Abstract
Background
Melanoma-associated antigen (MAGE)-A3 is a member of the family of cancer-testis antigens and has been found to be epigenetically regulated and aberrantly expressed in various cancer types. It has also been found that MAGE-A3 expression may correlate with an aggressive clinical course and with chemo-resistance. The objectives of this study were to assess the relationship between MAGE-A3 promoter methylation and expression and (1) gastric cancer patient survival and (2) its functional consequences in gastric cancer-derived cells.
Methods
Samples from two independent gastric cancer cohorts (including matched non-malignant gastric samples) were included in this study. MAGE-A3 methylation and mRNA expression levels were determined by methylation-specific PCR (MSP) and quantitative real-time PCR (qPCR), respectively. MAGE-A3 expression was knocked down in MKN1 gastric cancer-derived cells using miRNAs. In addition, in vitro cell proliferation, colony formation, apoptosis, cell cycle, drug treatment, immunohistochemistry and Western blot assays were performed.
Results
Clinical analysis of 223 primary patient-derived samples (ntumor = 161, nnormal = 62) showed a significant inverse correlation between MAGE-A3 promoter methylation and expression in the cancer samples (R = −0.63, p = 5.99e–19). A lower MAGE-A3 methylation level was found to be associated with a worse patient survival (HR: 1.5, 95 % CI: 1.02–2.37, p = 0.04). In addition, we found that miRNA-mediated knockdown of MAGE-A3 expression in MKN1 cells caused a reduction in its proliferation and colony forming capacities, respectively. Under stress conditions MAGE-A3 was found to regulate the expression of Bax and p21. MAGE-A3 knock down also led to an increase in Puma and Noxa expression, thus contributing to an enhanced docetaxel sensitivity in the gastric cancer-derived cells.
Conclusions
From our results we conclude that MAGE-A3 expression is regulated epigenetically by promoter methylation, and that its expression contributes to gastric cell proliferation and drug sensitivity. This study underscores the potential implications of MAGE-A3 as a therapeutic target and prognostic marker in gastric cancer patients.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
1 Introduction
MAGE-A3 (melanoma-associated antigen-A3) is a member of the MAGE-A family of cancer-testis (CT) antigens. The MAGE-A3 gene is located on chromosome Xq28. MAGE proteins are commonly expressed in germline cells but silenced in somatic cells [1]. MAGE-A3 expression is regulated by epigenetic mechanisms such as DNA promoter methylation and histone acetylation [2–4]. Aberrant expression of MAGE-A3 has been reported in a wide range of tumor types [5–8]. The differential expression of MAGE-A3 in cancer and normal tissues makes it an attractive target for anti-tumor vaccines [9, 10].
Although CT antigens have been shown to confer cell survival and transformation to an invasive phenotype [11], the biological functions of MAGE-A3 are still unclear. In vitro and in vivo studies, together with clinical data, indicate that MAGE-A3 plays an important role in tumor progression and metastasis in various types of cancers [12–15]. MAGE-A proteins have been shown to interact with the tumor suppressor p53 in sarcoma and non–small cell lung cancer-derived cell lines, thereby inhibiting p53 transactivation activity and conferring resistance to etoposide-induced apoptosis [16]. In addition, MAGE-A3 has been found to interact with several apoptotic proteins such as p21, Bax and survivin through p53-dependent and/or independent pathways, resulting in increased cell survival and tumor progression [14, 17–19].
Gastric cancer remains one of the leading causes of cancer-related death worldwide. Due to its asymptomatic characteristics, patients are often diagnosed at late stages of the disease [20–24]. This usually translates into a poor prognosis and, thus, warrants a further delineation of its underlying biological mechanisms. Recently, MAGE-A expression was reported as a molecular marker of gastric cancer [25]. Honda et al. investigated MAGE-A3 gene expression and promoter methylation in gastric cancer-derived cell lines and promoter methylation in primary gastric tumor specimens. Hypomethylation of the MAGE-A3 promoter was observed in 56 out of 84 (66 %) of the gastric cancers studied and a correlation was observed between hypomethylation and a higher incidence of lymph node metastasis. The authors suggested that MAGE-A3 may play a major role in tumor spread. However, no significant association with a worse survival was observed in cases with a hypomethylated MAGE-A3 promoter [2]. To date, the functional role of MAGE-A3 in gastric cancer development is still unclear and the prognostic significance of MAGE-A3 expression has not been studied yet.
In order to investigate the functional role of MAGE-A3 in gastric cancer development and progression, we assessed the effects MAGE-A3 expression knock down on gastric cancer cell proliferation, survival and chemoresistance. In addition, we examined MAGE-A3 gene expression and promoter methylation levels in 9 gastric cancer-derived cell lines, 161 primary gastric cancer samples and 62 normal gastric tissue samples. Putative associations between MAGE-A3 expression and methylation status with gastric cancer patient survival were assessed.
2 Materials and methods
2.1 Gastric cancer cohorts and genomic profiling
Two independent patient cohorts that have been reported previously were used for this study [26, 27]. Cohort 1 included 203 gastric tumors and 94 matched non-malignant gastric samples (“normals”) profiled for genomic methylation using an Infinium human methylation 27 K BeadChips platform (Illumina, San Diego, CA) [27]. The methylation microarray data have been deposited into the Gene Expression Omnibus (GEO) under accession number GSE30601 (Ntumor = 203, Nnormal = 94). Cohort 2 included 200 gastric tumors and 147 matched non-malignant gastric samples (“normals”) profiled for gene expression using an Affymetrix human genome U133 Plus 2.0 GeneChips platform (Affymetrix, Santa Clara, CA) [26]. The gene expression microarray data are available at GEO under accession number GSE15460 (Ntumor = 200, Nnormal = 147). In total 161 gastric tumors and 62 matched “normals” (i.e., 223 samples) were profiled on both platforms. All primary gastric tissues were obtained from the National University Hospital or National Cancer Centre Singapore tissue repositories with approvals from the respective institutional Research Ethics Review Committees, including signed patient informed consent. The tumor samples were histologically confirmed to contain cancer cells, with an average tumor cellularity of 40 %. Non-malignant samples (i.e., “normals”) refer to stomach tissues harvested from sites distant from the tumor and showing no visible evidence of tumor or intestinal metaplasia/dysplasia upon surgical assessment. The histopathological data and patient characteristics of both cohorts have been reported before [26, 27].
2.2 Cell lines and reagents
Gastric cancer-derived cell lines AGS and KATO III were purchased from the ATCC, USA, SNU620 and SNU638 from the Korean Cell Line Bank, Korea, OCUM1, NUGC3 and NUGC4 from the Health Science Research Resources Bank, Japan, and AZ521 and MKN1 from the DUKE NUS, Singapore. All 9 gastric cell lines were cultured at 37 °C in a humidified atmosphere containing 5 % CO2 and maintained in RPMI-1640 medium (Gibco; Grand Island, NY) containing 10 % heat inactivated fetal bovine serum (FBS, Gibco; Grand Island, NY) and 1 % penicillin/streptomycin (Gibco; Grand Island, NY).
