Abstract
Anaerobic lignocellulosic microbial consortia are known to be prodigiously efficient at converting lignocellulosic biomass to methane. In this study, the efficacy of anaerobic fungal consortia (AFC) from five different inocula, including Bubalus bubalis rumen fluid (RU), in degrading filter paper, microcrystalline cellulose, and rice straw (RS), was screened. The AFC from RU performed best in lignocellulosic material degradation and methane production; thus, RU was selected for further experiments. Consecutive batch subculturing (CBS) was performed in RU to enrich and stabilize the dominant and key microorganisms categorized as anaerobic fungi, using the addition of antibacterial agents to suppress the growth of untargeted bacteria. After the CBS, subculture E19 proved the most efficient, with RS degradation of 84% and a methane yield of 310 mL/g VSadded, representing 1.83- and 2.25-fold increases compared to the initial seed, respectively. The microbial community of E19 consisted of anaerobic fungi (uncultured Neocallimastigales, Anaeromyces sp., Orpinomyces sp., and Feramyces sp.) coexisting with anaerobic bacteria (streptomycin resistant Proteiniphilum acetatigenes), and methanogens. The E19 consortium was able to use various carbon sources (87.5%) and contained potential genes encoding enzymes involved in RS degradation. The microbial community of E19 was highly stable, making it a promising inoculum for biomass degradation, especially for anaerobic digestion to produce biogas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Issues of energy security and environmental problems from using fossil fuels are the main factor that requires the acceleration of the search for the renewable bioenergy from biomass. Biomass is arguably the most widespread renewable energy source and is increasingly used as a feedstock because of concerns over fossil fuel consumption, including global warming and human health. Understanding the chemical composition, properties, and ways to use biomass in a cost-effective and environmentally friendly way is therefore essential [1]. Among the various forms of bioenergy introduced to date, biogas has been one aspiration of playing an important role in the bio/circular/green (BCG) economy. This is due to it being a proven form of energy, having worldwide use, as well as being clean and cheap renewable energy. Furthermore, biogas is mainly composed of methane and carbon dioxide via anaerobic digestion (AD) of all types of biomasses.
Biogas is mainly produced from agricultural residues, energy crops (i.e., maize, grass, and sugar beet), animal waste (i.e., livestock manure), and some residues from food industries and biorefineries [2]. Thailand is an agricultural and agro-industrial country that annually generates a huge number of agricultural residues as a potential feedstock in biogas production. In 2018, a total amount of 116 million tons of agricultural residues was produced. After sugarcane (55.33%), rice straw (RS) is an important residue in Thailand (25.75%), followed by cassava pulp (9.05%), oil palm residues (5.62%), and maize straw (3.16%) [3]. Rice straw, as a source of renewable feedstock, consists of 32% cellulose, 18% hemicellulose, and 11% lignin but has not been fully exploited as its complex lignocellulosic structure makes biogas production difficult [4].
In AD, the hydrolytic step is a rate-limiting step of biogas production from agricultural residues due to the complex lignocellulosic structure of the plant cell wall; the enclosure of cellulose fibrils by lignin and hemicellulose makes degradation challenging [5]. Starter seeds from various sources can yield different amounts of methane depending on the hydrolytic enzyme activity. Lignocellulolytic microbial populations from cow rumen gave a methane yield of 223 mL/g COD [6], whereas those from mesophilic wastewater treatment, municipal wastewater treatment, and animal manure gave methane yields of 130–226 mL/g [7,8,9]. Anaerobic bacteria (AB), anaerobic fungi (AF), and methanogens (MT) are main microorganisms in degrading lignocellulosic biomass for biogas production. Hydrolytic AB such as Acetivibrio cellulolyticus, Bacteroides cellulosolvens, Butyrivibrio fibrisolvens, Clostridium acetobutylicum, C. cellulovorans, C. cellobioparum, C. cellulolyticum, C. josui, C. papyrosolvens, Ruminococcus albus, and R. flavefaciens, commonly found in soils, the rumen of ruminants, compost, wastewater, and wood-processing plants, are known degraders of lignocellulosic biomass. These AB can produce cellulase-complex enzymes and cellulosomes, converting cellulosic matter into hexose sugar [10]. Most AF found in the gastrointestinal tract of ruminants, including Orpinomyces, Neocallimastix, and Piromyces, produce a multi-enzyme complex as cellulosomes that can degrade cellulose, hemicellulose, and pectin in lignocellulosic biomass [11], as well as hexose and pentose sugars from hemicellulose [12].
