measured the total cell type-specific gene expression weighted by the cell type proportions, the scRNA-seq tend to understand the cell type composition and proportions in intact tissues is critical because certain cell types are more susceptible to tumorigenesis than others [11,12,13]. Though many studies have investigated abnormal gene at the tissue level, there is still a lack of comprehensive understanding of cell type-specific aberrant gene expression in LUAD at the cellular level.
In recent years, scRNA-seq technology has enabled cell type-specific transcriptome analyses. Thus, several studies have extensively characterized the LUAD TME at a single-cell resolution. Previous studies investigating lung cancer have focused primarily on the landscape of immune and infiltrating cell populations or have specifically identified novel cell subtypes and altered pathways [14,15,16]. These studies have unveiled distinct cellular and transcriptional modules that are associated with survival in lung cancer [17]. Additionally, they have shed light on the heterogeneity of the tumor microenvironment in early/advanced-stage LUAD harboring EGFR mutations [18, 19]. And other studies have highlighted the expansion of precursor exhausted T cells during anti-PD-1 therapy [20].
In this study, we use the scRNA-seq to characterize the differences and similarities of transcriptome expression in patients with LUAD by comparing with adjacent tissue, to determine the universal law of the aberrant gene expression at both cellular level and tissue level in LUAD. We integrated multi-omics data, such as the scRNA-seq, bulk mRNA-seq and genomic data collected from LUAD patients. Comprehensive molecular attributes associated with prognosis-related genes in the LUAD were revealed, serving as a resource for the cancer community to further delineate the underlying biology and address unmet clinical needs.
2 Materials and methods
2.1 Patient sample collection
Fresh tumor samples were collected from patients at West China Hospital (WCH). Written permission was obtained from each patient. The clinical parameters that were noted during recruitment, such as age, sex, smoking status, clinical subtype, and stage, are detailed in Supplementary Table S1.
2.2 Library preparation and sequencing
Freshly acquired resected tissues were washed with Hank’s balanced salt solution (HBSS) after the procedure, cut into smaller pieces on ice with collagenase I/IV in HBSS, and incubated for 30 min at 37 °C with manual shaking every 5 min. Following the passage of the digested tissues through a 40 µm nylon mesh filter, the cells that were still in suspension were centrifuged at 500× g for 5 min. The bombarded cells were suspended in red blood lysis solution after the supernatant was removed, and after being washed with HBSS, the cells were then resuspended in sorting buffer (0.04% BSA + PBS). Following the manufacturer’s recommendations, cell suspensions were immediately processed for single-cell RNA-seq after dead cells were removed by flow cytometry. The Chromium Single-cell 3′ Gel Bead, Chip and Library Kit v3 (10X Genomics) and the Chromium Single-cell Gene Expression Solution were used to produce single cells in accordance with the manufacturer’s instructions. The number of cells added to each channel varied between 8000 and 10,000. After the RNA was barcoded and the cells were lysed, the cells were separated into gel beads in emulsion in a chromium apparatus. Amplification, shearing, and attachment of the 5′ adaptor and sample index were subsequently performed. Libraries were sequenced at West China Hospital using the Illumina Nova-Seq 6000 platform.
2.3 Gene expression data, somatic mutation data, and clinical information
The mRNA expression profiles, somatic mutation data, and related clinical data of LUAD patients were obtained from the TCGA database via the University of California–Santa Cruz (UCSC) Xena browser (https://xena.ucsc.edu/). The somatic mutation data were analyzed via the “maftools” R package (version 2.2.10). The tumor mutation burden (TMB) scores were obtained by calculating the total number of mutations/exon length. The mRNA expression data and related clinical parameters used for the validation cohort were accessed from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov//geo//), which included the GSE30219 and GSE31210 datasets. Moreover, we performed log 2 transformations for all the mRNA expression data. When duplicate RNA expression values were found, we calculated and retained the average expression value. Patients whose survival time was ≤30 days were excluded from this study.
2.4 The scRNA-seq data clustering, dimension reduction, and cell annotation
We employ log normalization to standardize the merged data. Following this, we utilize the “FindVariableFeatures” function to identify the top 2000 genes with substantial variation. This identification is based on the variance stabilization transformation method. Additionally, all genes undergo scaling via the “ScaleData” function. Subsequently, we employ the “RunPCA” function to perform dimensionality reduction on the PCA for the previously selected 2000 highly variable genes. We choose a dimensionality of 20 (dims = 20). To cluster the cells, we apply the “FindNeighbors” and “FindClusters” functions, using a resolution of 0.5. These steps help identify the cell clusters. Lastly, we employ the “FindAllMarkers” function to identify the marker genes of the subgroups, using a log fold change (log2FC) of 0.25 and a minimum percentage (min. pct) of 0.25. We further filter the marker genes by utilizing an adjusted p value < 0.05.
2.5 Cell type pseudo-time and trajectory analysis
Pseudo-time and trajectory analyses of the scRNA-seq data were carried out with the ‘Monocle2’ package. In addition, differential expression analysis was performed between branches, and genes that exhibited differential expression levels were defined as branch-dependent or state-specific genes or marker genes. Then, the intracellular DEGs in cells with distinct differentiation states were defined according to the following criteria: |log2 FC| > 0.5 and FDR < 0.05. The “ClusterProfiler”, “org.Hs.eg.db”, “enrichplot” and “ggplot2” packages were used for GO annotation and KEGG enrichment analysis; only Biological Processes (BP) were extracted from the GO annotation.
2.6 Estimation of activity for diverse signatures and pathways
The GSVA algorithm was used to evaluate the relative activation status of a signature or pathway. GSVA scores for cancer hallmark pathways were calculated using predefined gene sets extracted from the “MSigDB” algorithms.
2.7 Based prognosis-related genes classifications of LUAD patients in the TCGA data
Unsupervised consensus clustering, an algorithm based on k-means machine learning, was utilized to explore the molecular classification of both TCGA cohorts based on the expression patterns of prognosis-related genes using the “ConsensusClusterPlus” package in R. The optimal number of clusters was determined by the relative change in the area under the CDF curves of the consensus score and consensus heatmap. K‒M survival analysis was subsequently performed to evaluate the prognosis of patients in the different subgroups.