2.3 Reverse transcription-PCR (RT-PCR)
Total RNA was isolated from cell lines using a RNeasy Plus Mini Kit (Qiagen, USA). Next, 5 μg of isolated RNA was reverse-transcribed and amplified using a GeneAmp® RNA PCR Core Kit (Applied Biosystems, USA). The reverse-transcribed product (cDNA) was amplified with primer sequences as listed in Supplementary Table S1. For MAGE-A3 the PCR conditions were as follows: preheat at 94 °C for 5 min, then 35 cycles of denaturation at 94 °C for 30 s, annealing at 64 °C for 30 s and extension at 72 °C for 60 s, followed by a final 5 min extension at 72 °C. For GAPDH the PCR conditions were as follows: preheat at 95 °C for 10 min, then 25 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 30 s, followed by a final 10 min extension at 72 °C. The RT-PCR products were separated on 2 % agarose gels and its bands were visualized and documented using a Gel Doc EZ system (Bio-Rad, USA).
2.4 Quantitative PCR (qPCR) analysis
Total RNA was extracted from cultured cells and reverse transcribed as outlined in Section 2.3. The cDNA obtained was analysed quantitatively using a Power SYBR Green PCR Master Mix (Applied Biosystems) on an ABI7300 Real-time PCR system. The primers used are listed in Supplementary Table 1. The cycling conditions were: 95 °C for 15 min, 40 cycles of 15 s at 94 °C, 30 s at 55 °C and 30 s at 72 °C. Ct values were generated using default analysis settings. Relative quantification (RQ) was calculated using the 2 –ΔΔCT method.
2.5 Bisulphite conversion and methylation-specific polymerase chain reaction (MSP)
DNA was extracted from gastric carcinoma-derived cell lines using a PuregeneTM DNA Isolation Kit (Gentra Systems, USA). Next, 500 ng of gastric carcinoma cell line DNA, positive control DNA (CpGenome Universal Methylated DNA, Chemicon, USA) and negative control human sperm DNA (HsD) were subjected to bisulphite conversion using an EZ DNA Methylation-Gold™ Kit (Zymo Research Corporation, USA) as per manufacturer’s protocol. The primers used for the MSP reactions are listed in Supplementary Table S2. The PCR reactions were performed as follows: preheating at 94 °C for 5 min, then 40 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C/56 °C for 60 s for methylated/unmethylated MAGE-A3, and extension at 72 °C for 60 s, followed by a final 7 min extension at 72 °C. The PCR products were separated on 2 % agarose gels.
2.6 Immunohistochemistry (IHC)
Cells were cytospun onto glass slides and fixed with 4 % formaldehyde for 10 min. Next, the slides where incubated with H2O2 (Sigma-Aldrich) for 10 min, blocked with 1 % BSA (Sigma-Aldrich) for 1 h and treated with 0.2 % triton X100 (Thermo Scientific) for 10 min. Subsequently, the slides were incubated with 6 μg anti-MAGE-A3 primary rabbit monoclonal antibody (Novus Biolab) at 4 °C overnight, after which the slides were incubated with an anti-rabbit antibody for 1 h and stained with 1:50 chromogen substrate (Dako) for 3 min. Counterstaining was performed with hematoxylin (Dako) for 1 min followed by ammonium hydroxide (Sigma-Aldrich) for 3 min. Finally, the slides were immersed serially in 70 % EtOH, 95 % EtOH and 100 % EtOH for 5 min and then immersed in histoclear (National Diagnosics) before microscopic analysis.
2.7 Drug treatment
Docetaxel (Aventis Pharma., France) was stored at 4 °C, and Trichostatin A (TSA) and 5-Aza-2’-deoxycytidine (5’aza) (Sigma-Aldrich; USA) were stored at −20 °C. . To assess the role of DNA promoter methylation on MAGE-A3 expression, AZ521, AGS, MKN1 and KATOIII cell lines were cultured at a density of 1 × 10 6 cells in 100 mm dishes. Next, the cells were treated with 5’aza (10 μM) for 72 h and replenished every 24 h with fresh medium, or with TSA (500 nM) for 24 h. In the combination treatment, cells were treated with 5’aza (10 μM) for 72 h and fresh medium was replenished every 24 h, followed by TSA (500 nM) for 24 h. Subsequently, MAGE-A3 expression was examined by RT-PCR. To determine the apoptotic effects of docetexel, the respective cell lines were co-treated with 10 μM of the caspase inhibitor ZVAD.fmk (Selleckchem).
2.8 miRNA transfections
Custom-designed MAGE-A3 primers were purchased from Invitrogen (Supplementary Table S3). miRNA double-strand fragments were ligated into appropriate vectors using Block-iTTM Pol II miR RNAi Expression Vector Kits (K4936-00, Invitrogen, Germany) following the manufacturer’s instructions. MAGE-A3 miRNA vectors and control miRNA vectors were stably transfected into MKN1 cells using Lipofectamine-2000 (Invitrogen, Germany) according to the manufacturer’s instructions.
2.9 MAGE-A3 knockdown verification
MAGE-A3 expression knockdown was verified using qPCR. To this end, a mastermix solution was prepared using DEPC treated water, a Taqman Universal Mastermix™ (Applied Biosystems, USA) and a gene Taqman expression Assay™ probe (Applied Biosystems, USA). 19 μl of the prepared mastermix solution was loaded into each well of a MicroAmp® 96-well fast plate (Applied Biosystems, USA) followed by 1 μl of cDNA. Next, PCR reactions were carried out at 95 °C for 30 s, followed by 35 cycles of 92 °C for 1 s and 72 °C for 30 s using a Fast 7500 machine (Applied Biosystems, USA). The expression of MAGE-A3 was normalized to the expression of GAPDH measured in the same RNA sample. Relative expression levels were calculated using the 2−ΔΔCt method.
2.10 Cell proliferation assay
Cells were seeded in 96-well plates at a density of 2 × 103 cells/well. Cell proliferation was measured using a BrdU incorporation assay (Roche, Germany) at 5 and 10 h according to the manufacturer’s protocol. The absorbance values measured correlate directly to the amount of DNA synthesis.
2.11 Soft agar colony formation assay
The ability to form colonies in soft agar at low cell densities was determined by seeding 1 × 105 cells into 6-well plates. Before seeding, the cells were suspended in RPMI-1640 medium containing 0.35 % agar and 10 % FBS. These suspensions were layered on RPMI-1640 containing 0.5 % agar and 10 % FBS and incubated for 2–3 weeks. Subsequently, the colonies were stained with 0.5 % crystal violet in 30 % ethanol, 3 % formaldehyde for 10 min at room temperature and imaged using a bright field microscope.