Although AD technology is applied on a full scale in many countries worldwide, it remains necessary to further enhance biogas production through transformative advancement in order to boost the microbial efficiency, methane productivity, and sustainability of AD systems. Recent biological innovations to enhance biogas production can be characterized by three approaches: biological pretreatment (upstream) [13], bioaugmentation (mainstream) [14], and improvement of effective inoculum as starter seed using an AD reactor [15]. A summary of studies involving biological approaches at a laboratory scale to boost methane production from straw at mesophilic temperatures is shown in Table 1. Biological pretreatments by aerobic fungi [16, 17], microbial consortium [18, 19], and bacteria [20] aim to increase the digestibility of biomass before feeding in AD. These strategies provide good results but tend to be time-consuming and require extra equipment. Bioaugmentation can enhance biogas production by inoculating the external sources of certain microorganisms (pure, co-culture, and microbial consortium) into the AD system [21,22,23,24,25,26,27,28,29]. Bioaugmentation is highly recommended to speed up biogas production. However, there are some limitations concerning the adaptability of bioaugmented microorganisms in new environments and their competitiveness with indigenous microorganisms, which result in the washout of bioaugmented cells from the AD system. The AD of lignocellulosic biomass without pretreatment requires effective lignocellulolytic microorganisms to accelerate the conversion to methane and save costs. Therefore, the increased number of adaptive and stabilized effective anaerobic microorganisms for starter seeds in the AD system is a major concern. The enriched and stabilized anaerobic microbial consortium could be a strategy for maintaining and prolonging the effectiveness of microorganisms in the system.
The rumen of ruminants is known as the natural inoculum for AD reactors in which the complex and synergistic microbial communities are extremely efficient in converting lignocellulose to volatile fatty acids (VFAs) and then to methane [30]. The lignocellulolytic microbial consortia can accomplish these processes better than individual species [21]. Anaerobic bacteria, AF, and MT involve biogas production, with AB being the most common in AD reactors [21,22,23,24], despite the fact that AF has a higher efficiency in lignocellulose degradation than AB [24, 26, 28]. The increase of the lignocellulolytic microbial population, especially AF in the microbial consortium, will accelerate lignocellulose degradation to VFAs and enhance biogas production. Besides, the mycorrhiza of AF can penetrate and help break the plant cell wall, facilitating hydrolysis. Anaerobic bacteria can degrade lignocellulosic material more rapidly than AF in separated cultures (mono-cultures) due to the different types of degradation mechanisms; the spores of AF take time to grow filamentous rhizoids (16–24 h) that can attach to the lignocellulosic material and degrade it later [31]. After filamentous rhizoids of AF grow and cover lignocellulosic biomass, cellulases are still active and can degrade the biomass more rapidly than AB during AD [32]. For co-culture, AF associated with MT can degrade wheat straw and RS better than only AF [29]. Recently, Ma et al. [24] found that AF co-cultured with MT could degrade lignocellulosic biomass and produce more methane than the co-culture of AB and MT.
In this study, to maximize the use of the lignocellulosic biomass from agriculture residues, the efficacy and potential of an enriched anaerobic lignocellulolytic fungal consortium (AFC) with high methane production from lignocellulosic biomass were established. Screening, selection, and enrichment techniques were used to establish the AFC under AD. Five different sources of seed inoculum were explored by sorting the sources with the best AFC potential and the highest activity in the degradation of lignocellulose and the production of methane. Both anaerobic Gram-positive and Gram-negative bacteria were removed by antibiotics during screening. Consecutive batch subcultures were cultivated until the enriched and stabilized. AFC was obtained, which was then stabilized AFC was characterized based on the biochemical methane potential (BMP), the metabolic diversity by Biolog®EcoPlate, the anaerobic microbial communities by using the ITS1 gene target for AF, the 16S rRNA gene target for AB including MT, and metagenomics shotgun sequencing for the microbial role and function.
2 Materials and methods
The effective AFC for RS conversion to methane was obtained by (1) screening of anaerobic lignocellulolytic fungi in wastewater sludge, animal manure, and the rumen fluid of swamp buffalo, (2) enriching and stabilizing the consortium by consecutive batch subculturing (CBS), (3) determining its efficacy for lignocellulosic biomass degradation and methane production by BMP and metabolic diversity, and (4) analyzing the microbial communities and functions by 16S/ITS gene target Illumina and metagenomic sequencing shotgun approaches. In this study, the substrate is expressed as per gram of volatile solids (g VS). The strategies and methods used are shown in Fig. 1.
2.1 Materials and growth medium
The lignocellulolytic microbial seed inocula were selected from five different seed sources in Thailand: cow (CM), goat (GM), and pig (PM) manure from livestock farms in Phitsanulok province, the swamp buffalo rumen fluid of Bubalus bubalis (RU) from a local farm in Phichit province, and microbial seed sludge from the anaerobic wastewater treatment system of a palm oil mill factory (MS), Eastern Palm Oil Co., Ltd., in Chonburi province. The inocula were preliminarily characterized by analysis of total solids (TS) and volatile solids (VS) using a standard technique according to the American Public Health Association (APHA) [33]. The seed biomass of CM, GM, and PM (presented as VS based on %TS) ranged from 76 to 80%, whereas the RU and MS values were 81 and 94%, respectively (Table S1).
The ability of these inocula to degrade cellulose was tested with various cellulosic substrates in basal growth medium [34]. The antibacterial agents penicillin and streptomycin (Sigma-Aldrich, Japan) were added to the medium to inhibit the growth of Gram-positive and Gram-negative bacteria, respectively. Whatman no. 1 filter paper (FP) (GE Healthcare UK Ltd., UK), representing amorphous cellulose, and microcrystalline cellulose (MCC) (Avicel PH-101; Sigma-Aldrich, Japan), representing crystalline cellulose, were used. Rice straw was used as the lignocellulosic substrate. It was prepared by drying at 50 °C for 48 h and cutting and sieving through a mesh with a pore size of 205–350 µm. The lignocellulose content of RS was analyzed by a Fibretherm® (C. Gerhardt GmbH & Co. KG, Germany) according to the Association of Official Agricultural Chemists’ (AOAC) method for neutral fiber [35]. Rice straw is composed of cellulose (35.66 ± 0.24%), hemicellulose (18.70 ± 0.16%), and lignin (8.62 ± 0.12%) as reported in previous study [15].