2.8 Cell-type identification by estimating relative subsets of RNA transcripts
The “CIBERSORT” algorithms is based on the input matrix of a gene expression file to accurately estimate the relative proportions of various cell subsets in tissues. Spearman correlation analysis was performed to explore the association between the risk score and infiltrating immune cells. The “ggplot2” package was used to visualize the differences in the abundances of immune cells and the results of the correlation analysis.
2.9 Establishment of the survival risk-related model
The R package “glmnet” was used to perform least absolute shrinkage and selection operator (LASSO) Cox regression and to predict the prognosis of patients based on genes. First, the trajectory of change in each independent variable was examined. Cross-validation is used to select the tuning parameter. The outcome is simply the least squares estimate when lambda is small. The cox.ph function of the R package “survival” was used to create the Cox proportional hazards model, which was subsequently used to evaluate the consistency and variability of the estimates generated by the Lasso Cox regression model. Patients were separated into high- and low-expression groups based on the median score. A Cox proportional hazards model was fitted with patient death as the outcome, and a hazard ratio was computed using the fitted model.
2.10 Survival analysis and immunotherapeutic evaluation
Survival analysis was performed by the R package survival. The hazard ratio (HR) was calculated by the Cox proportional hazards model, the 95% CI was reported, and the Kaplan–Meier survival curve was modeled by the “survfit” function. The “maxstat.test” function of the R package “maxstat”, in which all potential cutoff points were repeatedly tested to find the maximum rank statistic, was used to perform dichotomization of cell population infiltration or gene expression and then to divide the patients into two groups according to the selected maximum logarithm statistics. The two-sided long-rank test was used to compare Kaplan–Meier survival curves.
2.11 Quantitative real time-PCR (qRT-PCR)
RNA was extracted from the cells after flow cytometry sorting using RNA-easy Isolation Reagent (Vazyme #R701). An iScript™ Advanced cDNA Synthesis Kit (Bio-Rad) was used for reverse transcription, and iTaq™ Universal SYBR® Green Supermix (Bio-Rad) was utilized for qRT‒PCR in accordance with the manufacturer’s instructions. The qRT‒PCR program was as follows: initial denaturation for 30 s at 95 °C; 40 cycles of denaturation for 5 s at 95 °C; and 40 cycles of amplification for 30 s at 60 °C. The mRNA expression in the cells was calculated as 2−(∆∆Ct), and β-actin was used as the internal reference. The primer sequences are shown in Supplementary Table S2.
2.12 Cell lines and cell culture
The human lung cancer cell lines A549, H1975, HCC95, and PC-9 were obtained from the Shanghai Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were routinely cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS (PAA Laboratories GmbH, Pasching, Austria) at 37 °C in a humidified atmosphere of 5% CO2 in the air.
2.13 RNA interference
Short-interfering RNAs (siRNAs) specific for Cell Division Cycle 25C (CDC25C) were purchased from RiboBio (Guangzhou, China) and then transfected into lung cancer cells using Lipofectamine 2000 reagent (Invitrogen, Shanghai, China) according to the manufacturer’s protocol. Cells transfected with the corresponding scrambled siRNA were used as controls. The gene silencing effect was measured by Western blotting 48 h post transfection.
2.14 Western blotting
Proteins were extracted from cell lysates with RIPA buffer (Thermo Fisher Scientific, Waltham, MA, USA), separated by 10% SDS–PAGE and subsequently transferred onto PVDF membranes (Millipore, Billerica, MA, USA). Immunoblots were blocked with 5% BSA in TBS/Tween-20 and incubated with primary antibodies overnight at 4 °C. The following primary antibodies were used: β-catenin (Proteintech, Wuhan, China) and EPAS1 (Affinity Biosciences, Cincinnati, OH, USA).
2.15 Cell invasion and migration assays
Invasion and migration assays were performed using Corning chambers (Corning, Tewksbury, MA, USA) coated with Matrigel (for invasion assays) or without Matrigel (for migration assays) following the manufacturer’s protocol. The cells were suspended in media containing 2% FBS and seeded in the upper chambers, while media containing 20% FBS was placed in the lower chambers. After incubating for 24 or 48 h at 37 °C, the remaining cells on the upper surface were gently removed with a cotton swab. Then, cells that had invaded or migrated to the lower surface of the membrane were fixed with methanol and stained with hematoxylin and eosin. Cells in three random visual fields (at 100× magnification) were counted. All the experiments were repeated in triplicate independently.
2.16 Statistical analysis
The R programming language was used for all the statistical studies (version 4.2.0). For the correlation analysis, Spearman’s correlation was used. To examine the differences between these two risk groups, the Wilcoxon test was applied. p < 0.05 indicated statistical significance.
3 Results
3.1 Single-cell transcriptome atlas and cell typing in normal lung and LUAD tissues
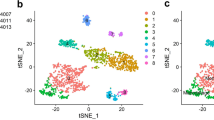
To understand the cellular diversity and molecular features of LUAD, we performed scRNA-seq on twelve libraries from 6 paired primary LUAD and adjacent normal samples (Figs. 1A and S1A). The detailed clinical and pathological information of the patients is provided in Supplementary Table 1. To perform the scRNA-seq, cells were dissociated, sorted for viability, and profiled using 10× Chromium Genomics protocols. As shown, cells grouped primarily by dataset were mixed after integration by the “Harmony” package, which provided well-integrated scRNA-seq data (Fig. S1B). After quality control, a total of 34,995 cells that met the inclusion criteria were selected for subsequent analysis. These cells included 18,231 and 16,764 cells from adjacent tissue and LUAD tissue, respectively (Fig. 1B). By characteristic canonical cell markers, eleven major cell types were detected and classified as tumor cells, epithelial cells other than carcinoma cells, immune cell types (T cells, B cells, myeloid cells, NK cells, mast cells, and plasma cells), and stromal cell types (fibroblasts and endothelial cells) (Fig. 1C).