2.12 Apoptosis assay under starvation
To examine the apoptotic role of MAGE-A3 under conditions of serum starvation, cells were starved in serum-free media for 24 and 48 h and re-suspended in phosphate buffered saline (PBS) with FITC-conjugated Annexin-V. For the detection of apoptotic cells, surface expression of phosphatidylserine was assessed using an Annexin V–Fluos kit (Roche, Germany). Flow cytometric analyses were conducted using a BD FACSCalibur 6.0 equipped with Cell Quest Pro Software 5.1.
2.13 Cell cycle analysis
Cells were seeded at a density of 2 × 106 cells in 100 mm dishes. After overnight incubation, the cells were treated for 24 h with either 10 nM docetaxel or docetaxel, in combination with 10 μM ZVAD.fmk, fixed with 70 % ethanol and stained with PI (0.1 % Triton X-100, 200 μg/ml RNase A, 20 μg/ml PI in PBS). In total 10,000 events per sample were analyzed by flow cytometry. Sub-G1 populations were considered as apoptotic cells and debris was eliminated from the analysis using a forward and side scatter plot. The flow cytometric analyses were conducted using a BD FACSCalibur 6.0 equipped with Cell Quest Pro Software 5.1.
2.14 Western blot analysis
Western blot analyses were performed using the following primary antibodies: anti-p21Waf1/Cip1 (12D1), anti-p53, anti-Bax, anti-Survivin, anti-Noxa, anti-Puma and anti-GAPDH (Cell Signaling, USA). The secondary antibodies used (anti-rabbit IgG, HRP-linked) were also purchased from Cell Signaling, USA. For Western blotting, cells were washed with ice cold PBS and resuspended in lysis buffer (CelLytic; Sigma-Aldrich, MO, USA), after which 20 μg protein was electrophoretically separated by 12 % SDS-PAGE and blotted. The protein signals on the blots were visualized using an ECL reagent (AmershamTM ECL Plus Western Blotting Detection System; GE Healthcare, UK), followed by exposure to a chemiluminescence film (Amersham HyperfilmTM ECL; GE Healthcare, UK). The Western blot analyses were repeated twice for each protein tested.
2.15 Statistical analyses
Cell Lines: χ 2 or Fisher exact tests (2-sided) were performed to compare the clinicopathological characteristics of the patients. These statistical analyses were performed using the SPSS 11.0 statistical software program (SPSS, USA). Patient Samples: differential analyses between tumor and normal samples were evaluated using a Student’s t-test, and associations between gene expression and DNA methylation levels were assessed using Pearson correlations. Survival curves were plotted according Kaplan-Meier and statistical significances were assessed using the Cox proportional hazards model adjusting for stage (AJCC6: Stage I and Stage II versus Stage III and Stage IV). Note: high and low expressing or methylated groups were determined using the median of respective distributions. These analyses were performed using the statistical package in R (www.r-project.org). Statistical significance was set at p-values of less than 0.05.
3 Results
3.1 MAGE-A3 gene expression is regulated by promoter methylation in gastric cancer-derived cells
The MAGE-A3 mRNA expression and promoter methylation levels were examined in nine gastric cancer-derived cell lines by qPCR and MSP, respectively (Supplementary Fig. S1a and b). Four cell lines were found to exhibit high MAGE-A3 expression levels. Additionally, in these cell lines the MAGE-A3 gene promoter was found to be unmethylated. Conversely, five gastric cancer-derived cell lines that showed MAGE-A3 gene promoter methylation exhibited intermediate or low MAGE-A3 mRNA expression levels (Table 1). These qPCR data were further supported by MAGE-A3 IHC staining of the gastric cancer-derived cell lines (Supplementary Fig. S2). These results indicate that MAGE-A3 expression is regulated predominantly by promoter methylation in gastric cancer-derived cells.
To determine whether global demethylation or histone acetylation could affect MAGE-A3 expression, two cell lines with a low or negligible MAGE-A3 expression (i.e., AZ521 and AGS) and two cell lines with a high MAGE-A3 expression (i.e., MKN1 and KATO III) were treated with the demethylating agent 5’azacytidine (5’aza) and/or the HDAC inhibitor TSA (Fig. 1). Demethylation of the MAGE-A3 gene promoter was observed in AZ521 and AGS cells after 5’aza treatment, which coincided with MAGE-A3 re-expression (Fig. 1). The expression of MAGE-A3 was, however, not affected by TSA treatment, suggesting that the epigenetic modulation of MAGE-A3 expression was mediated only by MAGE-A3 promoter methylation. Alterations in MAGE-A3 promoter methylation status or mRNA expression levels were not observed in MKN1 and KATOIII, i.e., these cells showed high MAGE-A3 expression levels and were unmethylated (Fig. 1).
MAGE-A3 expression is controlled by methylation. mRNA expressions of MAGE-A3 in cell lines treated with 5’aza (demethylating agent) and/or TSA (histone acetylation inhibitor). Cell lines with no MAGE-A3 expression (AZ521 and AGS) showed re-expression of MAGE-A3 after 5’aza treatment but not after TSA treatment. In contrast, cell lines with a high MAGE-A3 expression (MKN1 and KATO III) showed no change in MAGE-A3 expression after 5’AZA and TSA treatment. GAPDH expression was used as baseline. At least two independent experiments were performed for each cell line
3.2 Correlations between MAGE-A3 promoter methylation and expression in primary gastric cancers and patient survival
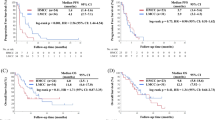
To investigate the relationship between MAGE-A3 expression and methylation in primary gastric cancers, we evaluated the respective data of a cohort of primary gastric cancer samples profiled on multiple genomic platforms from previous studies (see materials and methods). A significantly higher MAGE-A3 expression was observed in gastric cancer tissues compared to the adjacent normal gastric tissues (p = 5.25e-12, t-test) (Fig. 2a), while a lower MAGE-A3 methylation was found in the gastric cancer tissues (p = 9.26e-14, t-test) (Fig. 2b). Additionally, an inverse correlation between MAGE-A3 expression and its methylation status was found (R = −0.63, p = 5.99e-19, Pearson correlation) (Fig. 2c). These observations on primary clinical samples, together with the data from the gastric cancer-derived cell lines, indicate that promoter methylation may be the major regulator for MAGE-A3 expression in gastric cancer cells. In addition, we found that a lower MAGE-A3 promoter methylation level was significantly correlated with a poor patient survival (Hazard ratio, adjusted for stage: 1.62, 95 % CI: 1.05–2.51, p = 0.03) (Hazard ratio, unadjusted: 1.61, 95 % CI: 1.04–2.47, p = 0.03) (Fig. 2d). A significant difference was observed in unadjusted patient survival for MAGE-A3 expression, but no significant difference was observed when adjusted for stage (Hazard ratio, adjusted for stage: 0.66, 95 % CI: 0.43–1.02, p = 0.06) (Hazard ratio, unadjusted: 0.56, 95 % CI: 0.36–0.86, p = 0.008) (Fig. 2e).