2.2 Screening of anaerobic lignocellulolytic fungal consortium for lignocellulose conversion to methane
To select the effective AFC from five different seed inocula, screening was first performed using amorphous and crystalline cellulose. Subsequently, the seed inocula that showed cellulose degradation were further screened using the lignocellulosic biomass RS. The primary screening was carried out in two steps. First, the screening of AFC was performed in a Hungate tube (a working volume of 10 mL) containing 0.1 g of FP (5 × 40 mm) as the substrate, 0.5 g of each inoculum, the basal growth medium [34], 0.5 mL of 40 mg/mL streptomycin sulfate salt, and 40 mg/mL of penicillin G sodium salt. Nitrogen gas was flushed into the headspace of the vial and the liquid phase for 15 and 10 s, respectively, and the vial was then sealed with a butyl rubber septum and an aluminum cap for anaerobic conditions, followed by incubation at 37 °C for 7 days until the complete degradation of FP. Each inoculum had a treatment set that comprised 27 replicates with FP and a control set (no FP added), replicated thrice. Three replicates were randomly selected from the treatment set to measure FP removal after 7 days. Only three FP-degrading seed inocula (CM, GM, and RU) were found, and these were used in the following step. Then, the remaining FP-degrading treatment sets (24 replicates) for CM, GM, and RU were transferred to an MCC assay; a 10 mL working volume containing 0.5 g of the inoculum, 0.1 g of MCC, basal medium, and 0.5 mL of the antibiotics used earlier. The subculturing was performed under anaerobic conditions at 37 °C for 7 days (till completion of MCC degradation), using three replicates each for the treatment and control (no MCC added) experiments, and MCC removal was determined at the end of the experiment. For the secondary screening, the remaining 18 MCC-degrading replicates of each inoculum were transferred to an RS removal assay, a 10 mL working volume containing 0.5 g of the inoculum, 0.1 g of RS, the basal medium, and 0.5 mL of the antibiotics used earlier. The subcultures were incubated at 37 °C under anaerobic conditions for 14 days to complete RS degradation, using three replicates each for the treatment and control (no RS added) experiments, and RS removal was measured at the end of the experiment. The schematic diagrams of the screening experiment are shown in Figs. 1 and S1. During the primary and secondary screenings, biogas production was monitored daily using the pressure transducer by Theodorou et al. [36]. The methane content was determined via gas chromatography (Shimadzu, Class-GC 14B, Japan) as described by Panichnumsin et al. [37]. The biodegradation of substrates (VS removal) was determined at various time intervals. The calculation of substrate removal and the rate of degradation over time was done using Eqs. 1 and 2, respectively:
where VS is the volatile solids of the substrate, t0 is the initial time, and t is the time of degradation (d).
2.3 Enrichment of the anaerobic lignocellulolytic fungal consortium
The AFC from RU showed the highest RS removal efficiency and was selected for enrichment and stabilization using CBS, denoted by E1 − E19. The experiment was conducted in a 120-mL sealed vial with 0.5 g of RU, 0.5 g of RS (ISR of 1:1), 2.5 mL of the antibiotics used earlier (at similar concentrations), supplemented only to the first batch subculture, and the basal growth medium, filled to a 50 mL working volume at pH 7.0–7.2. The vial was flushed with nitrogen gas for anaerobic conditions and incubated at 37 °C. Each subculture was replicated seven times; three replicates were selected randomly to determine RS removal, pH, and the molecular characteristics at the end of the experiment (Fig. S1). For each batch subculture, biogas production was measured daily. The percentages of CO2 and CH4 in the biogas were analyzed using a gas chromatograph; biogas production stopped after 14 days of RS degradation. Two milliliters of culture was collected at the end of each batch culturing for chemical and microbial analysis. The chemical analysis of VS was performed to determine the solid removal of RS (Eq. 1). Subsequently, the new batch was subcultured in a vial using 24 mL of the pellet fraction from the previous batch (0.5 g of RU), 0.5 g of RS at ISR of 1:1, and the fresh basal medium, totaling a working volume of 50 mL adjusted to pH 7.0–7.2.
