Abstract
The objective of this study was to characterize the probiotic lactic acid bacteria to improve the quality of silage. A total of 42 LAB isolates were isolated from the date syrup, and the potent strains were characterized as Lactobacillus casei LC09 and L. lactis LL25. These two strains showed the ability to survive at low pH, stimulate gastric conditions, and be sensitive to most of the prominent antibiotics. LAB strains have adhesion ability (p < 0.001) on the surface of the HT-29 cell lines and γ-hemolytic type, and no zones developed around the blood agar medium. The cell-free extract of the selected LAB showed promising DPPH and ferric reducing power activity. Among the selected fungi, LC09 showed the maximum zone of inhibition against Fusarium oxysporum (26 ± 1 mm), Aspergillus niger (27 ± 2 mm), and Fusarium graminearum (26 ± 1 mm). The strain LC09 showed maximum activity against F. graminearum (29 ± 1 mm) and F. oxysporum (24 ± 2 mm). Acetic acid and lactic acid production are the prominent end products determined from the cell free extract of both LAB (p < 0.001). Lactic acid production was 5.3 ± 0.32 g/L and 4.7 ± 0.28 g/L for strains LC09 and LL25, whereas, acetic acid level was 0.39 ± 0.03 g/L and 0.27 ± 0.47 g/L for these LAB. Quail bush and date wastes were used for the preparation of silage, and LAB strains were inoculated and treated for 30 days. In vitro silage analysis showed that quail bush-date waste inoculated with LC09 and LL25 showed a decreased pH level at the end of fermentation (after 30 days) (p < 0.001). Lactic acid and acetic acid levels were increased in the experimental silage (p < 0.001). Thus, ensiled quail bush-date waste could promote silage quality, improve fibre digestion and reduce the growth of pathogenic bacteria and fungal strains.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Epiphytic microorganisms from the plant materials or additional inoculums are useful for the ensiling process and improving the quality of silage. Many microbial strains, mainly lactic acid bacteria (LAB), are used in this process to improve the silage quality. There are many factors that influence on the quality of silage. The supplementation of various microorganisms that produce increased amounts of lactic acid is of great interest because of their great effect on decreasing the pH value of the medium [1]. The impacts of LAB inoculants on the silage process are due to the production of various metabolites of interest that effectively inhibit the growth of pathogenic microorganisms. Hence, the ability of a potent strain to utilize various substrates from the forage plants and synthesize various metabolites can be an additional advantage in the competition with other microorganisms for food or other factors. These criteria are useful for the selection of the most prominent strains for this process [2]. The ensiling process is highly complex and involves interactions between various microbiological and chemical processes. Several bacterial genera are playing a potent role in the process of ensilage. LAB strains that are highly associated with silages are bacteria from the genera Streptococcus, Lactococcus, Enterococcus, Leuconostoc, Pediococcus and Lactobacillus [3]. Bacteria from these genera are able to compete for nutrients with other microorganisms, including, fungi, yeast, bacilli, enterobacteria, and clostridia. LAB produces various quality end products, and degrades various nutrients, and enhances the availability of nutrients to the ruminants [3]. LAB produces volatile fatty acids, which inhibit the growth of other microbes that support the growth of molds and yeast and initiate aerobic deterioration, which causes the nutritional value of fodders to decrease, induce spoilage, and generate heat [4].
The presence of bacteria in the fodder significantly determines the effect of the fermentation process. One of the common procedures to enhance LAB populations in silage is to increase the exogenous LAB population, and another way is to reduce the pathogenic microbial population in silage. The LAB supplement enhances silage quality by improving lactic acid content, inhibiting the growth of pathogenic microorganisms, and reducing the pH value of silage. The heterofermentative LAB species are considering for their antifungal activities and significantly improve the silage quality compared to homofermentative LAB strains. Lactic acid is an important substrate for yeast, reducing the aerobic stability and feeding-out of the silage. Inoculation with heterofermentative Lactobacillus buchneri and facultative heterofermentative Lactobacillus plantarum was evaluated during the ensiling of sugar cane. L. buchneri showed good capacity for reducing DM losses and effectively increasing aerobic stability [5]. Pedroso et al. [6] stated that Lb. buchneri improved aerobic stability in silage, however, L. plantarum negatively influenced silage. The LAB species such as, Lactobacillus brevis, Lactobacillus paracasei, Lactobacillus brevis buchneri and L. plantarum improved silage [7]. Inoculations with various strains that belong to the same sub species have yielded silages with distinct properties, suggesting that analyses should be done not only at the subspecies level but also at the strain level [8]. Most of the LAB produce antifungal and antibacterial compounds and therefore have the potential to inhibit the proliferation of fungi and bacteria. Heterofermentative and homofermentative LAB species synergistically act on each other and improve microbial quality in the silage. Moreover, under certain conditions, the epiphytic LAB population is not effective or the composition is inadequate for enhancing effective homolactic fermentation. Applications of LAB improve nutrient quality and are safe for all organisms [4].
Mycotoxin and fungal spoilage affected feed quality, nutrient composition, reduction in consumption, palatability, and dry matter of silage. The fungi from genera such as Fusarium, Pencillum and Aspergillus were contaminated with silage. The supplementation of LAB can improve the production of acetic acid, which inhibits the growth of molds and yeasts, and thus enhances the stability of the silage. The inoculated LAB species, such as L. paracasei and L. rhamnosus GG, could inhibit the proliferation of fungi and yeasts [9]. LAB synthesizes novel metabolites such as acetic acid, lactic acid, phenylacetic acid, cyclic dipeptides, 3-hydroxy propionaldehyde, and hydroxy fatty acids, and these metabolites are associated with antifungal activity [10]. LAB produces various secondary metabolites, including, diacetyl, organic acids, and antimicrobial secondary metabolites, and inhibits the growth of Aspergillus flavus [11]. LAB effectively degrades mycotoxins by enzymatic activity. L. paracasei, L. bulgaricus, and L. plantarum are used to improve the quality of fermentation in silage, and these LAB strains produce various hydrolytic enzymes [12]. LAB species such as Lactobacilli plantarum and L. casei enhanced silage quality by effectively preventing the proliferation of various pathogenic microbial flora by decreasing the pH value in the surrounding environment. Silage quality was mainly affected by bacteria such as Enterococcus hirae, E. mundtii, E. casseliflavus, and E. faecalis. These bacterial species wer broadly reported on fodder [13]. The pH of the forage and its level of butyric acid were considerably decreased by various LAB strains [14].