Statistical analysis of MAGE-A3 expression and methylation levels in patient samples. a Higher MAGE-A3 expression was found in gastric cancer tissues compared to normal gastric tissues (p = 5.25e-12, t-test). b Lower MAGE-A3 promoter methylation was found in gastric cancer tissues compared to normal gastric tissues (p = 9.28e-14, t-test). c An inverse correlation between MAGE-A3 promoter methylation and expression was found in gastric tumor tissues. The best-fit line reveals an inverse correlation of MAGE-A3 expression to its methylation (R = −0.63, p = 5.99e-19, Pearson correlation). d Lower MAGE-A3 methylation correlates with a poorer patient survival in gastric cancer (Hazard ratio, adjusted for stage: 1.62, 95 % CI: 1.05–2.51, p = 0.03) (Hazard ratio, unadjusted: 1.61, 95 % CI: 1.04–2.47, p = 0.03). e Higher MAGE-A3 expression shows a marginal association with a poorer patient survival in gastric cancer (Hazard ratio, adjusted for stage: 0.66, 95 % CI: 0.43–1.02, p = 0.06) (Hazard ratio, unadjusted: 0.56, 95 % CI: 0.36–0.86, p = 0.008)
3.3 Loss of MAGE-A3 expression reduces proliferation and colony formation of gastric cancer-derived cells
To further uncover the role of MAGE-A3 in gastric cancer, we set out to knock down its expression in MKN1 cells by targeted miRNA transfection. Two variant clones (MKN-V1 and -V2) were obtained that showed a near complete knockdown (Fig. 3a). To subsequently assess the role of MAGE-A3 in gastric cancer cell proliferation, BrdU incorporation and colony formation assays were performed in MAGE-A3 expressing MKN1 cells (MKN1-P) and its variant knock down (MKN1-V1) and scrambled miRNA control (MKN1-S) cells (Fig. 3b). Following treatment with BrdU for 5 and 10 h under standard culture conditions, the proliferation rate of MKN1-V1 cells was found to be significantly lower compared to that of the parental and scrambled controls (p = 0.027 and p = 0.011, respectively), hence suggesting a role of MAGE-A3 in gastric cancer cell proliferation.
MAGE-A3 promotes proliferation and colony formation of gastric cancer-derived cells. a Relative MAGE-A3 expression in MKN1-P, MKN1-S, MKN1-V1 and MKN1-V2 cells as detected by q-PCR. b Proliferation rate of MKN1-P, MKN1-S and MKN1-V1 cells as evaluated with a BrdU incorporation assay after 5 and 10 h. c Soft agar colony formation assay performed on MKN1-P, MKN1-S and MKN1-V1 cells. P parental control, S scrambled control, V1 and V2 knock down variant cells. At least two independent experiments were performed for each cell line. A p value < 0.05 was considered statistically significant
The colony forming capacities of MKN1-P, MKN1-S and MKN1-V1 cells were assessed by soft agar assay (1,000 cells/well in a 6-well plate incubated for 14–21 days). We found that the colony forming capacity was significantly reduced in the MKN1 MAGE-A3 knockdown variant (MKN1-V1) cells compared to the scrambled (MKN1-S) and parental control (MKN1-P) cells (p = 0.00057) (Fig. 3c). Together, these findings suggest that MAGE-A3 promotes the proliferation and colony formation of gastric cancer-derived cells.
3.4 Loss of MAGE-A3 expression reduces proliferation of gastric cancer-derived cells under stress conditions
To investigate whether MAGE-A3 may contribute to the proliferation of gastric cancer cells under nutrient deprivation, MKN1-P, MKN1-S and MKN1-V1 cells were starved in serum-free media for up to 48 h, after which the effect on apoptosis was investigated using Annexin-V staining. We found that the MKN1-V1 cells exhibited an increased apoptotic rate compared to the MKN1-P and MKN-S cells (p = 0.025 and p = 0.009, respectively) (Fig. 4a). This result suggests that MAGE-A3 may promote the proliferation of gastric cancer cells under conditions of stress.
MAGE-A3 promotes cell viability under starvation. a Starvation assay performed on MKN1-P, MKN1-S and MKN1-V1 cells by culturing in serum-free medium for 24 and 48 h, respectively. The percentage of Annexin-V positive apoptotic cells was detected by flow-cytometric analysis. b Expression of apoptosis related proteins in MKN1-P, MKN1-S and MKN1-V1 cells after 48 h of serum starvation examined by Western blot analysis. P parental control, S scrambled control, V1 knock down variant cells. At least two independent experiments were performed for each cell line. The Western blot images were quantified using ImageJ and the relative expression levels of Bax, survivin, p21 and p53 were normalized to GAPDH values. A p value <0.05 was considered statistically significant
MKN1 cells have been reported to express mutant p53 harbouring a V143A amino acid substitution [28]. This mutation results in activation of target genes involved in cell cycle arrest and DNA repair, but is deficient in driving the apoptotic machinery and, thus, protects cells from cell death [29, 30]. We found that V143A mutant p53 expression was reduced in the MAGE-A3 knockdown MKN-V1 cells grown under standard culture conditions, as well as after 48 h of serum deprivation, compared to MKN1-P and MKN1-S cells (Fig. 4b). The pro-apoptotic proteins p21 and Bax were also found to be up-regulated in the MKN1-V1 cells (Fig. 4b). These observations are consistent with the reduced proliferation and increased apoptosis observed in MKN-V1 cells under stress conditions. Interestingly, the expression of the anti-apoptotic protein survivin was found to be increased in MKN-V1 cells (Fig. 4b). Together, these data suggest that MAGE-A3 may play a role in gastric cancer cell proliferation in response to starvation via the regulation of p21 and Bax.