To assess the stability of the AFC and the microbial community profile, the DNA information was analyzed. DNA samples of the initial buffalo rumen (RU0) inoculum and each subculture batch were evaluated for AB and AF community patterns using B-ARISA and F-ARISA, respectively [38]. The DNA fragments were examined by QIAxcel (QIAGEN, Germany). The fragment of B-ARISA and F-ARISA from the enriched and stabilized AFC was detected in separated bands by 2% agarose gel, and the dominant bands were cut and cloned into pGEM®-T Easy (Promega, USA). The PCR product was inserted into the plasmid, which was then transferred into competent cells of E. coli from kits. The selected clones were sent to a laboratory (1st BASE Pte. Ltd., Singapore) for Sanger sequencing. The BLAST program of NCBI was used to identify the microorganisms. The enriched and stabilized consortium was determined by the quantitative gene targets of AB, AF, and MT using real-time PCR. Moreover, the enriched and stabilized AFC culture was investigated by BMP assay, Biolog®EcoPlate, 16S rRNA and ITS1 sequencing, and shotgun metagenomic sequencing to determine the degradation performance and methane production performance, metabolic diversity, and classify the microbial communities and functions.
2.4 Quantitative determination of anaerobic fungi, anaerobic bacteria, and methanogens
The enriched and stabilized AFC was used to prepare the standard DNA via real-time PCR to quantify the AF, using the primer and PCR conditions as described in Kittelmann et al. [39]. Briefly, 2 mL of AFC sample was centrifuged at 5000 rpm, 22 °C, for 5 min to collect the pellet, which was purified using the Monarch Total RNA Miniprep Kit (New England Biolabs Inc., USA), following the manufacturer’s instructions. The DNA quality and quantity were measured using a NanoDrop®ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., USA). The PCR product of approximately 120 bp was cloned into pGEM®-T Easy and inserted into a plasmid, followed by transferal to competent cells of E. coli from kits. Ten selected clones were sent to the laboratory for sequencing of the nucleotide base pairs. Identification was done using the BLAST program of NCBI. One clone of AF was selected as the standard anaerobic fungal gene for quantitative real-time PCR (qPCR). The standards of AB and MT gene targets were prepared as those of the AF, using different specific primers (Table S2).
The enriched AFC microbial community was measured by qPCR and then compared to the original seed RU0 and the anaerobic lignocellulosic microbial consortium (ALMC), to which antibiotics were not added during screening and enrichment [15]. Briefly, 2 mL of RU0, ALMC, and AFC samples were extracted for DNA, which was then used to determine the quantities of AF, AB, and MT by qPCR with specific primers. The PCR conditions are described in Yu et al. [40] and Kittelmann et al. [39].
2.5 Potential for methane production from rice straw
The BMP was used to determine the potential of the enriched and stabilized AFC for the conversion of RS to methane. The BMP assay was set up in a 120-mL vial with a 50 mL working volume containing 1.0 g of RS, 1.0 g of the inoculum (ISR of 1:1, by weight), and the basal medium, with the pH adjusted to 7.2. The vial was flushed with N2 to obtain anaerobic conditions and incubated at 37 °C. The assay was done in triplicate. Each set had five vials for the determination of pH, VS, and VFAs at days 4, 7, 14, and 40; biogas and methane production were measured daily. The VS removal and rates were determined using Eqs. 1 and 2. The biogas composition in the gas phase and the VFA content in the liquid phase was measured as described by Panichnumsin et al. [37]. The methane production was analyzed using a BMP assay, and the cumulative CH4 potential data were fitted by a modified Gompertz equation [41], as shown in Eq. 3. The maximum methane production rate was determined and compared with other experimental results [7, 15, 42, 43]. All kinetic data of the enriched and stabilized AFC were checked and fitted by using Solver in the Microsoft Excel 365 software. Equation 3 is as follows:
where M is the volume of cumulative methane production (mL), Rm is the maximum specific methane potential rate (mL/d), P is the methane production potential (mL), λ is the period of the lag phase (d), t is the time of microbial degradation (d), and e is 2.718.
2.6 Metabolic diversity
The enriched and stabilized AFC was assayed for its abilities to use the different carbon sources on Biolog®EcoPlate (Biolog Inc., USA). The metabolic diversity of the enriched and stabilized AFC was determined using the same technique as mentioned by Thongbunrod and Chaiprasert [15].
2.7 Microbial community and functions
The enriched and stabilized AFC was analyzed for its microbial community and functions in converting lignocellulose to methane by 16S rRNA and ITS1 gene target amplicon and metagenomic shotgun sequencing analysis.
2.7.1 Taxonomy of the microbial community by metagenomics analysis
The DNA of the enriched and stabilized AFC sample was prepared in the barcode of 16S rRNA gene and ITS1 libraries for Illumina sequencing, as described by Li et al. [44]; this was done in the Anaerobic Microbiology and Biotechnology (AMB) group of the Swedish University of Agricultural Sciences (SLU). The PCR products were sequenced at the Swedish Institute for Infectious Disease Control in Solna (Stockholm, Sweden). The obtained raw DNA sequencing data of the 16S rRNA gene of RU0 can be found under the BioProject accession number PRJNA609587 [15]. The 16S rRNA gene and ITS1 region sequencing of the enriched and stabilized AFC for this study are stored under the BioProject accession number PRJNA666510. The derived pair-end reads of the 16S rRNA gene and ITS1 were processed and analyzed using the Mothur pipeline tutorial on Galaxy (https://usegalaxy.org/), as described by Schloss et al. [45]. The ITS1 region was analyzed to determine the anaerobic fungal community, using three databases: Silva V132 (20,246 eukaryote sequences of full-length sequences and taxonomy references) [46], UNITE V8.2 (12,664 fungal sequences) [47], and the anaerobic fungi ITS region database version 3.5 (705 anaerobic fungal sequences) [48]; files can be downloaded at https://anaerobicfungi.org/tools/.