Inoculation of silage with LAB strains not only improved the quality of the animal feed, but also improved animal performance, absorbance of various mycotoxins, reducing greenhouse gas emissions, protein availability, and N availability. At optimum temperature (28 °C), fodders like ryegrass showed good results from the LAB strains L. plantarum, P. acidilactici, and P. pentosaceus. However, at higher temperature levels (45 °C), P. acidilactici improved silage quality [15]. Several LAB strains are commercially available, and these are used to improve forage fermentation by enhancing lactic acid production. The supplementation of Lactobacillus with silage showed positive impacts like being less expensive than enzyme formulation, non-corrosive to farm machinery, and eco-friendly [16]. The impacts of facultative heterofermentative and homofermentative LAB have been studied, and improved beneficial effects were reported previously [17]. Inoculation of fodders with LAB during the process of ensiling very much improves the quality of silage by their ability to ferment ethanol and acetic acid using heterofermentative pathways or ferment soluble sugars to lactic acid using homofermentative pathways. The macromolecules, including, 1,2-propanediol are produced by some LAB species, such as Lactobacillus buchneri which uses heterofermentative pathways to ferment ethanol and acetic acid and improved aerobic stability of the silage [18]. Halophytes are plants that survive at very high concentrations of soil and water and have various physiological functions that enable them to withstand the high concentrations of salt. Plants from the genus Atriplex lentiformis (quail bush) grow under abiotic and biotic stress and are used as fodder along with other nutrient supplements. In Saudi Arabia, various saltbushes, including, Atriple semibaccata, A. canescens, A. tatarica, A. leucoclada, A. halimus, A glauca, A. farinose, A. dimorphostegia and A. coriacea were reported [19].
Agro-industrial by-products were used as the raw material for silage preparation. These include date palm, wheat bran, olive cake, tomato pulp, grape marc, citrus pulm, and banana waste. These traditional preparations of silage practice replace the traditional feedstuffs and concentrations [20]. Agro-industrial waste by-products of plants are promising ingredients that are capable of providing protein, energy, and other nutrients, as well as bioactive compounds such as nondigestible polysaccharides, polyphenols, and bioactive peptides for livestock. These agro-industrial waste by-products can be fed to livestock after the modification process. The present study was performed to analyze the ensiling properties and nutrient quality of quail bush and date palm to be effectively used for feeding ruminants to improve their health benefits. To improve the silage quality, Lactobacillus strains were inoculated, and beneficial effects were analyzed. We hypothesize that LAB can improve the nutritional value and anaerobic fermentation quality of mixed quail bush and date palm by preventing the growth of pathogenic bacteria.
2 Materials and methods
2.1 Isolation of lactic acid bacteria from date syrup
Date syrup samples were serially diluted using sterile double distilled water. The samples were spread on MRS agar medium (Himedia, Mumbai, India) (pH 6.5) supplemented with l-cysteine hydrochloride (0.12 g/L) to suppress the growth of non-lactic acid bacteria. The culture plates were incubated for 72 h at 30 ± 2 °C. Then the isolated colonies were further purified using the same culture medium, and the pure colonies were maintained on MRS agar slants. The LAB strains were preserved in glycerol (50%) for long-term storage.
2.2 Characterization of LAB
A total of 42 LAB isolates were isolated from the date syrup, and the potent strains (LC09 and L. lactis LL25) were characterized. These two strains were characterized by morphological, physiological, and biochemical tests and 16S rDNA sequencing. The isolates were individually cultured in nutrient broth medium (Himeida, Mumbai, India) for 18 h. It was centrifuged at 10,000 rpm for 10 min. Genomic DNA of the selected strains was performed using a commercial kit (Merck, Germany) and used as a template for polymerase chain reaction (PCR). Forward (5′-GAGAGTTTGATCCTGG-3′) and reverse (5′-TACCGCGGCTGCTGGCAC-3′) primers were selected as described earlier. The primers, template, DNA polymerase, and DTP mix were used to amplify 16S rDNA using a thermocycler PCR machine. The amplified gene was purified and sequenced using Applied Biosystems [21, 22].
2.3 Analysis of antibiotic susceptibility
The selected bacterial strains, L. casei LC09 and L. lactis LL25, were cultured in MRS broth (Hiemdia, Mumbai, India). The culture medium was sterilized using an autoclave at 121 °C for 20 min. Then the medium was cooled and inoculated with the strains LC09 and LL25. The inoculums were diluted using physiological saline for appropriate density and spread on Petri plates with MRS agar medium. Then the antibiotics such as streptomycin (10 µg), bacitracin (10 µg), erythromycin (10 µg), chloramphenicol (20 µg), amoxicillin (20 µg), kanamycin (20 µg), and penicillin (10 µg) (Himedia, Mumbai, India) were loaded. Plates were incubated at 32 ± 2 °C, and antibiotic resistance was analyzed. The values from all determinations are expressed as the mean ± standard deviation.
2.4 Tolerance to low pH values
This experiment was performed as suggested by Leite et al. [23] with a few modifications. Briefly, 2.0 mL of 18 h culture at 32 ± 2 °C was harvested by centrifugation (10,000 rpm for 10 min). The LAB pellet obtained after centrifugation was suspended in phosphate buffered saline (pH 6.5, 0.1 M) and the optical density was observed at 600 nm. Then the sample was appropriately diluted, and the final optical density of the sample was 0.60 ± 0.20 at 600 nm. Then the pH of the medium was adjusted between 2.0 and 4.0 with 1 N hydrochloric acid and incubated at 32 ± 2 °C for 3 h. The pH tolerance of the LAB was evaluated by enumerating the bacterial cells using MRS agar medium. Then the percentage of the LAB survival rate was evaluated.