3.5 Loss of MAGE-A3 expression modulates the cell cycle and increases the sensitivity to docetaxel
To investigate the impact of MAGE-A3 expression loss on drug sensitivity, MAGE-A3 knockdown MKN1-V1 cells, as well as control MKN1-P and MKN1-S cells, were treated with 10 nM docetaxel for 24 h, after which cell cycle profiles were analyzed using flow cytometry (Fig. 5a). We found that MAGE-A3 knockdown MKN1 cells exhibited a 30 % sub-G1 population after docetaxel treatment, whereas a 20 % sub-G1 population was observed in the parental cells (p < 0.01). Additionally, we found that treatment with a caspase inhibitor, ZVAD.fmk, resulted in a reduction of the docetaxel effects, hence confirming the apoptotic induction in docetaxel-treated cells. These data indicate that knock down of MAGE-A3 expression modulates both the cell cycle kinetics and the docetaxel sensitivity of MKN1 gastric cancer-derived cells.
Silencing of MAGE-A3 modulates the cell cycle profile and increases the sensitivity to docetaxel. a Cell cycle analysis of MKN1-P, MKN1-S and MKN1-V1 cells followed by docetaxel treatment at 10 nM ± ZVAD for 6 h b Western blot analysis of apoptosis related proteins in MKN1-P, MKN1-S and MKN1-V1 cells in response to docetaxel treatment at 10 nM for 24 h incubation. P parental control, S scrambled control, V1 knock down variant cells. At least two independent experiments were performed for each cell line. A multiple t-test was performed between MKN-1 S (scrambled control) and MKN-V1 (MAGE-A3 knockdown variant) to compare the variation across the sub-G1, G1, S and G2 phases of the cell cycle. A p value <0.05 was considered statistically significant
As reported above, we found that MAGE-A3 knockdown leads to increases in Bax, p21 and survivin expression (Fig. 5b). Additionally, we analyzed the expression of Puma and Noxa. We found that MAGE-A3 expression knockdown in MKN1 cells resulted in increases in Puma and Noxa expression compared to the parental counterparts. More interestingly, we found that docetaxel treatment caused a further increase in Puma and Noxa expression in MAGE-A3 knockdown MKN1 cells. A decrease in p53 expression was observed in MAGE-A3 knockdown MKN1 cells, which was rescued upon docetaxel treatment. The expression of p53 expression remained, however, the same in the control parental variants of the MKN1 cells, irrespective of docetaxel treatment.
4 Discussion
In the past, MAGE proteins have been associated with poor clinical outcomes, including increased tumor growth, metastasis and resistance to treatment. Recent studies have shown that MAGE-A and MAGE-A3 interfere with apoptosis and promote cell proliferation by regulating p53 and p53-responsive genes, either directly or indirectly [16–18, 31]. Marcar et al. showed that MAGE-A interacts with the core DNA-binding domains in p53 and, by doing so, may interfere with its transcriptional activity. The authors reported that loss of MAGE-A expression resulted in an up-regulation of p53 activity, thereby increasing the expression of several p53-responsive genes including p21 [17]. Nardiello et al. reported that silencing of MAGE-A resulted in loss of p53 ubiquitination and stabilization, which led to an increased expression of pro-apoptotic Bax and a reduced expression of survivin [18].
Our study highlights a pro-proliferative role of MAGE-A3 in gastric cancer-derived cells that express a mutant form of p53 (V143A). In primary gastric lesions, an increasing frequency of p53 abnormalities occurs as the gastric mucosa progresses from gastritis, through intestinal mataplasia, dysplasia and early stage gastric cancer to advanced invasive gastric cancer. The cancer genome atlas (TCGA) database indicates that 50 % of the gastric cancers shows p53 mutations. Also, p53 mutations were found to be more common in intestinal gastric cancers compared to the diffuse type [32]. Since wild-type p53 is an important denominator of the G1-S cell cycle checkpoint, p53 mutations are considered pivotal events in the neoplastic transition from intestinal metaplasia to gastric carcinoma [33]. In the present study, we found that loss of MAGE-A3 expression led to reduced levels of mutant p53 expression in cells grown under standard culture conditions as well as under serum starvation. Consistent with this, MAGE-A3 knockdown cells exhibited a reduced proliferation under standard culture conditions, as well as under limiting cell density and serum starvation conditions. Interestingly, increased levels of the pro-apoptotic proteins Bax and p21 were observed in MAGE-A3 knockdown cells grown under standard culture conditions. These data suggest that loss of MAGE-A3 expression enhances the sensitivity of gastric cancer cells to cell death stimuli through down-regulation of pro-survival mutant p53 protein, as well as through up-regulation of pro-apoptotic proteins, possibly via p53-independent pathways. Although both Bax and p21 are transcriptional targets of p53, they have also been reported to be regulated through other, p53-independent, mechanisms. These include direct induction of p21 expression by the activating enhancer binding protein 4 (AP4) [34], and inhibition of Bax expression by the nuclear factor-kappa B (NF-κB) [35, 36]. Although current evidence points to the involvement of MAGE proteins in transcription factor complexes regulating gene expression [37–39], it is currently unknown whether MAGE-A3 physically interacts with these transcription factors.
Increased expression of members of the MAGE-A family has been associated with resistance to chemotherapy in patients with various types of cancer [40–44]. Taxanes, such as docetaxel and paclitaxel, can affect the cell cycle by arresting cells in the G1 phase [45] or the G2/M phase [46], resulting in cell death. Induction of apoptosis via docetaxel is tightly regulated by cell cycle control genes, including p53, Bax and p21 [47]. Over-expression of the pro-apoptotic Bax protein has also been shown to induce apoptosis and to confer paclitaxel sensitivity to cancer cells [48]. In MAGE-A3 knockdown cells we found that the basal expression levels of Bax and p21 were increased even in the absence of treatment, suggesting a regulatory role of MAGE-A3. We also observed a contributory role of Noxa and Puma, which may at least partly explain the enhanced sensitivity of MAGE-A3 knockdown cells to docetaxel-induced apoptosis. Although the p53 expression levels were found to be down-regulated in MAGE-A3 knockdown cells, docetaxel treatment rescued the expression of mutant p53 in these cells to levels similar to those in parental cells treated with docetaxel. Despite the increased mutant (pro-survival) p53 level, however, MAGE-A3 knockdown cells exhibited an enhanced docetaxel sensitivity as indicated by an accumulation of cells in the sub-G1 phase. To verify whether the increase in sub-G1 population upon docetaxel treatment was mediated by activation of a caspase-induced apoptotic program, MKN1 cells were co-treated with the caspase inhibitor ZVAD.fmk. Although a reduction in sub-G1 population was observed after this treatment, the significant amount of cells that remained in the sub-G1 phase suggest a non-apoptotic mechanism of docetaxel sensitivity that is independent of caspase activation. Therefore, it can be inferred that the increased docetaxel sensitivity in cells that had lost MAGE-A3 expression arises both from apoptotic and non-apoptotic mechanisms in a p53-independent manner. It would be interesting to address the role of MAGE proteins in p53-independent pathways that confer survival. Of note, knockdown of MAGE-A3 in MKN1 cells also resulted in a high expression of survivin. Survivin is a multi potent inhibitor of apoptosis and its expression rescues cells from apoptosis induced by Bax over-expression [36, 49]. Survivin also regulates cell cycle progression through transcription repression of p21 and inhibits apoptosis through blocking caspase activation [50–52]. Moreover, expression of survivin in cancer cells is usually associated with resistance to a broad spectrum of chemotherapeutic drugs [53]. This could explain why increased expression of pro-apoptotic Bax, p21 and Puma in untreated MAGE-A3 knockdown cells does not correlate with an increase in overall apoptosis. Taken together, we conclude that MAGE-A3 exerts a hyper-proliferative role in gastric cancer-derived cells.