2.7.2 Gene potential and functional role analysis using metagenomic shotgun sequencing
The raw read of the enriched and stabilized AFC was deposited under BioProject accession number PRJNA666510 and analyzed following the Galaxy training material for shotgun metagenomics data [49], assembled by the metaSPAdes tool [50]; the taxonomic information was generated by MetaPhlAn2, and the pathways and lignocellulolytic genes were classified using HUMAn2 (Galaxy Version 0.11.1.0) [51]. The websites uniport (https://www.uniprot.org) and Cazy (http://www.cazy.org) were used to find the enzymes and functions of the genes.
For AF, the eukaryotic metagenomics data were classified by using the BUSCO tool (http://busco.ezlab.org) and the discovered genes by the MetaEuk Easy Predict tools [52] and then was predicted as Neocallimastix californiae G1 by Augustus [53]. Genes were classified by BlastP in NCBI to find enzymes involved in biodegradation. The enzymes from dominant AB, AF, and MT involved in degradation, fermentation, and methanogenesis were used to describe the functions linked to lignocellulose degradation, using RS as a substrate for methane production.
2.8 Statistical analysis
One-way analysis of variance (ANOVA) was used to compare the FP and MCC removal for each seed inocula, biogas production, RS removal at each CBS cycle for AFC enrichment and stabilization, and RS removal and biogas production at various incubation times. Values are presented as mean. Tukey’s tests were used for multiple comparisons with a level of significance at P < 0.05.
3 Results
3.1 Screening and selection of anaerobic fungal consortia from various sources
The five different seed sources were sequentially screened for FP (amorphous cellulose), MCC (crystalline cellulose), and RS degradation, with the addition of antibiotics (penicillin and streptomycin) during incubation. The PM and MS inocula were unable to degrade FP and were dropped. CM, GM, and RU could degrade FP, MCC, and RS, as presented in Table 2. The AFC from RU showed the highest rate of biodegradation for FP, MCC, and RS; it degraded MCC at a lower rate (2.0 to 2.5 times) than FP. The FP, MCC, and RS degradation rates were 107, 54, and 27 mg/d for RU, 72, 30, and 22 mg/d for CM, and 85, 34, and 24 mg/d for GM, respectively. The rates of methane production from RS degradation by RU, CM, and GM were 5.97, 4.14, and 4.02 mL/d, with methane production of 49, 40, and 38 mL, respectively.
3.2 Enrichment and stabilization of the anaerobic fungal consortium for methane production
This study aimed to enrich and stabilize the dominant AFC for high lignocellulosic biomass degradation and methane production. Using RS enrichment by the CBS technique, the selected AFC from RU was applied to establish the stabilized microbial consortium. Removal of RS, production of biogas, and the microbial community were investigated for each cycle of CBS by the DNA fragment bands pattern of AF (F-ARISA) and AB (B-ARISA). This AFC inoculum was checked to confirm the existence of AF by primer ITS1 and qPCR. Accordingly, 19 cycles of CBS (E19) were performed to enrich and stabilize the dominant and key microorganisms in RU. This inoculum (E19) was used for the BMP assay to monitor the stability of the microbial community.
3.2.1 Methane production and RS degradation
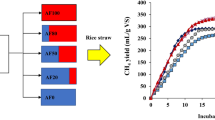
The CBS cycles (E1 − E19) were classified into three phases, namely acclimatization, intermediate, and stabilized phases, based on the profiles of RS removal, biogas production, and DNA fragment, as shown in Fig. 2. For the acclimatization phase (E1 − E6), RS removal was slightly increased from 48 to 55%, while biogas and methane production increased from 96 to 168 mL and from 48 to 89 mL, respectively (Fig. 2a). For the intermediate phase (E7 − E13), a higher increase in RS removal (60 to 83%), biogas production (179 to 294 mL), and methane production (91 to 145 mL) was found. The stabilized phase (E14 − E19) varied minimally (< 1%) in RS removal, biogas production, and methane production, with E19 showing values of 84%, 310 mL, and 152 mL, respectively.
3.2.2 DNA fragment band pattern
We found no effects of the addition of antibiotics on the AF based on the DNA fragment band pattern obtained by F-ARISA during enrichment and stabilization (Fig. S2a–c). In addition, the DNA fragment band pattern of AB, obtained by B-ARISA, was observed (Fig. S2d–f). Anaerobic bacteria were not detected after antibiotics addition. The DNA fragment band patterns of AF and AB were also investigated in all three CBS phases (acclimatization, intermediate, and stabilized phases). Anaerobic bacteria were undetected in E1 (with antibiotics) and E2 but were detected in E3 − E19. No changes in the DNA fragment band patterns of the anaerobic fungal community were observed (Fig. 2b). It was noticed that at least two groups of anaerobic lignocellulolytic fungi were successfully enriched and stabilized during the 19 cycles of CBS. For the AB community (Fig. 2c), the DNA fragment band pattern varied in the acclimatization (E1 − E6) and intermediate (E7 − E13) phases, but no shift was observed in the stabilization phase (E14 − E19). The dominant DNA fragment bands (306, 313, 330, 339, and 720 bp) from E19, by B-ARISA were cut, cloned, and then sequenced for identification. Analysis using BLAST from NCBI showed these fragments belonged to the Thermotogales, Bacilliles, Frankiales, Bacteroidales, and Clostridiales order, with similarities of 84–98%. For AF, two dominant DNA fragment bands (380 and 400 bp) were cloned, sequenced, and identified as uncultured fungi.