2.5 Bile salt tolerance test
In this method, 18 h cultures of Lactobacillus strains (LC09 and LL25) were grown at 32 ± 2 °C and harvested by centrifugation (10,000 rpm for 10 min). The pellets were suspended in PBS (pH 6.5, 0.1 M), and the cell density was measured at 600 nm using a UV visible spectrophotometer. The optical density of the culture was adjusted to 0.50 ± 0.05 at 600 nm using sterile double distilled water. Then the oxygall concentration of the culture was adjusted to 0.3%, 0.4% and 0.5% (w/v). Then the culture was incubated for 3 h at 32 ± 2 °C. It was spread on MRS agar medium, and viable cell counts were made using an automated colony counter. The bacterial survival rate (%) was calculated as described previously [23].
2.6 Adherence test
The adhering ability of the tested bacterial strains to the intestine was determined in vitro using HT-29 cells [24]. In this method, a monolayer culture of HT-29 cells was prepared in a tissue culture plate at a 5 × 105 cells/mL concentration. Then the monolayer was washed with phosphate buffered saline (PBS) (pH 7.2, 0.1 M). In the meantime, the overnight culture of LAB strains was individually harvested by centrifugation at 10,000 rpm for 10 min. The pellets were suspended in PBS (pH 6.5, 0.1 M), and the cell density was measured at 600 nm using a UV visible spectrophotometer. The final LAB concentration was maintained at 109 CFU/mL and was added to the HT-29 monolayer with RPMI-1640 medium. The microtiter culture plate was incubated at 37 ± 2 °C for 2 h in 5% CO2. Then the monolayer was washed three times with phosphate buffered saline (pH 7.2, 0.1 M) and finally fixed in methanol (Merck, Germany) for 15 min. After complete fixation, the monolayer was washed again with PBS, air dried and stained with Giemsa stain. Then the microtitre plate was examined under a bright field microscope. The adhesion property was analyzed by examining viable bacterial counts. MRS agar plate was prepared and 0.1 mL sample was spread on MRS medium. The plate was incubated at 32 ± 2 °C for 24 h for the development of colonies. The percentage of LAB adhesion was finally calculated.
2.7 Hemolytic activity test
The selected bacterial strains were plated on MRS agar plates (Himedia, Mumbai, India) with defibrinated sheep blood. The culture plate was incubated at 32 ± 2 °C for 48 h. The positive control (Staphylococcus aureus ATCC6538) was used to check hemolysis. A clear zone around the colonies indicates β-hemolysis, a greenish zone around the colonies indicates α-hemolysis, and no zone around the colonies indicates γ-hemolysis or no hemolysis [23].
2.8 Antioxidant properties of Lactobacillus strains
2.8.1 2,2-Diphenyl-2-picrylhydrazyl Assay
In this study, the free-radical-scavenging properties of the screened LAB from date syrup samples were evaluated using the DPPH method with Trolox as the standard. DPPH was dissolved in ethanol and prepared at a 0.2 mmol/L concentration. The cell-free extract (50 -100 µL) was mixed with a DPPH solution (500 µL) and incubated for 30 min in the dark at room temperature. The sample was centrifuged at 5000 rpm for 5 min, and the absorbance of the sample was measured using a microtiter plate reader.
2.8.2 Ferric reducing power assay
The working reagent was prepared by adding 2 mL of 2,4,6-tri(2-pyridyl)-triazine (TPTZ) (10 mM/L) with 2 mL of Iron(III) chloride hexahydrate (20 mM/L) to 40 mL of acetate buffer (300 mM, pH 3.6) (Sigma-Aldrich, USA). The solution was equilibrated, incubated at 37 °C, and stored in the dark. The cell free extracts (50—100 µL) was added to microtiter plate and mixed with FRAPS solution (200 µL) and incubated for 30 min at room temperature in a dark. The absorbance of the sample was measured using a UV–visible spectrophotometer at 595 nm against a blank. FeSO4 was used as the standard curve, and the final results were expressed as mg/mL Fe2+ [25].
2.9 Antifungal properties of LAB strains
The antifungal properties of Lactobacillus species were determined by the agar spot method against Aspergillus niger, Aspergillus flavus, Fusarium oxysporum, Fusarium graminearum, and Rhizopus microsporus. In this method, 20 mL of sterilized MRS agar (Himedia, Mumbai, India) medium was carefully poured into the Petri dishes. Then 50 µL of the selected bacterial strain was spotted on it. The plates were incubated at 32 ± 2 °C. Then, 100 µL of bacterial suspensions were added to MRS soft agar medium on the same Petri plates. Then it was incubated for 3 days at 32 °C, and antifungal activity was measured. In the control experiments, only fungal strains were incorporated. Experiments were performed in triplicate, and an average value was considered for analysis [16].
2.10 Organic acid production
The LAB strains were grown in the culture medium with the following composition (g/L): fructose, 10.0; glucose, 10.0; malic acid, 2.0; sorbitol, 5.0; MnSO4.H2O, 0.05; MgSO4.7H2O, 0.2; EDTA, 0.1; K2HPO4, 1.0; Tween 80, 1 mL; and ammonium citrate, 2.0; (pH 6.2). After 24 h of culture, the cell free extract was subjected to the determination of organic acid. Lactic acid and acetic acid production were analyzed by high-performance liquid chromatography (HPLC) [26].
2.11 Pectinase activity of LAB
Pectinase activity of the bacterial strain was screened as suggested by Varghese et al. [27]. The selected bacterial strains were cultured on pectin-agar medium and incubated for 24 h at 37 °C in an incubator. After 24 h of incubation, Gram’s iodine solution was flooded and incubated for 20 min to determine a zone of clearance due to the hydrolysis of pectin.