The current study also shows a strong association between silencing of MAGE-A3 expression and MAGE-A3 promoter methylation in primary patient-derived gastric cancer samples and in gastric cancer-derived cell lines. MAGE-A3 expression was found to be restored by treatment of the cells with a demethylating agent, but not with an HDAC inhibitor, suggesting that promoter DNA methylation is the predominant mechanism by which MAGE-A3 expression is regulated in gastric cancer cells. The observed effects of MAGE-A3 expression on gastric cancer-derived cell proliferation and chemoresistance are supported by our clinical findings that the MAGE-A3 expression and methylation status differed significantly between cancer and non-cancer tissues, and that low methylation levels were associated with a poor survival. Our findings underscore the results from an earlier study by Honda et al. indicating that MAGE-A3 hypomethylation correlated with a worse survival of gastric cancer patients [2]. Hence, MAGE-A3 hypomethylation may serve as a useful prognostic factor for gastric cancer patients.
5 Conclusion
We observed an inverse correlation between MAGE-A3 expression and methylation in primary gastric cancer specimens, where lower MAGE-A3 methylation was found to be associated with a worse survival. In vitro, MAGE-A3 expression was found to be regulated by promoter methylation. Additionally, we found that MAGE-A3 expression knockdown led to an increased sensitivity to cellular stress and docetaxel treatment by regulating the expression of cell cycle and apoptosis related genes. Taken together, this study underscores the potential implications of MAGE-A3 as a therapeutic target and a prognostic biomarker in gastric cancer patients.
References
P. Chomez, O. De Backer, M. Bertrand, E. De Plaen, T. Boon, S. Lucas, An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 61, 5544–5551 (2001)
T. Honda, G. Tamura, T. Waki, S. Kawata, M. Terashima, S. Nishizuka, T. Motoyama, Demethylation of MAGE promoters during gastric cancer progression. Br. J. Cancer 90, 838–843 (2004). doi:10.1038/sj.bjc.6601600
F. Wischnewski, K. Pantel, H. Schwarzenbach, Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, −A2, −A3, and -A12 in human cancer cells. Mol. Cancer Res. MCR 4, 339–349 (2006). doi:10.1158/1541-7786.MCR-05-0229
A. Moreno-Bost, S. Szmania, K. Stone, T. Garg, A. Hoerring, J. Szymonifka, J. Shaughnessy Jr., B. Barlogie, H.G. Prentice, F. van Rhee, Epigenetic modulation of MAGE-A3 antigen expression in multiple myeloma following treatment with the demethylation agent 5-azacitidine and the histone deacetlyase inhibitor MGCD0103. Cytotherapy 13, 618–628 (2011). doi:10.3109/14653249.2010.529893
R.A. Wascher, P.J. Bostick, K.T. Huynh, R. Turner, K. Qi, A.E. Giuliano, D.S. Hoon, Detection of MAGE-A3 in breast cancer patients’ sentinel lymph nodes. Br. J. Cancer 85, 1340–1346 (2001). doi:10.1054/bjoc.2001.2079
W. Sienel, C. Varwerk, A. Linder, D. Kaiser, M. Teschner, M. Delire, G. Stamatis, B. Passlick, Melanoma associated antigen (MAGE)-A3 expression in Stages I and II non-small cell lung cancer: results of a multi-center study. Eur. J. Cardiothorac. Surg. Off. J. Eur. J. Cardiothoracic Surg. 25, 131–134 (2004)
E.J. Jung, M.A. Kim, H.S. Lee, H.K. Yang, Y.M. Lee, B.L. Lee, W.H. Kim, Expression of family A melanoma antigen in human gastric carcinoma. Anticancer Res. 25, 2105–2111 (2005)
H.M. Shantha Kumara, M.J. Grieco, O.L. Caballero, T. Su, A. Ahmed, E. Ritter, S. Gnjatic, V. Cekic, L.J. Old, A.J. Simpson, C. Cordon-Cardo, R.L. Whelan, MAGE-A3 is highly expressed in a subset of colorectal cancer patients. Cancer Immun. 12, 16 (2012)
P.G. Coulie, V. Karanikas, C. Lurquin, D. Colau, T. Connerotte, T. Hanagiri, A. Van Pel, S. Lucas, D. Godelaine, C. Lonchay, M. Marchand, N. Van Baren, T. Boon, Cytolytic T-cell responses of cancer patients vaccinated with a MAGE antigen. Immunol. Rev. 188, 33–42 (2002)
M.J. Scanlan, A.O. Gure, A.A. Jungbluth, L.J. Old, Y.T. Chen, Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol. Rev. 188, 22–32 (2002)
A.J. Simpson, O.L. Caballero, A. Jungbluth, Y.T. Chen, L.J. Old, Cancer/testis antigens, gametogenesis and cancer. Nat. Rev. Cancer 5, 615–625 (2005)
C. Roeder, B. Schuler-Thurner, S. Berchtold, G. Vieth, P. Driesch, G. Schuler, M. Luftl, MAGE-A3 is a frequent tumor antigen of metastasized melanoma. Arch. Dermatol. Res. 296, 314–319 (2005). doi:10.1007/s00403-004-0527-7
S. Abdul-Rasool, S.H. Kidson, E. Panieri, D. Dent, K. Pillay, G.S. Hanekom, An evaluation of molecular markers for improved detection of breast cancer metastases in sentinel nodes. J. Clin. Pathol. 59, 289–297 (2006). doi:10.1136/jcp.2005.028357
W. Liu, S. Cheng, S.L. Asa, S. Ezzat, The melanoma-associated antigen A3 mediates fibronectin-controlled cancer progression and metastasis. Cancer Res. 68, 8104–8112 (2008)
T. Bujas, Z. Marusic, M. Peric Balja, A. Mijic, B. Kruslin, D. Tomas, MAGE-A3/4 and NY-ESO-1 antigens expression in metastatic esophageal squamous cell carcinoma. Eur. J. Histochem. 55, e7 (2011). doi:10.4081/ejh.2011.e7