3.2.3 Number of microorganisms
The uncultured fungi (380 and 400 bp fragments) were further investigated using the ITS1 primer and qPCR techniques. Primers targeting the small subunit (SSU) and the ITS1 region of AF [39] were used. Eight clones of various fragments (110–119 bp) of this region were amplified and then cloned into E. coli competent cells for identification (Table S3) according to the NCBI database. Clones 2 and 4 were uncultured fungi with 90.00–92.50% similarity, clones 1 and 5 were uncultured Neocallimastigales with 93.81–96.43% similarity, clones 6–8 and clone 3 were Orpinomyces sp. with 92.79–100.00% similarity and Anaeromyces sp. with 99.12% similarity, respectively. This means that the primer was specific to AF and could identify four groups of AF in the stabilized AFC. Besides, this specific primer could be applied to amplify these four groups of AF in any samples. Therefore, one plasmid of eight clones can be a representative DNA standard for qPCR to determine the number of AF in a consortium. In this experiment, clone 5 was selected as a DNA standard to monitor and compare the number of AF in the enriched and stabilized AFC (E19) with RU0 and ALMC from a previous study.
The numbers of AB, AF, and MT from RU0, as determined by qPCR, were 3.6 × 109, 5.1 × 103, and 1.7 × 105 copies/mL, respectively, whereas the numbers of enriched and stabilized ALMC and E19 were 5.8 × 1010, 7.3 × 102, 2.9 × 106, and 3.8 × 108, 9.6 × 104, and 3.2 × 106 copies/mL, respectively (Table S4). The number of AF in ALMC (no antibiotics added) decreased compared to RU0. Anaerobic bacteria grew faster than AF and competed for substrates with fungi. The addition of antibiotics, however, eliminated AB during the screening stage and the first CBS cycle, allowing the enrichment of AF during the consecutive subculturing. Antibiotics did not affect MT, whose number in AFC (106 copies/mL) was close to that in ALMC (no antibiotics).
3.3 Metabolic diversity profiles by EcoPlate
The metabolic diversity of E19 was further investigated for the use of various carbon substrates. The active stage of E19 (14 days) was assayed for its metabolic diversity. Biolog® EcoPlate contained 31 different types of carbon sources with five groups of biochemical reagents: 10 carbohydrates, 9 carboxylic acids, 6 amino acids, 2 amines and amides, and 4 polymers. The results are shown in Table S5. Out of 31 different carbon sources, 28 carbon sources were used: carbohydrates (10/10), carboxylic acids (7/9), amino acids (6/6), polymers (3/4), and amines and amides (2/2). E19 showed high functional diversity at 90.3%, covering five groups of essential nutrients. Carbohydrate substrates such as β-methyl-D-glucoside, D-cellobiose, D-xylose, glucose-1-phosphate, i-erythritol, N-acetyl-D-glucosamine, α-D-lactose, pyruvic acid methyl ester, and D, L-α-glycerol phosphate were rapidly degraded within 24 h, whereas D-mannitol (mono hexose sugar alcohol) was used up within 48 h. E19 could degrade D-cellobiose (two molecules of hexose sugar), D-xylose (mono pentose sugar), and pyruvic acid methyl ester within 24 h, and L-phenylalanine and 4-hydroxy benzoic acid (derivative compounds of lignin) within 24–96 h. Amino acids such as L-arginine, L-asparagine, L-phenylalanine, L-serine, L-threonine, and glycyl-L glutamic acid were degraded within 24–96 h. Polymers such as tween 40, tween 80, and glycogen were degraded rapidly within 24 h. E19 also used carboxylic and acetic acids within 24–96 h.
3.4 Potential methane production
The potential of the enriched and stabilized consortium E19 for RS conversion to methane was investigated (Fig. 3). Values of pH and VFAs were determined to monitor AD stability. RS removal and methane production were also investigated for AD performance throughout the experiment. The pH values at 0, 4, 7, 14, and 40 days of AD were 7.21 ± 0.02, 6.93 ± 0.11, 7.13 ± 0.16, 7.24 ± 0.11, and 7.41 ± 0.14, respectively (Fig. 3a). Acetic, propionic, and butyric acids were the main intermediate VFA products during AD of RS. The concentration of acetic acid peaked at 805 mg/L (4 d) and gradually decreased to 750 mg/L at 7 days, 350 mg/L at 14 days, and 20 mg/L at 40 days. Propionic acid concentrations of 300 and 20 mg/L were found at 4–14 days and 40 days, respectively. Concentrations of butyric acid stood at 400 mg/L at 4–7 days, 300 mg/L at 14 days, and 20 mg/L at 40 days. At the end of the incubation period (40 days), less accumulation of propionic and butyric acids was found in the system. In addition, RS removal of 35, 52, 84, and 86% and biogas production of 290, 426, 655, and 690 mL were found at 4, 7, 14, and 40 days, respectively (Fig. 3b).