Single-cell transcriptome atlas and cell typing in adjacent normal tissues and LUAD tissues. A Scheme of the overall study design. B TSNE plots of the major LUAD cell populations, and each point depicts a single cell, colored according to cell types. C The Vlnplot shows the relative expression of the top 3 genes in each cell type. The canonical markers for each cell type are color-coded and shown on the top. D The KEGG functional enrichment analysis of DEGs of the LUAD. E Feature plot shows that the up-regulated gene score and down-regulated gene score, scoring of each cell type was calculated by the “Addmodulescore” algorithms. F Dot plots that depict how the relative expression of particular genes varies at bulk RNA-seq. The size displays the values for average expression in the cell types, colors are marked as percent expression in the cell types
To explore the pathogenic mechanisms underlying LUAD progression. We matched these genes with the sequence data of LUAD-related mRNAs in the TCGA database, GSE140343 dataset and GSE31210 dataset, and only common mRNAs were identified [5, 21] (Fig. S1C and D). Considering the cutoff criteria (see Methods), a large number of DEGs were separately identified, and only the common DEGs of the three cohorts consisting of 603 upregulated and 537 downregulated DEGs were selected as credible DEGs. The KEGG enrichment pathway showed that these upregulated genes were significantly associated with signaling pathways related to tumor progression and metabolic abnormalities in cancer, such as the non-small cell lung cancer pathway, glycolysis/gluconeogenesis-related pathway, and p53 signaling/PI3K-AKT signaling pathway. Likewise, the downregulated genes were highly enriched for the pathways responsible for extracellular matrix (ECM) remodeling, cell adhesion, phagocytosis, and the immune response (Fig. 1D). To further investigate differential gene expression levels at the cellular level, the most significant DEGs were mapped into single cells using the “AddmoduleScore” algorithm [22, 23]. The results indicated that the differentially expressed genes identified from the bulk RNA-seq data were expressed in multiple cell types, upregulated genes are more likely to be highly expressed in tumor cells, basal cell and macrophage cell, but down-regulated genes were not significantly expressed in unique any cell types (Fig. 1E). The same results suggested that the DEGs identified via bulk RNA-seq tended to be expressed predominantly in different cell types; for example, the upregulated gene ADAMDEC1 was expressed in myeloid cells, and SNRK was the downregulated gene that was most highly expressed in T cells and NK cells (Fig. 1F).
These observations strongly suggest that the pathogenesis of LUAD is linked to cell type-specific reprogramming. Our results indicate that LUAD is associated not only with the dysregulation of tumor proliferative signals but also with abnormalities in the immune microenvironment, suggesting the important role of microenvironmental abnormalities in the progression of LUAD.
3.2 Cell type-specific aberrant gene expression in LUAD and adjacent normal tissues
Bulk RNA-seq experiments typically measure total gene expression from heterogeneous tissues but lack cell-level heterogeneity [24]. To simultaneously define gene expression changes at the global and cellular levels, we also compared the proportions of different subpopulations between the two groups. Similarly, the proportions of cells in the two groups were significantly different (Fig. 2A and B). Next, to identify gene dysregulation in LUAD at the level of cell type specificity, we detected DEGs in each cell type between LUAD and adjacent normal samples. Several hundred to thousands of DEGs were detected in each cell type (Fig. 2C). There were more upregulated genes than downregulated genes in all cell types except for tumor cells and endothelial cells. Notably, each cell type contained several cell type-specific upregulated and downregulated genes (Fig. 2D). The results of the functional enrichment analysis were consistent with the DEG results of the bulk RNA-seq pathway enrichment analysis [25]. indicating that the terms oxidative phosphorylation pathway, glycolysis/gluconeogenesis, PI3K-AKT signaling pathway, ECM organization, and immune reaction were enriched among the DEGs in both the non-immune and immune cell types (Fig. 2E). We further mapped the glycolysis/gluconeogenesis pathway score across cell types and found that this pathway score was significantly higher in macrophages and AT2 cells than other cell types (Fig. 2F). Moreover, the two cell types exhibited consistent patterns in that the glycolysis/gluconeogenesis pathway was more active in the tumor tissue than in the adjacent tissue (Fig. 2G). By further analyzing the DEGs of macrophages, we identified the oxidative phosphorylation pathway and glycolysis/gluconeogenesis pathway, which confirmed the previous finding that the Warburg effect results in metabolic reprogramming in macrophages (Fig. 2H) [26,27,28]. These differences reflect cellular heterogeneity in gene expression changes, further suggesting that investigating gene expression changes in each cell type in LUAD patients is important.
Aberrant gene expression profiles in cell type-specific manners in LUAD. A TSNE plots of the major LUAD cell populations. B Stacked bar plots showing the frequencies of cell types between the tumor and adjacent tissue. C Differential genes in each cell type between tumor and NAT of single cell. Each panel represents a subgroup. the horizontal axis represents significance, the vertical axis represents log2FC, with positive There is a negative. The two horizontal dotted lines in the middle are the thresholds of log2FC. Only when the p-value < 0.05 and log2FC > 0.3 which marked as the single cell DEGs. D The radar plot showed the number of different genes identified in different cell types. The up-regulated gene is marked as red, and the down-regulated gene is marked as blue color. E Functional analysis of differentially expressed genes (DEGs) based on aberrant gene expression profiles in cell type. F Feature plot shows that the glycolysis/gluconeogenesis score of each cell type. G Vlnplot shows that the glycolysis/gluconeogenesis score of AT2 cells and macrophages between adjacent tissue and tumor. ns p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. H The GSVA enrichment pathways analysis of the DEGs of the macrophages
3.3 Plasticity of lung epithelial cells and their developmental trajectories into tumor cells
Previous studies have shown that AT2 cells and club cells can both develop into LUAD cells, the key molecular events governing the cell-fate transition during progression from normal to cancer cells are critical for tumor progression [29, 30]. Therefore, we organized AT2 cells, tumor cells according to their developmental trajectory. Here, we further classified 5460 AT2 cells and grouped them into 5 subpopulations, AT1-like 0 cells, AT1-like 1 cells, AT1-like 2 cells, AT1-like 3 cells, and AT1-like 4 cells (Fig. 3A). We further assessed the malignancy of AT2 cells, the results of the “inferCNV” analysis indicated that AT1-like0 cells exhibited a reduced number of copy number alterations and a relatively low CNV score. indicating that AT1-like0 can be regarded as the normal AT2 cells (Figs. 3B and S2A) [31, 32]. We quantified oncogenic signal strength by pathway target gene signature expression and found highly variable activities for Epithelial-Mesenchymal Transition (EMT), Transforming growth factor beta (TGF-β), JAK-STAT pathway, hypoxia, and phosphoinositide 3-kinase (PI3K) /Akt signaling pathway signaling across the different epithelial cell subgroups, which are implicated in these cell types and play important roles in tumor progression (Fig. 3C).