2.12 Silage preparation
The whole quail bush (Atriplex suberecta) was collected from the wild and used for the preparation of silage. Date waste was collected from the date processing industry. The Lactobacillus strains, L. casei LC09 and L. lactis LL25 isolated from date syrup, were used and the culture was incubated for 24 h in MRS broth medium (Himedia, Mumbai, India). The growth was monitored by using a VU – Visible spectrophotometer at 700 nm, and the optical density was adjusted (0.50 ± 0.07) using physiological saline. Quail bush and date were used at various ratios (1:1, 1:2, 2:1, 1:3, 3:1, 1:0, and 0:1). Then 1.0 kg of fresh matter was mixed with 1.0 mL of L. casei LC09 and L. lactis LL25. Silage preparation was carried out in small plastic bags containing 1.0 kg fodder. Bush was cut into about 10-mm length and carefully packed into the plastic bags. All bags were vacuumized and stored at 30 ± 2 °C for 30 days.
2.13 Analysis of compost maturity
The silage sample (100 g) was mixed with 300 mL of deionized water and extracted for 24 h at 4 °C. It was filtered using cheesecloth, followed by filter paper, and the filtered sample was stored at 0 °C. Organic acid (lactic acid and acidic acid) levels were evaluated by the HPLC method as described previously by an Agilent 1260 HPLC system (Agilent, USA). The pH of the sample was evaluated using a digital pH meter. Crude fibre and organic matter were detected by the AOAC method [28]. Crude protein was determined by Lowry’s method, and organic matters were tested as described previously [29]. Acid detergent fiber and neutral detergent fiber were estimated as suggested previously [2]. To determine aerobic stability, sampling (100 g) was made using a sterile container (n = 3) and subjected to testing aerobic stability at room temperature (28 ± 2 °C). To avoid particle (dust) contamination and drying, a layer of film with some holes was applied to cover the neck of the container. An empty container was used to determine the room temperature. Aerobic stability of the silage is defined as the time it took for the temperature in the silage to increase above ambient temperature.
2.14 Statistical analysis
A one way analysis of variance was performed, and a p-value < 0.05 was considered statistically significant.
3 Results and discussion
3.1 Characterization of lactic acid bacteria
The selected LAB was analyzed based on macroscopic analysis and culture characteristics. A total of 42 LAB isolates were observed on MRS agar plates, and all colonies were white pinhead colonies. The biochemical characters of two of the most active Lactobacillus strains were selected for 16S rDNA sequencing. The characterization of the isolated date syrup samples was carried out using the 16S rDNA gene using PCR analysis. The two isolated LAB were identified as Lactobacillus casei and L. lactis. The probiotic properties of these two bacterial strains were analyzed to fulfil some criteria of probiotics, such as survival ability at low pH values, presence of high bile salt concentrations, ability to strongly adhere intestinal cells, and non-pathogenic strains [30].
3.2 Antibiotic sensitivity of LAB strains
The selected bacterial strains LC09 and LL25 were sensitive against all tested commercial antibiotics, however, the sensitivity pattern differs between the tested strains (Table 1). The antibiotic-sensitivity test revealed that L. casei LC09 and L. lactis LL25 are highly safe, which is the initial step to analyzing their probiotic features. Increased amounts of probiotics in dietary feed supplements can effectively establish a potential reservoir of various antibiotic-resistant genes in the gut and transfer them to pathogenic organisms that commonly share the same intestinal habitat. Hence, it is an essential task to evaluate the antibiotic resistance of any isolated LAB to avoid any serious clinical implications. The selected bacterial strains were highly susceptible to tetracycline, penicillin, chloramphenicol, and ampicillin. These findings were in accordance with the observations by Arasu et al. [4] who revealed that Lactobacillus are highly sensitive to beta-lactam antibiotics, such as penicillin, ampicillin, and broad-spectrum antibiotics tetracycline. Antibiotic susceptibility varied based on the type of isolate. Lactobacillus strains were susceptible to gentamicin [31] and erythromycin [32].
3.3 LAB strains tolerate low pH
In the present study, the LAB strains (LC09 and LL25) were subjected for various pH tolerance (pH 2.0 – 4.0). The isolated strains LC09 and LL25 showed potential surviving rate of 93.9 ± 2.1% and 98.1 ± 3.8% after exposure to pH 3.0. At pH 2.0, the isolated strains were unable to survive after 3 h of exposure. At p 4.0, the survival rate improved. LC09 and LL25 showed a surviving rate of 90.3 ± 3.1% and 98.6 ± 3.6%, respectively. The influence of LAB survival rate in relation to pH is described in Fig. 1. The selected bacterial strains survived at low pH. The potent probiotic strains should survive at a low pH value (pH 3.0) in the stomach, which was considered a major criteria for the screening of any probiotic bacteria [16]. The survival rate of L. casei LC09 and L. lactis LL25 varied based on pH values and viability was reduced, but viability was observed at low pH values. Previous results also showed that incubation of bacterial strains at very low pH (< 2.0) affected the viability of the bacterial cells [33]. The present finding was similar to the results of Arasu et al. [16] revealed the adaptation of LAB strains in relation to the surviving environment. The potent probiotic strains should survive at a low pH value (pH 3.0) in the stomach, as previously suggested for the screening of any probiotic bacteria [4]. In the present study, the selected strains survive at low pH values (pH 3.0–4.0). The survival rates of L. casei LC09 and L. lactis LL25 varied based on the present observation. Previous studies revealed that incubation of bacterial strains at very low pH (< 2.0) affected the viability of the bacterial cells [29]. The present finding was similar to the results of Valan Arasu et al. [16] revealed the adaptation of LAB strains in relation with the surviving environment (low pH).
3.4 Bile salt tolerance of LAB strains
The bacterial strains LC09 and LL25 were incubated with various bile concentrations (0.3%—0.5%) and bile salt tolerance was determined. As described in Fig. 2, the selected strains from date syrub survived at the selected concentrations of bile salt after 3 h of incubation. The strains, LC09 and LL25 showed maximum survival rate of 93 ± 3.1% and 96 ± 2.9%, respectively, at a 0.3% bile salt concentration. Moreover, survival rates slowly declined at 0.4% and 0.5% concentrations. The survival ability of the Lactobacillus strains in bile salt concentrations supports the organism’s rapid colonization and increased metabolic activity in the small intestine of the host species [34]. The bile salt concentration of humans varied between 0.3% and 0.5%. However these bile salt concentrations varied based on the secretion of pancreatic enzymes and diet composition [35]. The increased bile salt tolerance revealed the application of LAB strains in feed applications.