The findings for VFAs were related to RS removal and biogas production. At 14 days of incubation, RS removal gradually increased from 35 to 84%, as was reflected by the decrease in accumulated VFAs, the intermediate compounds of biogas production. In the same period, methane production gradually increased from 124 to 310 mL with the conversion of acetic acid (from 805 to 350 mg/L) and butyric acid (from 400 to 290 mg/L) to methane at a neutral pH. Propionic acid was constant at 300 mg/L for 4 to 14 days, decreasing to 20 mg/L by the end of the experiment (40 days). RS was slightly degraded from 84 to 86% with decreasing acetic acid (from 350 to 20 mg/L), propionic acid (from 300 to 23 mg/L), and butyric acid (from 290 to 25 mg/L) values, owing to the conversion of VFAs to smaller molecules for methane production (336 mL at 40 days). During AD, no impact of VFAs on methane production was found. It was noticed that E19 displayed a good balance between non-MT (acid production) and MT (acid consumption), resulting in less VFA accumulation and maintenance of a neutral pH.
The kinetic parameters of methane production from RS by E19 and RU0 at 14 and 40 days of AD are shown in Table 3. The modified Gompertz model described by Eko and Chaiprasert [54] was used to estimate the maximum CH4 production, the maximum rate of CH4 production, and the duration of the lag phase. A close relationship between the experimental data (the BMP assay) and the fitted curve of the Gompertz equation was indicated by R2 > 0.99. The results indicated that E19 was highly efficient in degrading RS for CH4 production. The methane rate, yield, and selectivity of E19 and RU0 were compared. A lag phase was not found during RS degradation by E19 and RU0. Methane production by E19 plateaued at 14 days, with just a slight increase afterward. RS removal of 84%, CH4 production of 310 mL, a maximum CH4 production rate of 32 mL/d, CH4 yield of 310 mL/g VSadded, and CH4 selectivity of 370 mL/g VSremoved were obtained from E19 at 14 days. When compared to RU0, RS removal by E19 increased 1.82-fold, cumulative CH4 production, CH4 yield increased 2.25-fold, and the rate of CH4 production was 2.10-fold faster. At the end of the experiment (40 days), RS removal and methane production of E19 were 84% and 336 mL/g VSadded, whereas RU0 showed values of 50% and 147 mL/g VSadded, respectively. Also, E19 could accelerate the degradation of lignocellulosic biomass and the production of methane in a shorter period than RU0 during AD.
3.5 Microbial community and functions of anaerobic fungal consortium
The community and functions of AB, MT, and AF in E19 were analyzed by 16S rRNA/ITS1 gene and metagenomic sequencing shotgun. The taxonomic profiles for phylum, genus, and species levels were classified, including gene functions and pathways in converting lignocellulosic biomass to methane.
3.5.1 Taxonomic identification
The E19, RU0, and ALMC microbial consortia were investigated by the 16S rRNA gene sequencing technique to study the structure and communities of AB and archaea (Fig. 4a). The results showed bacterial populations of 99.70% in E19, 99.98% in ALMC, and 99.90% in RU0, with archaeal populations of 0.3%, 0.2%, and 0.1%, respectively. RU0 comprised Proteobacteria (46%), Bacteroides (31%), and Firmicutes (15%), ALMC comprised Synergistetes (30%), Bacteroides (22%), and Firmicutes (19%), and E19, comprised Synergistetes (32%), Bacteroides (20%), Firmicutes (12%), Spirochaetes (8%), and Proteobacteria (5%). Proteobacteria were absent in ALMC and E19, whereas Synergistetes was enriched by up to 30–32% relative to RU0, which had less than 0.1%.
The archaeal communities of RU0, ALMC, and E19 are shown in Fig. 4b. Three groups of archaea were found in RU0, namely Methanobrevibacterium (52%), Methanosphaecra (4%), and unclassified Euryarchaeota (44%). The E19 consisted of the phyla Euryarchaeota (62%), Crenarchaeotes (13%), and unclassified archaea (25%). The archaeal diversity of E19 was higher than RU0. Considering the phylum Euryarchaeota, these MT were affiliated with Methanosaeta (13%), Methanomethylovorans (6%), Methanofollis (6%), Methanoculleus (6%), Methanobrevibacter (6%), Methanobacterium (6%), and the unclassified genus (19%). The archaea in ALMC were classified into Methanosaeta (23.1%), Methanofollis (23.1%), Methanospirillum (15.4%), Methanobacteriaum (7.7%), and Methanoculleus (7.7%). Most of the MT were hydrogenotrophic methanogens (HMT). Only the acetoclastic methanogens (AMT) of the genus Methanosaeta could produce methane from acetate. The other five genera, mostly hydrogenotrophic methanogenesis, could use H2/CO2 for methane production.