ScRNA-seq analysis reveals the status and invasive trajectory of LUAD cells. A TSNE plots of show the subpopulations of the AT2 cells, which are named as AT2-like cells (0–4). B The CNV score of each subpopulation, the “inferCNV” was used to calculate the CNV score. C Gene set variation analysis enrichment of DEGs among the subpopulations. D Reconstruction of a trajectory reveals the branched structure and the normal cell-to-tumor cell path. The single-cell trajectory reconstructed by Monocle contains six cell types. E Heatmap depicting genes in a branch-dependent manner for pseudo-time trajectory. Each row represents the dynamic expression of a gene. The heatmap center represents the root of the trajectory, and proceeding to the left follows the kinetic curve from the root along the trajectory to the tumor cell. F Representative gene expression levels of different marker genes. The size of each dot represents relative expression levels. G qRT-PCR validation of the differentiation genes along the pseudo-time trajectory. ns p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001
To identify the key molecular events governing the cell fate transition during progression from normal to cancer cells, we selected cell clusters that closely resembled those of AT2-like cells and tumor cells and then tracked the changes in gene expression along the trajectory. We performed pseudo-time analysis based on Monocle and observed nonrandom expression patterns (Fig. 3D) [33]. The transcriptional states in the trajectory revealed progression-associated changes in tumors. Tumor cells gathered on one end, while AT2-like cells were on the other branches. Then, we ordered these genes according to pseudo-time and reconstructed a diffusion map (Fig. 3E). We identified 356 DEGs that exhibited dynamic expression over pseudo-time (q < 0.05) and classified them into four gene modules (modules1–4). In contrast, the genes upregulated in tumors are involved in metabolism, ribosomal activity, or MHC class II molecule expression, which suggests that these activities are essential during tumor progression (Fig. 3E). Notably, the genes significantly upregulated in tumor cells included ELF3, ERRFI1, LRRK2, LDHA, and LPCAT1, and the downregulated genes S100A10, S100A9, and MRPS6, consistent with the results of the present study (Fig. 3G). The results of the experiment confirmed that these genes were expressed throughout the pseudo-time trajectory (Fig. 3F). These findings suggest that genes that change in pseudo-time trajectory are important genes involved in tumor cell evolution and are risk genes for tumorigenesis.
3.4 Molecular subtype 4 is associated with a worse prognosis in LUAD patients
Bulk RNA-seq measures the average expression of genes, and the scRNA-seq enabled cell type-specific transcriptome profiling and cell type-specific aberrant gene expression. We identified statistically significant genes for survival analysis from the DEGs for tissue, cell type-specific aberrant genes for cell type, and pseudo-time differentiation genes of normal cell to tumor cell (Fig. 4A). Based on their cumulative distribution function and functional delta area, we chose k = 4, where tumorigenesis- and tumor progression-related genes appeared to be stably clustered [34] (Fig. 4B). we obtained 4 tumor subtypes, designated molecular subtype1, subtype2, subtype3, and subtype4 (MS1, MS2, MS3, MS4). Patients with MS1, MS2, and MS3 had better prognoses, whereas MS4 had the poorest overall survival (Fig. 4C). Next, we compared the DEGs between MS4 and the other subtypes, a large number of differential genes were identified between subtypes (Fig. 4D). The enrichment pathways of the common up-regulated down-regulated genes result show that cytokine-cytokine receptor interaction and natural killer cell mediated cytotoxicity immune regulation and immune resistance pathways were downregulated in MS4, which is likely to be an immune-suppressed tumor (Fig. 4F). Taken together, the common DEGs identified at both bulk and single-cell resolutions can be used to predict the prognosis of LUAD patients and resemble an immunosuppressive TME.
Identification and validation of the DEGs-based classification of LUAD patients. A significant genes for survival analysis from the DEGs via bulk RNA-seq, cell type-specific aberrant genes via scRNA-seq, and pseudo-time differentiation genes. B CDF curves of the consensus score (k = 2–7) in the TCGA-LUAD cohorts. C Consensus clustering matrix for the ideal cluster size in the TCGA-LUAD cohort, k = 4 which was represented by the matrix. D Kaplan-Meier survival analyses of the patients in four subgroups of TCGA-LUAD cohort. E The number of differential genes in the four molecular subtypes. F GSEA analysis of chemokine signaling pathway, cytokine-cytokine receptor interaction pathway, natural killer cell-mediated cytotoxicity pathway, and p53 signaling pathway among four subgroups
3.5 Association of tumor molecular subtypes with mutational status
To examine the mutation frequency among the four molecular subtypes, we first compared their molecular gene mutation features. The top 20 genes were more frequently mutated in each subtype. TP53 and TTN were the most common genes mutated in MS1, MS3 and TP53, TTN and EGFR were the most common genes mutated in MS2. Inversely, KRAS and KEAP1 were most frequently mutated in MS4. The KRAS gene mutations promote immunosuppression (Fig. 5A). A high TMB and high mutation-allele tumor heterogeneity (MATH) are correlated with increased anticancer immunity. Therefore, we calculated the TMB and number of mutations in each patient using the Mutect2 processed mutation dataset of LUAD patients and analyzed the same data for all subtypes. As shown in (Fig. 5B), patients in MS1 and MS4 had significantly greater TMB; conversely, patients in MS4 had a lower TMB. Similar trends were also observed for the MATH score, as MS4 had a lower MATH score. The results suggested that immune subtype can predict the TMB and somatic mutation rate in LUAD patients and confirmed that MS4 immunosuppressive subtypes determine the efficacy of immunotherapy.