3.5 Adhesion ability of LAB
The adhesion property of the selected LAB strains from date syrup on HT-29 cell lines was evaluated by microscopic observation after Giemsa staining. The present finding showed that the selected strains adhered very strongly on the surface of the HT-29 cell lines. The number of adhered bacteria on the cell lines was evaluated by plating method. The LAB strain LC09 showed maximum adhesion ability (93 ± 3.9%), followed by LL25 (89.1 ± 2.6%). The present finding revealed that the strains firmly adhered to the mucus layer of the intestine. The selected probiotic LAB must be able to firmly adhere to the mucus layer of the intestine in order to be established and colonized in the intestine [36]. In the study both isolates showed excellent ability to colonize and adhere on the intestinal cells. The present findings were similar to the properties of LAB strains, which found that the adherence property is mainly strain specific mainly due to the variation in the receptors for microbial adhesions on mucus and also based on bacterial origin. The screened Lactobacillus sp. from date syrup showed potent binding activity, strongly adhered to the cell lines, and auto-aggregation took place between LAB and HT-29 cells. Muryany et al. [37] reported auto-aggregative pattern of bacterial cell attachment with cell lines (HT-29) which was screened from the fermented fish.
3.6 Hemolytic test
The selected bacterial strains did not show any hemolytic signs on the blood agar medium. The selected two strains were γ-hemolytic (non-hemolytic) and no zones developed around the colonies. Hemolytic properties is an important virulent factor among pathogenic bacteria. The present finding was similar to previous report that showed bacteria from the genus Lactobacillus show no haemolytic property [38]. Bacteria exhibiting hemolytic activity can induce the release of haemoglobin by disrupting red blood cells. These hemolytic bacteria produce hemolysins, and beta haemolytic strains produce cytotoxic phospholipase. The hemolytic property of Bacillus strains was determined among probiotics, and 58% of probiotic strains exhibited haemolytic activity [39].
3.7 Antioxidant activity of CFE from the LAB
The antioxidant properties of the cell free extract (CFE) of the selected LAB strains were analyzed. Figure 3a and b depict the antioxidant properties of strains LC09 and LL25. The strain LC09 exhibited better DPPH antioxidant activity than the strain LL25 (Fig. 3a). Also, the CFE of the strain exhibited 2.1 ± 0.3 mM FRAP/µg protein at 100 µL concentration (Fig. 3b). Bacteria from the genus Lactobacillus are widely used as natural antioxidants that effectively prevent cellular damage due to oxidative stress caused by free radicals in the host species. These Lactobacillus species metabolized flavonoids and phenolic compounds and this metabolizing ability was species specific [40]. The increased amount of flavonoids and phenols during the enzymatic hydrolysis of LAB increased its antioxidant properties [16]. The present finding showed that the selected Lactobacillus spp. from date syrup had different abilities to metabolize flavonoids and phenolic compounds and contributed various degrees of antioxidant properties. The higher concentrations of phenol and flavonoids in the date syrup may be due to the activity of Lactobacillus sp. The 1,1-diphenyl-2-picryl-hydrazine (DPPH) and ferric reducing ability of plasma (FRAP) assays analyze the antioxidant properties of the CFE to scavenge and reduce various radical compounds [28]. The antioxidant power of any sample is based on the total phenol and total flavonoid of the extract. Based on the present findings, the selected strains from date syrup showed significant scavenging activity in DPPH and reduction activity in the FRAP assay. During microbial fermentation, the amount of phenolic content and flavonoids in the medium increased due to the metabolic activities of Lactobacillus sp. [24, 40].
3.8 Antifungal activity
The antifungal properties of the selected bacterial strains were evaluated by the agar overlay method against A. niger, A. flavus, F. oxysporum, F. graminearum and R. microsporus. Among the selected LAB strains, LC09 showed maximum zone of inhibition against F. oxysporus (26 ± 1 mm), A. niger (27 ± 2 mm) and F. graminearum (26 ± 1 mm) after 3 days of incubation. The strain LL25 showed the highest zone of inhibition against F. graminearum (29 ± 1 mm) and F. oxysporus (24 ± 2 mm) (Table 2). Lactic acid bacteria produce various organic acids, including lactic acid. The enhanced production of various organic acids critically reduces the pH of the culture medium. The reduced acidic pH inhibited the growth of pathogens by reducing their intracellular pH, leading to an arrest of cell functions. These inhibitory functions of LAB are associated with antibacterial and antifungal organic acid production [41]. Lactobacillus species can be effectively used in various agricultural practices for controlling the growth of mycotoxigenic fungi [42].
3.9 Organic acids production
Acetic acid and lactic acid production were end products determined from the CFE. Lactic acid production was 5.3 ± 0.32 g/L and 4.7 ± 0.28 g/L for strains LC09 and LL25, respectively. Acetic acid production was 0.39 ± 0.03 g/L and 0.27 ± 0.47 g/L for strains LC09 and LL25. The bacterial strains from the genus Lactobacillus specifically metabolize the oligosaccharides using fructofuranosidase enzyme. These organisms degraded long-chain inulin and oligofructose and utilized them for metabolic processes. Lactic acid was considered the major end product. Acetic acid production in the culture medium by the LAB strains may be the result of the biochemical pathway, including, citrate metabolism, heterofermentative pathway, and degradation product of lactic acid. Antibacterial organic acid production was reported in lactic acid bacteria [43] and the organic acid exhibited antifungal activity against Penicillium nordicum [44].
3.10 Pectinolyic property of LAB
In plant tissues, pectic polysaccharides are common components; thus, LAB isolated from the fruit syrup could harbour pectinolytic activity. The strain LC09 showed a 28 ± 3 mm zone of clearance on the pectin-agar plate, whereas LL25 showed a 19 ± 1 mm zone around the colony. Vidhyasagar et al. [45] reported the degradation of pectin by LAB strains screened from various fermented foods. Among the strains screened, L. plantarum subsp. argentoratensis, Leuc. lactis and P. pentosaceus hydrolyzed pectin significantly. Chatterjee et al. [46] reported that the available pectin effectively improved titratable acidity and bacterial growth in Bifidobacterium and LAB cultures. The pectin degrading LAB species have wide applications in feed formulation and fermented food production [47].