M. Monte, M. Simonatto, L.Y. Peche, D.R. Bublik, S. Gobessi, M.A. Pierotti, M. Rodolfo, C. Schneider, MAGE-A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc. Natl. Acad. Sci. U. S. A. 103, 11160–11165 (2006). doi:10.1073/pnas.0510834103
L. Marcar, N.J. Maclaine, T.R. Hupp, D.W. Meek, Mage-A cancer/testis antigens inhibit p53 function by blocking its interaction with chromatin. Cancer Res. 70, 10362–10370 (2010). doi:10.1158/0008-5472.CAN-10-1341
T. Nardiello, A.A. Jungbluth, A. Mei, M. Diliberto, X. Huang, A. Dabrowski, V.C. Andrade, R. Wasserstrum, S. Ely, R. Niesvizky, R. Pearse, M. Coleman, D.S. Jayabalan, N. Bhardwaj, L.J. Old, S. Chen-Kiang, H.J. Cho, MAGE-A inhibits apoptosis in proliferating myeloma cells through repression of Bax and maintenance of survivin. Clin. Cancer Res. 17, 4309–4319 (2011). doi:10.1158/1078-0432.CCR-10-1820
D. Atanackovic, Y. Hildebrandt, A. Jadczak, Y. Cao, T. Luetkens, S. Meyer, S. Kobold, K. Bartels, C. Pabst, N. Lajmi, M. Gordic, T. Stahl, A.R. Zander, C. Bokemeyer, N. Kroger, Cancer-testis antigens MAGE-C1/CT7 and MAGE-A3 promote the survival of multiple myeloma cells. Haematologica 95, 785–793 (2010). doi:10.3324/haematol.2009.014464
G. Maconi, G. Manes, G.B. Porro, Role of symptoms in diagnosis and outcome of gastric cancer. World J. Gastroenterol. 14, 1149–1155 (2008)
R. Siegel, D. Naishadham, A. Jemal, Cancer statistics. CA Cancer J. Clin. 63, 11–30 (2013). doi:10.3322/caac.21166
X. Xu, X. Chang, Z. Li, J. Wang, P. Deng, X. Zhu, J. Liu, C. Zhang, S. Chen, D. Dai, Berrant SOX11 promoter methylation is associated with poor prognosis in gastric cancer. Cell. Oncol. 38, 183–194 (2015). doi:10.1007/s13402-015-0219-7
A. Yousuf, M.Y. Bhat, A.A. Pandith, D. Afroze, N.P. Khan, K. Alam, P. Shah, M.A. Shah, S. Mudassar, MGMT gene silencing by promoter hypermethylation in gastric cancer in a high incidence area. Cell. Oncol. 37, 245–252 (2014). doi:10.1007/s13402-014-0179-3
M. Labots, T.E. Buffart, J.C. Haan, N.C. van Grieken, M. Tijssen, C.J. van de Velde, H.I. Grabsch, B. Ylstra, B. Carvalho, R.J. Fijneman, H.M. Verheul, G.A. Meijer, High-level copy number gains of established and potential drug target genes in gastric cancer as a lead for treatment development and selection. Cell. Oncol. 37, 41–52 (2014). doi:10.1007/s13402-013-0162-4
T.B. Lee, S.C. Lim, Y.S. Moon, C.H. Choi, Melanoma antigen gene family A as a molecular marker of gastric and colorectal cancers. Oncol. Rep. 30, 234–238 (2013). doi:10.3892/or.2013.2458
C.H. Ooi, T. Ivanova, J. Wu, M. Lee, I.B. Tan, J. Tao, L. Ward, J.H. Koo, V. Gopalakrishnan, Y. Zhu, L.L. Cheng, J. Lee, S.Y. Rha, H.C. Chung, K. Ganesan, J. So, K.C. Soo, D. Lim, W.H. Chan, W.K. Wong, D. Bowtell, K.G. Yeoh, H. Grabsch, A. Boussioutas, P. Tan, Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 5, e1000676 (2009). doi:10.1371/journal.pgen.1000676
H. Zouridis, N. Deng, T. Ivanova, Y. Zhu, B. Wong, D. Huang, Y.H. Wu, Y. Wu, I.B. Tan, N. Liem, V. Gopalakrishnan, Q. Luo, J. Wu, M. Lee, W.P. Yong, L.K. Goh, B.T. Teh, S. Rozen, P. Tan, Methylation subtypes and large-scale epigenetic alterations in gastric cancer. Sci. Transl. Med. 4, 156ra140 (2012). doi:10.1126/scitranslmed.3004504
Y. Yamada, T. Yoshida, K. Hayashi, T. Sekiya, J. Yokota, S. Hirohashi, K. Nakatani, H. Nakano, T. Sugimura, M. Terada, p53 gene mutations in gastric cancer metastases and in gastric cancer cell lines derived from metastases. Cancer Res. 51, 5800–5805 (1991)
Y.C. Lin, F.F. Wang, Mechanisms underlying the pro-survival pathway of p53 in suppressing mitotic death induced by adriamycin. Cell. Signal. 20, 258–267 (2008). doi:10.1016/j.cellsig.2007.10.017
P.K. Lo, S.Z. Huang, H.C. Chen, F.F. Wang, The prosurvival activity of p53 protects cells from UV-induced apoptosis by inhibiting c-Jun NH2-terminal kinase activity and mitochondrial death signaling. Cancer Res. 64, 8736–8745 (2004). doi:10.1158/0008-5472.CAN-04-2584
B. Yang, S.M. O’Herrin, J. Wu, S. Reagan-Shaw, Y. Ma, K.M. Bhat, C. Gravekamp, V. Setaluri, N. Peters, F.M. Hoffmann, H. Peng, A.V. Ivanov, A.J. Simpson, B.J. Longley, MAGE-A, mMage-b, and MAGE-C proteins form complexes with KAP1 and suppress p53-dependent apoptosis in MAGE-positive cell lines. Cancer Res. 67, 9954–9962 (2007). doi:10.1158/0008-5472.CAN-07-1478
C.M. Fenoglio-Preiser, J. Wang, G.N. Stemmermann, A. Noffsinger, TP53 and gastric carcinoma: a review. Hum. Mutat. 21, 258–270 (2003). doi:10.1002/humu.10180
R.A. Busuttil, G.V. Zapparoli, S. Haupt, C. Fennell, S.Q. Wong, J.M. Pang, E.A. Takeno, C. Mitchell, N. Di Costanzo, S. Fox, Y. Haupt, A. Dobrovic, A. Boussioutas, Role of p53 in the progression of gastric cancer. Oncotarget 5, 12016–12026 (2014)
R. Jackstadt, P. Jung, H. Hermeking, AP4 directly downregulates p16 and p21 to suppress senescence and mediate transformation. Cell Death Dis. 4, e775 (2013). doi:10.1038/cddis.2013.282
M. Bentires-Alj, E. Dejardin, P. Viatour, C. Van Lint, B. Froesch, J.C. Reed, M.P. Merville, V. Bours, Inhibition of the NF-kappa B transcription factor increases Bax expression in cancer cell lines. Oncogene 20, 2805–2813 (2001). doi:10.1038/sj.onc.1204343