Association between immune subtypes and TMB and mutation. A Heatmap showing the top 20 gene mutation frequencies in the high and low-risk group in the LUAD. B, C The Tumor Mutation Burden (TMB) score and Mutant-Allele Tumor Heterogeneity (MATH) in the different subgroups in the LUAD. ns p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
3.6 The immune landscape of LUAD patients in different immune subgroups
The relationships between patient subgroups and immune infiltration in LUAD patients were investigated. Thus, we used the “CIBERSORT” method for cell-type deconvolution and the Wilcoxon test to compare the distribution of 28 infiltrating immune cells in different LUAD molecular subgroups [35] (Fig. 6A). In particular, we found that M1 macrophages and resting memory CD8+ T cells were markedly decreased in MS4, which was concordant with previous observations linking MS4 to an immunosuppressive microenvironment (Fig. 6B). Given the importance of immune checkpoints (ICPs) and immunogenic cell death (ICD) modulators in cancer immunity, we next analyzed their expression levels among different subtypes, eighteen ICP-related genes were detected were differentially expressed between the immune subtypes. For instance, CD200, CD274, CD40, CD70, CTLA4, HAVCR2, IDO1, LAG3, NRP1, PDCD1, PDCD1LG2, TIGIT, TNFRSF25, TNFRSF4, TNFRSF8, TNFRSF9, TNFSF4 and VSIR were significantly upregulated in MS4 in the TCGA cohort. Five ICD-related genes were detected were differentially expressed among the immune subtypes, while ANXA1, CXCL10, FPR1, MET and PANX1were overexpressed in the MS4. Taken together, our comprehensive analysis revealed that MS4 was significantly correlated with patient prognosis and TME characteristics.
Association between immune subtypes and ICPs and ICD modulators. A “CIBERSORT” analysis identifying the relative infiltration level of immune cell populations in two LRIs subgroups of LUAD samples in the TCGA cohort. B, C Different cell types of proportion among distinct subtype in TCGA cohorts. ns p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. C, D Association between immune subtypes and ICPs and ICD modulators. Differential expression of ICP genes among the four LUAD subgroups. ns p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001
3.7 CDC25C gene can be a distinct prognostic biomarker which indicates immune infiltration levels and response to immunotherapy
To further to find the key genes for the immunosuppression subtypes, the LASSO Cox algorithm was used to identify the most robust prognostic genes among the candidate key genes. Overall, by performing least absolute shrinkage and selection operator (LASSO) Cox regression analysis, 5 genes (MSLNL, LINC01116, CDC25C, ATP8B3, and FAM133A) that met the criterion of p < 0.05 were retained for further analysis (Fig. 7A). Furthermore, a prognostic risk score model was constructed using the 5 genes. Based on two independent cohort Kaplan–Meier (KM) survival analyses, patients in the high-risk subgroup had significantly worse OS than did those in the low-risk subgroup (p < 0.0001, p < 0.0001) (Fig. 7B). We found that EMT pathways up-regulated in the high-risk group and MTORC1 signaling down-regulated.
Identification and validation of the immunosuppressive-related genes classification of LUAD patients. A Construction of immunosuppressive-related genes of subtype 4 on clinical outcome for LUAD patients. B Kaplan–Meier curves immunosuppressive-related genes scores for patients with high and low risk in the TCGA-LUAD cohort. C GSEA analysis of chemokine signaling pathway, MTORC1 pathway, and Epithelial-Mesenchymal transition pathway between high or low risk. D Detecting the level of CDC25C in MRC-5, A549, NCI-H1975, NCI-HCC95, PC-9 by qRT-PCR. E Western-Blot detection of EMT-related markers in A549 after knockdown of CDC25C. F Invasion and migration assay to detect the ability of migration and invasion in A549 cells after CDC25C knockdown. G The lymphocyte infiltration signature score and TCR richness level stratified by expression of CDC25C gene. ns p ≥ 0.05; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 H Overall survival and treatment response of the patients stratified by expression of CDC25C gene
Previous studies have confirmed that the CDC25C participates in regulating G2/M progression and mediating DNA damage repair [36, 37]. Then, we extracted whole RNA from human embryo lung fibroblasts (MRC-5) and four lung cancer cell lines (A549, NCI-H1975, NCI-HCC95, and PC-9) to detect the basal expression of CDC25C in LUAD. The results showed that the mRNA level of CDC25C was greater in lung cancer cell lines than in normal cells, and CDC25C expression was greater in lung adenocarcinoma than in lung squamous cell carcinoma (Fig. 7D). Thus, we selected the A549 cell line, which had the highest expression, for further investigation. After knocking down CDC25C in A549 cells, we evaluated the effect of knocking down CDC25C on cell migration and invasion, and knocking down CDC25C inhibited the migration and invasion of A549 cells (Fig. 7E). Since the EMT pathway influences tumor migration and invasion, we performed a protein immunoblot analysis on the expression of EMT markers. As shown, with the knockdown of CDC25C, E-cadherin was upregulated, while N-cadherin and Snail were downregulated (Fig. 7F). Based on these results, we preliminarily demonstrated that CDC25C can promote the metastasis of lung cancer. Compared with patients with high and low CDC25C expression, patients with high CDC25C expression had fewer effective therapies. Further survival analysis revealed that CDC25C expression was significantly correlated with OS in patients receiving immunotherapy. These findings suggested the reliability of the immunotherapy efficacy evaluation and prognostic model, which can be applied to diverse LUAD patients.