3.11 LAB improved silage quality
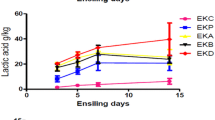
The chemical composition of silage was analyzed after 30 days. Lactic acid and acetic acid levels were increased in the experimental silage (p < 0.001). The pH of the silage decreased considerably compared to the control (p < 0.001). Compared with the control silage, the experimental silage improved fermentation quality after 30 h. Whole quail bush and date wastes were tested at various ratios, and a 1:3 ratio was most suitable. At this concentration, DM (%) improved significantly compared to the control and the pH value declined to 3.94 ± 0.01. Also, acetic acid and lactic acid production were high at this concentration (p < 0.001) (Table 3). In addition, the CP, OM, and NDF percentages increased at this ratio, which was considered the optimum level (p < 0.001). The crude protein content improved in the experimental silage due to LAB activities. The amount of NDF decreased significantly in the silage inoculated with LAB strains (Table 4). The objectives of silage preparation and additional inoculation with the selected LAB strains are to enhance the production of organic acids, decrease pH levels, and effectively inhibit the proliferation of pathogenic bacteria [48]. In recent years, several Lactobacillus strains have been used to inoculate the ensilaged forage to reduce the effect of native epiphytic LAB. They may be based on various properties, such as their ability to effectively dominate the naturally available microflora in the forages and to synthesize organic acids such as acetic acid and lactic acid to reduce the pH value of the silage. In our study, a decreased pH value was observed; correspondingly, the lactic acid and acetic acid amounts were improved in the silage. Inoculating silage with potent LAB effectively improved the silage quality by promoting the multiplication of LAB, inhibiting various pathogenic bacteria and considerably decreasing the pH level of the medium, which reflected the quality of the silage. The CP level was found to be higher in the experimental groups than in the control silage. The present finding confirmed that the supplemented LAB increased lactic acid production and improved the silage quality of natural biomass. The inoculated lactic acid bacteria improved alfalfa silage [49] and corn silage [50] and reduced the growth of pathogenic bacteria and fungi.
3.12 Aerobic stability of silage
The variation in aerobic stability of the mixture of silage prepared at various compositions of quail bush and date waste is described in Table 5. Silage temperature increased up to 100 h of aerobic exposure and decreased thereafter. The temperature variance is varied based on the composition of the fodder and date waste. After 80 h of aerobic exposure, temperature rose slowly in almost all silages and reached its maximum after 100 h of treatment (p < 0.001). Heterofermentative LAB strains have a good ability to maintain the aerobic stability of silage. The present finding indicates that the addition of LAB inoculum improves the aerobic stability of silage. In the case of homofermentative LAB inoculums, poor aerobic silage stability was reported [51, 52]. Rapid aerobic deterioration of silage material is mainly caused by the unrestricted growth of pathogenic bacteria and yeasts, which utilize organic acid and increase the pH value of the silage and temperature [53]. The present finding supports the application of the selected hetero-fermentative LAB strains as silage inoculants to enhance the production of organic acids, thus reduce deterioration activity.
4 Conclusions
The characterized two LAB isolates, Lactobacillus casei LC09 and L. lactis LL25, from date were resistant at low pH values and also in a stimulated gastric environment. These two strains produced lactic acid and acetic acid and inhibited the growth of fungi and pathogenic bacteria. These strains enhanced silage quality in quail bush-date waste silage after 30 days. The silage ensiled with Lactobacillus species showed a protective role from aerobic deterioration of silage material because of the presence of organic acids and the reduced level of pathogenic bacteria and fungi. The date syrup-derived LAB strains could be useful for the preparation of silage inoculants to improve ruminant feed.
Data availability
The data used to support the findings of this study are available from the corresponding author upon reasonable request.
References
Soundharrajan I, Kim D, Kuppusamy P et al (2019) Probiotic and Triticale silage fermentation potential of Pediococcus pentosaceus and Lactobacillus brevis and their impacts on pathogenic bacteria. Microorganisms 7:318. https://doi.org/10.3390/microorganisms7090318
Ilavenil S, Vijayakumar M, Kim DH et al (2016) Assessment of probiotic, antifungal and cholesterol lowering properties of Pediococcus pentosaceus KCC-23 isolated from Italian ryegrass. J Sci Food Agric 96(2):593–601. https://doi.org/10.1002/jsfa.7128
Lahtinen S, Ouwehand AC, Salminen S, von Wright A (eds) (2011) Lactic acid bacteria: microbiological and functional aspects. Crc Press
Arasu MV, Kim DH, Kim PI et al (2014) In vitro antifungal, probiotic and antioxidant properties of novel Lactobacillus plantarum K46 isolated from fermented sesame leaf. Ann Microbiol 64:1333–1346. https://doi.org/10.1007/s13213-013-0777-8
Roth APDTP, Reis RA, Siqueira GR et al (2010) Sugarcane silage production treated with additives at different times post burning. R Bras Zootec 39:88–96. https://doi.org/10.1590/S1516-35982010000100012
Pedroso ADF, Nussio LG, Loures DRS et al (2008) Fermentation, losses, and aerobic stability of sugarcane silages treated with chemical or bacterial additives. Scientia Agricola 65:589–594. https://doi.org/10.1590/S0103-90162008000600004