K. Vazquez-Santillan, J. Melendez-Zajgla, L. Jimenez-Hernandez, G. Martinez-Ruiz, V. Maldonado, NF-kappaB signaling in cancer stem cells: a promising therapeutic target? Cell. Oncol. 38, 327–339 (2015). doi:10.1007/s13402-015-0236-6
T.Z. Xiao, N. Bhatia, R. Urrutia, G.A. Lomberk, A. Simpson, B.J. Longley, MAGE I transcription factors regulate KAP1 and KRAB domain zinc finger transcription factor mediated gene repression. PLoS ONE 6, e23747 (2011). doi:10.1371/journal.pone.0023747
Q. Liu, S. Su, A.J. Blackwelder, J.T. Minges, E.M. Wilson, Gain in transcriptional activity by primate-specific coevolution of melanoma antigen-A11 and its interaction site in androgen receptor. J. Biol. Chem. 286, 29951–29963 (2011). doi:10.1074/jbc.M111.244715
S. Su, A.J. Blackwelder, G. Grossman, J.T. Minges, L. Yuan, S.L. Young, E.M. Wilson, Primate-specific melanoma antigen-A11 regulates isoform-specific human progesterone receptor-B transactivation. J. Biol. Chem. 287, 34809–34824 (2012). doi:10.1074/jbc.M112.372797
T. Suzuki, K. Yoshida, Y. Wada, Y. Hamai, K. Sentani, N. Oue, W. Yasui, Melanoma-associated antigen-A1 expression predicts resistance to docetaxel and paclitaxel in advanced and recurrent gastric cancer. Oncol. Rep. 18, 329–336 (2007)
S.D. Weeraratne, V. Amani, A. Neiss, N. Teider, D.K. Scott, S.L. Pomeroy, Y.J. Cho, miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol 13, 165–175 (2011). doi:10.1093/neuonc/noq179
M.M. Makhlouf, Melanoma antigen encoding gene-A3 expression, its clinical significance, and its risk of developing acute myeloid leukemia. J. Investig. Med. 59, 1131–1136 (2011). doi:10.231/JIM.0b013e31822a2360
S. Hartmann, U. Kriegebaum, N. Kuchler, G. Lessner, R.C. Brands, C. Linz, T. Schneider, A.C. Kubler, U.D. Muller-Richter, Efficacy of cetuximab and panitumumab in oral squamous cell carcinoma cell lines: prognostic value of MAGE-A subgroups for treatment success. J. Craniomaxillofac. Surg. 41, 623–629 (2013). doi:10.1016/j.jcms.2012.12.006
C. Kasuga, Y. Nakahara, S. Ueda, C. Hawkins, M.D. Taylor, C.A. Smith, J.T. Rutka, J. Neurosurg, Expression of MAGE and GAGE genes in medulloblastoma and modulation of resistance to chemotherapy. J. Neurosurg. Pediatr. 1, 305–313 (2008). doi:10.3171/PED/2008/1/4/305
D.A. Brito, C.L. Rieder, The ability to survive mitosis in the presence of microtubule poisons differs significantly between human nontransformed (RPE-1) and cancer (U2OS, HeLa) cells. Cell Motil. Cytoskeleton 66, 437–447 (2009). doi:10.1002/cm.20316
K.E. Gascoigne, S.S. Taylor, How do anti-mitotic drugs kill cancer cells? J. Cell Sci. 122, 2579–2585 (2009). doi:10.1242/jcs.039719
L.G. Wang, X.M. Liu, W. Kreis, D.R. Budman, The effect of antimicrotubule agents on signal transduction pathways of apoptosis: a review. Cancer Chemother. Pharmacol. 44, 355–361 (1999)
Y.T. Tai, T. Strobel, D. Kufe, S.A. Cannistra, In vivo cytotoxicity of ovarian cancer cells through tumor-selective expression of the BAX gene. Cancer Res. 59, 2121–2126 (1999)
K.W. Sommer, C.J. Schamberger, G.E. Schmidt, S. Sasgary, C. Cerni, Inhibitor of apoptosis protein (IAP) survivin is upregulated by oncogenic c-H-Ras. Oncogene 22, 4266–4280 (2003). doi:10.1038/sj.onc.1206509
S. Shin, B.J. Sung, Y.S. Cho, H.J. Kim, N.C. Ha, J.I. Hwang, C.W. Chung, Y.K. Jung, B.H. Oh, An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and −7. Biochemistry 40, 1117–1123 (2001)
M. Pennati, M. Binda, G. Colella, M. Zoppe, M. Folini, S. Vignati, A. Valentini, L. Citti, M. De Cesare, G. Pratesi, M. Giacca, M.G. Daidone, N. Zaffaroni, Ribozyme-mediated inhibition of survivin expression increases spontaneous and drug-induced apoptosis and decreases the tumorigenic potential of human prostate cancer cells. Oncogene 23, 386–394 (2004). doi:10.1038/sj.onc.1207071
L. Tang, X. Ling, W. Liu, G.M. Das, F. Li, Transcriptional inhibition of p21WAF1/CIP1 gene (CDKN1) expression by survivin is at least partially p53-dependent: evidence for survivin acting as a transcription factor or co-factor. Biochem. Biophys. Res. Commun. 421, 249–254 (2012). doi:10.1016/j.bbrc.2012.03.147
F. Li, G. Ambrosini, E.Y. Chu, J. Plescia, S. Tognin, P.C. Marchisio, D.C. Altieri, Control of apoptosis and mitotic spindle checkpoint by survivin. Nature 396, 580–584 (1998). doi:10.1038/25141
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Financial support
This work was supported by a NMRC Translational Clinical Research Grant, a NMRC Individual Research Grant and Duke-NUS Graduate Medical School Faculty Funds sponsored by the Ministry of Health, Ministry of Education and Ministry of Trade, Singapore and A*STAR.
Conflict of interest
The authors have no conflict of interest to report with respect to this work.
Additional information
Chen Xie and Vinod Vijay Subhash contributed equally to this work as co-first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 2117 kb)
Rights and permissions
About this article
Cite this article
Xie, C., Subhash, V.V., Datta, A. et al. Melanoma associated antigen (MAGE)-A3 promotes cell proliferation and chemotherapeutic drug resistance in gastric cancer. Cell Oncol. 39, 175–186 (2016). https://doi.org/10.1007/s13402-015-0261-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-015-0261-5