4 Discussion
The tumor microenvironment (TME) has a significant impact on therapeutic efficacy in tumor, The immunosuppression TME leads to the development and advancement of tumors, as well as resistance to chemotherapy and immunotherapy [38,39,40]. The scRNA-seq technology has also been applied in lung cancer research to investigate heterogeneity, metastasis, drug ineffectiveness and resistance [41, 42]. In this study, we investigated the cellular landscape of LUAD and adjacent tissues using scRNA-seq data, we evaluated cell type-specific aberrant gene expression and gene expression changes along the pseudo-time trajectory. The effectiveness of these DEGs in the diagnosis and prognosis of LUAD was explored by integration with bulk RNA-seq datasets. In addition, we classified four distinct molecular subtypes of LUAD and identified that the MS4 immunosuppressive subtype was correlated with poor survival outcomes. This subtype was also associated with a more frequent KRAS mutation, higher TMB, and upregulation of PD-L1 expression. We found that the gene CDC25C can be distinct prognostic biomarkers, immune infiltration levels, and responses to immunotherapy in LUAD, our prognostic model might be a potential biomarker for LUAD patient risk stratification and treatment response prediction.
Bulk transcriptomes measure the overall gene expression in diverse tissues, thereby concealing the expression of each cell type, proportions, and the interactions in the tumor microenvironment [43, 44]. Single-cell sequencing has provided the opportunity to study the heterogeneity of tumorigenesis at the cellular level [18, 45]. Interestingly, we identified more prognosis-related genes from 6 paired primary LUAD patients via single-cell data than through bulk RNA-seq data. We observe that pathway associated with the oxidative phosphorylation pathway, non-small cell lung cancer pathway, p53 signaling pathway, and PI3K-AKT signaling pathway were both enriched at bulk tissue level and the cellular type of level. Our results confirmed that the integration of bulk RNA-seq with single cells can bridge the shortcomings of individual techniques and provide a comprehensive understanding of lung cancer progression [46, 47].
Currently, to further confirm the effectiveness of our typing analysis, LUAD samples were divided into four molecular subtypes based on prognostic risk genes. The results showed that the four molecular subtypes had significantly different prognostic risk and that MS4 was associated with a worse prognosis than other subtypes. pathway analysis of DEGs between MS4 and other subtype revealed that the immune response and inflammatory pathway were more active in other subtypes, implying that MS4 tend to be an immunosuppressive microenvironment. Inversely, KRAS and KEAP1 were most frequently mutated in MS4 with lower TMB score and MATH score using genomics mutational status. The results showed that we have found a subtype with poor prognosis and suppressive tumor microenvironment [48, 49].
The immune suppressive mechanisms in the tumor microenvironment are very complex and involve multiple aspects such as genes, metabolism, and inflammation. In order to gain a deeper comprehension of the molecular mechanisms underlying immunosuppression in LUAD, our research was centered on genes that are highly expressed in the immunosuppressive subtypes. Based on the LASSO Cox regression analysis results, we found that MSLNL, LINC01116, CDC25C, ATP8B3, and FAM133A can be distinct prognostic biomarkers, immune infiltration levels, and responses to immunotherapy in LUAD. Our analysis results that high expression of CDC25C affects the epithelial-mesenchymal transition pathway. CDC25C have been regarded as participates in regulating G2/M progression and in mediating DNA damage repair which play an important role in the cell cycle [50, 51]. In this study we concluded that CDC25C expression affects lung cancer cell invasion and migration, and knockdown the CDC25C, E-cadherin was upregulated, while N-cadherin and snail were downregulated.
This study has several limitations. One limitation is that the tissue samples used for single-cell sequencing and bulk RNA-seq were not the same surgical specimens, so there might be potential differences in the biological complexities of these parts. the other is that our findings could be further validated and extended with patient-derived xenografts (PDX) approaches, preferably through the integration of genomics, transcriptomics, and proteomics, to comprehensively capture feature on the regulation of gene expression [52, 53].
5 Conclusion
Our study unveiled the detailed landscape of LUAD together with bulk and single-cellRNA transcriptome data contributed to understanding TME heterogeneity. We believe that will contribute to a more reliable interpretation of LUAD development at the single-cell level and provide valid biomarkers for the diagnosis and prognosis of LUAD.
Data availability
The experimental data that support the findings of this study are available in Figshare with the identifier https://doi.org/10.6084/m9.figshare.25027181.v1.
Abbreviations
- NSCLC:
-
Non-small cell lung cancer
- LUAD:
-
Lung Adenocarcinoma
- GEO:
-
Gene Expression Omnibus Database
- TCGA:
-
The Cancer Genome Atlas
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- ScRNA-seq:
-
Single-cell RNA sequencing
- TME:
-
Tumor microenvironment
- CNV:
-
Copy number variations
- EMT:
-
Epithelial-mesenchymal transition
- PDX:
-
patient-derived xenografts
References
The global challenge of cancer. Nat. Cancer 1, 1–2 (2020)