Ávila CLS, Schwan RF, Pinto JC, Carvalho BF (2011) Potential use of native microorganisms strains of forage for silage production. In: II Symposium on Forage Quality and Conservation. Zopollatto M, Daniel LLP, Nussio LG, Sa Neto A (eds) Fundacao de Estudos Agrarios Luiz de Queiroz (FEALQ), Piracicaba, Brazil, pp 25–44
Ávila CLDS, Valeriano AR, Pinto JC et al (2010) Chemical and microbiological characteristics of sugar cane silages treated with microbial inoculants. Braz J Ani Sci 39:25–32. https://doi.org/10.1590/S1516-35982010000100004
Siedler S, Rau MH, Bidstrup S et al (2020) Competitive exclusion is a major bioprotective mechanism of lactobacilli against fungal spoilage in fermented milk products. Appl Env Microbiol 86(7):e02312-e2319. https://doi.org/10.1128/AEM.02312-19
Axel C, Zannini E, Arendt EK (2017) Mold spoilage of bread and its biopreservation: A review of current strategies for bread shelf life extension. Cri Rev Food Sci Nut 57(16):3528–3542. https://doi.org/10.1080/10408398.2016.1147417
Parappilly SJ, Idicula DV, Chandran A, Mathil Radhakrishnan K et al (2021) Antifungal activity of human gut lactic acid bacteria against aflatoxigenic Aspergillus flavus MTCC 2798 and their potential application as food biopreservative. J Food Safety 41(6):e12942. https://doi.org/10.1111/jfs.12942
Crowley S, Mahony J, van Sinderen D (2013) Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives. Tren Food Sci Technol 33(2):93–109. https://doi.org/10.1016/j.tifs.2013.07.004
Contreras-Govea FE, Muck RE, Broderick GA, Weimer PJ (2013) Lactobacillus plantarum effects on silage fermentation and in vitro microbial yield. Ani Feed Sci Technol 179(1–4):61–68. https://doi.org/10.1016/j.anifeedsci.2012.11.008
Neres MA, Zambom MA, Fernandes T et al (2013) Microbiological profile and aerobic stability of Tifton 85 bermudagrass silage with different additives. R Bras Zootec 42:381–387. https://doi.org/10.1590/S1516-35982013000600001
Soundharrajan I, Kim DH, Srisesharam S et al (2017) Application of customised bacterial inoculants for grass haylage production and its effectiveness on nutrient composition and fermentation quality of haylage. 3 Biotech 7:1–9. https://doi.org/10.1007/s13205-017-0965-5
Valan Arasu M, Jung MW, Kim DH et al (2015) Identification and phylogenetic characterization of novel Lactobacillus plantarum species and their metabolite profiles in grass silage. Ann Microbiol 65:15–25. https://doi.org/10.1007/s13213-014-0830-2
Oliveira AS, Weinberg ZG, Ogunade IM et al (2017) Meta-analysis of effects of inoculation with homofermentative and facultative heterofermentative lactic acid bacteria on silage fermentation, aerobic stability, and the performance of dairy cows. J Dairy Sci 100(6):4587–4603. https://doi.org/10.3168/jds.2016-11815
Muck RE, Nadeau EMG, McAllister TA et al (2018) Silage review: Recent advances and future uses of silage additives. J Dairy Sci 101(5):3980–4000. https://doi.org/10.3168/jds.2017-13839
Al-Turki TA, Omer S, Ghafoor A (2000) A synopsis of the genus Atriplex L.(Chenopodiaceae) in Saudi Arabia. Feddes Repertorium 111(5–6):261–293. https://doi.org/10.1002/fedr.20001110503
Vasta V, Nudda A, Cannas A et al (2008) Alternative feed resources and their effects on the quality of meat and milk from small ruminants. Ani Feed Sci Technol 147(1–3):223–246. https://doi.org/10.1016/j.anifeedsci.2007.09.020
Oh NS, Lee JY, Oh S et al (2016) Improved functionality of fermented milk is mediated by the synbiotic interaction between Cudrania tricuspidata leaf extract and Lactobacillus gasseri strains. Appl Microbiol Biotechnol 100(13):5919–5932. https://doi.org/10.1007/s00253-016-7414-y
Tadesse BT, Tesfaye A, Muleta D et al (2018) Isolation and molecular identification of lactic acid bacteria using 16s rRNA genes from fermented Teff (Eragrostis tef (Zucc.)) dough. Int J Food Sci 2018:8510620. https://doi.org/10.1155/2018/8510620
Leite AM, Miguel MAL, Peixoto RS et al (2015) Probiotic potential of selected lactic acid bacteria strains isolated from Brazilian kefir grains. J Dairy Sci 98(6):3622–3632. https://doi.org/10.3168/jds.2014-9265
Kim JH, Baik SH (2019) Probiotic properties of Lactobacillus strains with high cinnamoyl esterase activity isolated from jeot-gal, a high-salt fermented seafood. Ann Microbiol 69:407–417. https://doi.org/10.1007/s13213-018-1424-1
Thaipong K, Boonprakob U, Crosby K et al (2006) Comparison of ABTS, DPPH, FRAP, and ORAC assays for estimating antioxidant activity from guava fruit extracts. J Food Compos Anal 19(6–7):669–675. https://doi.org/10.1016/j.jfca.2006.01.003
Bolsen KK, Lin C, Brent BE et al (1992) Effect of silage additives on the microbial succession and fermentation process of alfalfa and corn silages. J Dairy Sci 75(11):3066–3083. https://doi.org/10.3168/jds.S0022-0302(92)78070-9
Varghese LK, Rizvi AF, Gupta AK (2013) Isolation, screening and biochemical characterization of pectinolytic microorganism from soil sample of Raipur city. J Biol Chem Res 30(2):636–643
Thiex N, Novotny L, Crawford A (2012) Determination of ash in animal feed: AOAC official method 942.05 revisited. J AOAC Int 95(5):1392–1397. https://doi.org/10.5740/jaoacint.12-129
Horwitz W, Latimer GW (2000) Official methods of analysis of AOAC International (vol 1, p 17). Gaithersburg: AOAC international
Pringsulaka O, Rueangyotchanthana K, Suwannasai N et al (2015) In vitro screening of lactic acid bacteria for multi-strain probiotics. Livest Sci 174:66–73. https://doi.org/10.1016/j.livsci.2015.01.016
Guo H, Pan L, Li L et al (2017) Characterization of antibiotic resistance genes from Lactobacillus isolated from traditional dairy products. J Food Sci 82(3):724–730. https://doi.org/10.1111/1750-3841.13645
Gad GFM, Abdel-Hamid AM, Farag ZSH (2014) Antibiotic resistance in lactic acid bacteria isolated from some pharmaceutical and dairy products. Braz J Microbiol 45:25–33. https://doi.org/10.1590/S1517-83822014000100005
Gao Y, Li D, Liu S, Liu Y (2012) Probiotic potential of L. sake C2 isolated from traditional Chinese fermented cabbage. Eur Food Res Technol 234:45–51. https://doi.org/10.1007/s00217-011-1608-4
Nawaz AN, Jagadeesh KS, Krishnaraj PU (2017) Isolation and screening of lactic acid bacteria for acidic pH and bile tolerance. Int J Curr Microbiol Appl Sci 6(7):3975–3980
Archer AC, Halami PM (2015) Probiotic attributes of Lactobacillus fermentum isolated from human feces and dairy products. Appl Microbiol Biotechnol 99:8113–8123. https://doi.org/10.1007/s00253-015-6679-x
Shokryazdan P, Sieo CC, Kalavathy R et al (2014) Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. BioMed Res Int 2014, Article 927268. https://doi.org/10.1155/2014/927268
Muryany I, Lian HH, Ina-Salwany ARG et al (2018) Adhesion ability and cytotoxic evaluation of Lactobacillus strains isolated from Malaysian fermented fish (pekasam) on Ht-29 and Ccd-18Co intestinal cells. Sains Malays 47:2391–2399. https://doi.org/10.17576/jsm-2018-4710-15
Padmavathi T, Bhargavi R, Priyanka PR et al (2018) Screening of potential probiotic lactic acid bacteria and production of amylase and its partial purification. J Genet Eng Biotechnol 16(2):357–362. https://doi.org/10.1016/j.jgeb.2018.03.005
Deng F, Chen Y, Sun T et al (2021) Antimicrobial resistance, virulence characteristics and genotypes of Bacillus spp. from probiotic products of diverse origins. Food Res Int 139:109949. https://doi.org/10.1016/j.foodres.2020.109949
Muñoz R, de Las Rivas B, de Felipe FL et al (2017) Biotransformation of phenolics by Lactobacillus plantarum in fermented foods. In: Fermented foods in health and disease prevention (pp 63–83). Academic Press. https://doi.org/10.1016/B978-0-12-802309-9.00004-2
Son SH, Jeon HL, Jeon EB et al (2017) Potential probiotic Lactobacillus plantarum Ln4 from kimchi: Evaluation of β-galactosidase and antioxidant activities. LWT Food Sci Technol 85:181–186. https://doi.org/10.1016/j.lwt.2017.07.018
Deepa N, Rakesh S, Sreenivasa MY (2018) Morphological, pathological and mycotoxicological variations among Fusarium verticillioides isolated from cereals. 3 Biotech 8:1–10. https://doi.org/10.1007/s13205-018-1136-z
Bangar SP, Suri S, Trif M, Ozogul F (2022) Organic acids production from lactic acid bacteria: A preservation approach. Food Biosci 46:101615. https://doi.org/10.1016/j.fbio.2022.101615
Guimarães A, Venancio A, Abrunhosa L (2018) Antifungal effect of organic acids from lactic acid bacteria on Penicillium nordicum. Food Addit Contam: Part A 35(9):1803–1818. https://doi.org/10.1080/19440049.2018.1500718
Vidhyasagar V, Saraniya A, Jeevaratnam K (2013) Identification of pectin degrading lactic acid bacteria from fermented food sources. Int J Adv Life Sci 6(1):8–12
Chatterjee E, Manuel GAS, Hassan S (2016) Effect of fruit pectin on growth of lactic acid bacteria. J Probio Health 4(2):1000147
Garcia C, Guerin M, Souidi K, Remize F (2020) Lactic fermented fruit or vegetable juices: Past, present and future. Beverages 6(1):8. https://doi.org/10.3390/beverages6010008
Drouin P, Tremblay J, Chaucheyras-Durand F (2019) Dynamic succession of microbiota during ensiling of whole plant corn following inoculation with Lactobacillus buchneri and Lactobacillus hilgardii alone or in combination. Microorganisms 7(12):595. https://doi.org/10.3390/microorganisms7120595
Silva VP, Pereira OG, Leandro ES et al (2020) Selection of lactic acid bacteria from alfalfa silage and its effects as inoculant on silage fermentation. Agriculture 10(11):518. https://doi.org/10.3390/agriculture10110518
Li J, Wang W, Chen S et al (2021) Effect of lactic acid bacteria on the fermentation quality and mycotoxins concentrations of corn silage infested with mycotoxigenic fungi. Toxins 13(10):699. https://doi.org/10.3390/toxins13100699
Weinberg ZG, Ashbell G, Hen Y, Azrieli A (1993) The effect of applying lactic acid bacteria at ensiling on the aerobic stability of silages. J Appl Microbiol 75(6):512–518. https://doi.org/10.1111/j.1365-2672.1993.tb01588.x
Filya I (2003) The effect of Lactobacillus buchneri, with or without homofermentative lactic acid bacteria, on the fermentation, aerobic stability and ruminal degradability of wheat, sorghum and maize silages. J Appl Microbiol 95(5):1080–1086. https://doi.org/10.1046/j.1365-2672.2003.02081.x
Joo YH, Kim DH, Paradhipta DH et al (2018) Effect of microbial inoculants on fermentation quality and aerobic stability of sweet potato vine silage. Asian-Australasian J Anim Sci 31(12):1897. https://doi.org/10.5713/ajas.18.0264
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSP2024R190), King Saud University, Riyadh, Saudi Arabia
Author information
Authors and Affiliations
Contributions
DAA: methodology, formal analysis, TAS: methodology, data curation, review, MSE: validation, review, editing, project administration, P.V: methodology, investigation, original draft.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alfarraj, D.A., Sathya, T.A., Elshikh, M.S. et al. Nutritive value and aerobic stability of whole quail bush and date waste silage ensiled at different compositions and the role of hetero-fermentative lactic acid bacteria. Biomass Conv. Bioref. (2024). https://doi.org/10.1007/s13399-024-05744-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13399-024-05744-6