A. Leiter, R.R. Veluswamy, J.P. Wisnivesky, The global burden of lung cancer: current status and future trends. Nat. Rev. Clin. Oncol. 20, 624–639 (2023)
H. Sung, J. Ferlay, R.L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, F. Bray, Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021)
J. Chen, H. Yang, A.S.M. Teo, L.B. Amer, F.G. Sherbaf, C.Q. Tan, J.J.S. Alvarez, B. Lu, J.Q. Lim, A. Takano, et al, Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 52, 177–186 (2020)
J.Y. Xu, C. Zhang, X. Wang, L. Zhai, Y. Ma, Y. Mao, K. Qian, C. Sun, Z. Liu, S. Jiang, et al, Integrative proteomic characterization of human lung adenocarcinoma. Cell 182, 245–261 e217 (2020)
M. Miyake, S. Hori, Y. Morizawa, Y. Tatsumi, Y. Nakai, S. Anai, K. Torimoto, K. Aoki, N. Tanaka, K. Shimada, et al, CXCL1-mediated interaction of cancer cells with tumor-associated macrophages and cancer-associated fibroblasts promotes tumor progression in human bladder cancer. Neoplasia 18, 636–646 (2016)
Y. Liang, Y. Tan, B. Guan, B. Guo, M. Xia, J. Li, Y. Shi, Z. Yu, Q. Zhang, D. Liu, et al, Single-cell atlases link macrophages and CD8(+) T-cell subpopulations to disease progression and immunotherapy response in urothelial carcinoma. Theranostics 12, 7745–7759 (2022)
L. Gonzalez-Silva, L. Quevedo, I. Varela, Tumor functional heterogeneity unraveled by scRNA-seq technologies. Trends Cancer 6, 13–19 (2020)
M.L. Metzker, Sequencing technologies - the next generation. Nat. Rev. Genet. 11, 31–46 (2010)
D. Sun, X. Guan, A.E. Moran, L.Y. Wu, D.Z. Qian, P. Schedin, M.S. Dai, A.V. Danilov, J.J. Alumkal, A.C. Adey, et al, Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat. Biotechnol. 40, 527–538 (2022)
A.S. Thind, I. Monga, P.K. Thakur, P. Kumari, K. Dindhoria, M. Krzak, M. Ranson, B. Ashford, Demystifying emerging bulk RNA-Seq applications: the application and utility of bioinformatic methodology. Brief Bioinform. 22, bbab259 (2021)
B. Jew, M. Alvarez, E. Rahmani, Z. Miao, A. Ko, K.M. Garske, J.H. Sul, K.H. Pietilainen, P. Pajukanta, E. Halperin, Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 11, 1971 (2020)
J. Liao, J. Qian, Y. Fang, Z. Chen, X. Zhuang, N. Zhang, X. Shao, Y. Hu, P. Yang, J. Cheng, et al, De novo analysis of bulk RNA-seq data at spatially resolved single-cell resolution. Nat. Commun. 13, 6498 (2022)
X. Guo, Y. Zhang, L. Zheng, C. Zheng, J. Song, Q. Zhang, B. Kang, Z. Liu, L. Jin, R. Xing, et al, Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24, 978–985 (2018)
D. Lambrechts, E. Wauters, B. Boeckx, S. Aibar, D. Nittner, O. Burton, A. Bassez, H. Decaluwe, A. Pircher, K. Van den Eynde, et al, Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med. 24, 1277–1289 (2018)
J.D. Klement, P.S. Redd, C. Lu, A.D. Merting, D.B. Poschel, D. Yang, N.M. Savage, G. Zhou, D.H. Munn, P.G. Fallon, K. Liu, Tumor PD-L1 engages myeloid PD-1 to suppress type I interferon to impair cytotoxic T lymphocyte recruitment. Cancer Cell 41, 620–636 e629 (2023)
L. Zhang, Y. Zhang, C. Wang, Y. Yang, Y. Ni, Z. Wang, T. Song, M. Yao, Z. Liu, N. Chao, et al, Integrated single-cell RNA sequencing analysis reveals distinct cellular and transcriptional modules associated with survival in lung cancer. Signal Transduct. Target. Ther. 7, 9 (2022)
F. Wu, J. Fan, Y. He, A. Xiong, J. Yu, Y. Li, Y. Zhang, W. Zhao, F. Zhou, W. Li, et al, Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat. Commun. 12, 2540 (2021)
D. He, D. Wang, P. Lu, N. Yang, Z. Xue, X. Zhu, P. Zhang, G. Fan, Single-cell RNA sequencing reveals heterogeneous tumor and immune cell populations in early-stage lung adenocarcinomas harboring EGFR mutations. Oncogene 40, 355–368 (2021)
B. Liu, X. Hu, K. Feng, R. Gao, Z. Xue, S. Zhang, Y. Zhang, E. Corse, Y. Hu, W. Han, Z. Zhang, Temporal single-cell tracing reveals clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nat. Cancer 3, 108–121 (2022)
H. Okayama, T. Kohno, Y. Ishii, Y. Shimada, K. Shiraishi, R. Iwakawa, K. Furuta, K. Tsuta, T. Shibata, S. Yamamoto, et al, Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 72, 100–111 (2012)
Y. Hao, T. Stuart, M.H. Kowalski, S. Choudhary, P. Hoffman, A. Hartman, A. Srivastava, G. Molla, S. Madad, C. Fernandez-Granda, R. Satija, Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. (2023)
Y. Hao, S. Hao, E. Andersen-Nissen, W.M. Mauck 3rd, S. Zheng, A. Butler, M.J. Lee, A.J. Wilk, C. Darby, M. Zager, et al, Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529 (2021)
L.A. Huuki-Myers, K.D. Montgomery, S.H. Kwon, S.C. Page, S.C. Hicks, K.R. Maynard, Collado-Torres L: data-driven identification of total RNA expression genes for estimation of RNA abundance in heterogeneous cell types highlighted in brain tissue. Genome Biol. 24, 233 (2023)
S. Hanzelmann, R. Castelo, J. Guinney, GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013)
C.B. Thompson, K.H. Vousden, R.S. Johnson, W.H. Koppenol, H. Sies, Z. Lu, L.W.S. Finley, C. Frezza, J. Kim, Z. Hu, C.R. Bartman, A century of the Warburg effect. Nat. Metab. 5, 1840–1843 (2023)
P. Icard, S. Shulman, D. Farhat, J.M. Steyaert, M. Alifano, H. Lincet, How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist. Updat. 38, 1–11 (2018)
M. Fedele, R. Sgarra, S. Battista, L. Cerchia, G. Manfioletti, The epithelial-mesenchymal transition at the crossroads between metabolism and tumor progression. Int. J. Mol. Sci. 23, 800 (2022)
P. Sarode, S. Mansouri, A. Karger, M.B. Schaefer, F. Grimminger, W. Seeger, R. Savai, Epithelial cell plasticity defines heterogeneity in lung cancer. Cell. Signal. 65, 109463 (2020)
W.K. Cheung, D.X. Nguyen, Lineage factors and differentiation states in lung cancer progression. Oncogene 34, 5771–5780 (2015)
A.P. Patel, I. Tirosh, J.J. Trombetta, A.K. Shalek, S.M. Gillespie, H. Wakimoto, D.P. Cahill, B.V. Nahed, W.T. Curry, R.L. Martuza, et al, Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401 (2014)
I. Tirosh, B. Izar, S.M. Prakadan, M.H. Wadsworth 2nd, D. Treacy, J.J. Trombetta, A. Rotem, C. Rodman, C. Lian, G. Murphy, et al, Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016)
C. Trapnell, D. Cacchiarelli, J. Grimsby, P. Pokharel, S. Li, M. Morse, N.J. Lennon, K.J. Livak, T.S. Mikkelsen, J.L. Rinn, The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014)
M.D. Wilkerson, D.N. Hayes, ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010)
A.M. Newman, C.L. Liu, M.R. Green, A.J. Gentles, W. Feng, Y. Xu, C.D. Hoang, M. Diehn, A.A. Alizadeh, Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015)
J. Al-Matouq, T.R. Holmes, L.A. Hansen, CDC25B and CDC25C overexpression in nonmelanoma skin cancer suppresses cell death. Mol. Carcinog 58, 1691–1700 (2019)
N. Xu, Y. Ren, Y. Bao, X. Shen, J. Kang, N. Wang, Z. Wang, X. Han, Z. Li, J. Zuo, et al, PUF60 promotes cell cycle and lung cancer progression by regulating alternative splicing of CDC25C. Cell Rep. 42, 113041 (2023)
J.C. Marine, S.J. Dawson, M.A. Dawson, Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 20, 743–756 (2020)
G. Morad, B.A. Helmink, P. Sharma, J.A. Wargo, Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 184, 5309–5337 (2021)
Y. Yuan, Spatial heterogeneity in the tumor microenvironment. Cold Spring Harb. Perspect. Med. 6, a026583 (2016)
L. Sun, X. Kang, C. Wang, R. Wang, G. Yang, W. Jiang, Q. Wu, Y. Wang, Y. Wu, J. Gao, et al., Single-cell and spatial dissection of precancerous lesions underlying the initiation process of oral squamous cell carcinoma. Cell Discov. 9, 28 (2023)
J. Qi, H. Sun, Y. Zhang, Z. Wang, Z. Xun, Z. Li, X. Ding, R. Bao, L. Hong, W. Jia, et al, Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nat. Commun. 13, 1742 (2022)
M. Kumar, R.R. Bowers, J.R. Delaney, Single-cell analysis of copy-number alterations in serous ovarian cancer reveals substantial heterogeneity in both low- and high-grade tumors. Cell Cycle 19, 3154–3166 (2020)
Y. Luo, T. Tao, R. Tao, G. Huang, S. Wu, Single-cell transcriptome comparison of bladder cancer reveals its ecosystem. Front. Oncol. 12, 818147 (2022)
W.J. Chen, H. Cao, J.W. Cao, L. Zuo, F.J. Qu, D. Xu, H. Zhang, H.Y. Gong, J.X. Chen, J.Q. Ye, et al, Heterogeneity of tumor microenvironment is associated with clinical prognosis of non-clear cell renal cell carcinoma: a single-cell genomics study. Cell Death Dis. 13, 50 (2022)
S. Wang, G. Fan, L. Li, Y. He, N. Lou, T. Xie, L. Dai, R. Gao, M. Yang, Y. Shi, X. Han, Integrative analyses of bulk and single-cell RNA-seq identified cancer-associated fibroblasts-related signature as a prognostic factor for immunotherapy in NSCLC. Cancer Immunol. Immunother. 72, 2423–2442 (2023)
P. Ma, H.M. Amemiya, L.L. He, S.J. Gandhi, R. Nicol, R.P. Bhattacharyya, C.S. Smillie, D.T. Hung, Bacterial droplet-based single-cell RNA-seq reveals antibiotic-associated heterogeneous cellular states. Cell 186, 877–891 e814 (2023)
O.E. Rahma, F.S. Hodi, The intersection between tumor angiogenesis and immune suppression. Clin. Cancer Res. 25, 5449–5457 (2019)
S. Matsuda, A. Revandkar, T.D. Dubash, A. Ravi, B.S. Wittner, M. Lin, R. Morris, R. Burr, H. Guo, K. Seeger, et al, TGF-beta in the microenvironment induces a physiologically occurring immune-suppressive senescent state. Cell Rep. 42, 112129 (2023)
S. St Clair, J.J. Manfredi, The dual specificity phosphatase Cdc25C is a direct target for transcriptional repression by the tumor suppressor p53. Cell Cycle 5, 709–713 (2006)
K. Liu, M. Zheng, R. Lu, J. Du, Q. Zhao, Z. Li, Y. Li, S. Zhang, The role of CDC25C in cell cycle regulation and clinical cancer therapy: a systematic review. Cancer Cell Int. 20, 213 (2020)
A. Golebiewska, A.C. Hau, A. Oudin, D. Stieber, Y.A. Yabo, V. Baus, V. Barthelemy, E. Klein, S. Bougnaud, O. Keunen, et al, Patient-derived organoids and orthotopic xenografts of primary and recurrent gliomas represent relevant patient avatars for precision oncology. Acta Neuropathol. 140, 919–949 (2020)
E.R. Zanella, E. Grassi, L. Trusolino, Towards precision oncology with patient-derived xenografts. Nat. Rev. Clin. Oncol. 19, 719–732 (2022)
Acknowledgements
Thanks to the patients who provided clinical data for this medical study.
Funding
This research was supported by the Clinical Research Incubation Project of West China Hospital of Sichuan University (2018HXFH012) and the National Natural Science Foundation of China (82173251).
Author information
Authors and Affiliations
Contributions
LZ conceived the project and designed the experiments. SQM, YLW and NNC were involved in the analysis of the experiments and wrote the paper. SQM performed the bioinformatic analysis. YLW, NNC and LYZ contributed to the experiments and analyzed the data. All the authors discussed the results and reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of West China Hospital, Sichuan University. Ethical approval for this study was provided by the Ethics Committee on Biomedical research, West China Hospital of Sichuan University on 18/12/2020.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mao, S., Wang, Y., Chao, N. et al. Integrated analysis of single-cell RNA-seq and bulk RNA-seq reveals immune suppression subtypes and establishes a novel signature for determining the prognosis in lung adenocarcinoma. Cell Oncol. (2024). https://doi.org/10.1007/s13402-024-00948-4
Accepted:
Published:
DOI: https://doi.org/10.1007/s13402-024-00948-4