The structure and community of AF in the E19 microbial consortium were investigated using the high-throughput ITS1 region amplicon technique. The pair-end reads of 82,478 × 301 bp were processed by the Mothur pipeline resulting in 78,560 reads. A threshold of 0.03 was applied with an average length of 456 bp. To date, only a few databases for the classification of AF can be accessed. E19 showed an AF number of 9.60 × 104 copies/mL. The ITS1 sequence was classified using three different databases, the Silva V132, Unite V8.2, and AF V3.5 databases (Fig. 4c). The Silva V132 database showed low taxonomic clusters, accounting for 4% of eukaryotes and 96% of unknown phyla. The Unite V8.2 database revealed 34.557% unknown phyla, 59.359% unclassified phyla, 4.004% Ascomycota, 2.049% Basidiomycota, 0.003% Mucoromycota, 0.024% Neocallimastigomycota, and 0.004% Rozellomycota. However, when using the AF V3.5 database, we found 100% classified AF, providing more details for the taxonomic classification. The phylum with the highest abundance was Neocallimastigomycota (93%). The genus Feramyces (7%) was also classified in this database. The classification of AF in ALMC showed similar results, with Neocallimastigomycota (94%) as the most abundant phylum and Feramyces (6%) as the most abundant genus.
3.5.2 Anaerobic microbial community and functional roles
We investigated the functions of E19 with the aim to boost RS degradation. The metagenomic shotgun sequencing analysis demonstrated the functional roles of the anaerobic microbial consortium using the Galaxy pipeline. A total of 43,264,498 quality reads of 150 bp (6.48 GB) with a 49% GC content were assembled by the metaSPAdes assembler for metagenomic datasets (Galaxy Version 3.9.0) into 914,083 scaffolds (565 MB), ranging in size from 618 to 1,243,548 bp (N50 = 899). This assembly scaffold was used to classify the abundance of AB, AF, and archaea by MetaPhlAn2, and the gene family and pathway abundance by HUMaN2.
The taxonomic classification of AB and archaea via MetaPhlAn2 is shown as a pie chart (Fig. 5). Anaerobic fungi were not found, most likely because of the low DNA concentrations or the small AF database in this tool; therefore, BUSCOs were used to classify the gene abundance of AF in E19. Anaerobic bacteria and archaea in E19 accounted for 62 and 38% of the taxonomic abundance, respectively. The abundances of hydrolytic and fermentative microorganisms such as Bacteroidetes, Synergistetes, Spirochaetes, Firmicutes, and Proteobacteria were 37, 7.5, 5.8, 4.9, and 4.7%, respectively. The dominant phylum Bacteroidetes was classified into Proteiniphilum acetatigenes (12%), Bacteroides sp. (20%), and Sphingobacterium sp. (3%). The second dominant taxon, Pyramidobacter piscolens (7%) was classified into Synergistetes, and the third one, Sphaerochaeta sp. (5%) was classified into Spirochaetes. Firmicutes and Proteobacteria species accounted for less than 1% of the total bacteria. Four species accounted for approximately 34% of MT; two HMT species: Methanoculleus bourgensis (14%) and Methanoculleus marisnigri (13%), one AMT: Methanosaeta harundinacea (5%), and HMT and AMT species: Methanosarcina mazei (2%).
3.5.3 Gene family abundance
Two species of AB (P. piscolens and P. acetatigenes) and three species of MT (M. bourgensis, M. harundinacea, and M. marisnigri) were analyzed for their functions by HUMaN2. In total, 5776 genes were found, 1835 in P. piscolens, 1104 in P. acetatigenes, 1478 in M. bourgensis, 1466 in M. harundinacea, and 741 in M. marisnigri. The BUSCO application was used to classify the gene abundance of fungi in E19. This application is used for quality assessments of genetic and phylogenomic predictions to classify genes from metagenomic data. The assembly of E19 was identified as complete, duplicated, fragmented, and missing genes, enabling a like-for-like quality comparison of different data sets by the current version of BUSCO (V. 4.1.2) on the Galaxy website, with lineage data sets of 549 fungal species, containing proteins and data of 758 expected genes in BUSCOs. Fungal genes accounted for 102/758 of the complete genes (13.5%). The complete genes could be separated into 53/102 and 49/102, respectively, with complete and single-copy genes (7%) and complete and duplicate genes (6.5%). In addition, 43/758 were fragment genes (5.7%), and 613/758 were missing genes (80.8%), as most of the available datasets were for aerobic fungi. However, based on the complete genes in E19, we could predict the functions of AF in E19 using InterPro (https://www.ebi.ac.uk/interpro/), by searching the gene functions and adding possible pathways. From the fungal database, gene GH31, which is involved in cell wall degradation, was found in E19.
In the assembly of E19, we discovered the genes of eukaryotic metagenomics contigs using the MetaEuk Easy Predict tools, and found 37 sequences, which were predicted, using Augustus, to belong to AF such as Neocallimastix californiae G1. Sixteen protein sequences, including the glycoside hydrolase family 11 protein (GH11), endo-1,4-beta-xylanase, and cellulosome enzyme (dockerin type I), were predicted to belong to N. californiae G1 and be involved in biomass degradation, based on the NCBI BlastP tool. A hypothetical protein close to GH5, GH11, and endo-1,4-beta-xylanase was found in seven sequences. A leucine-rich repeat protein close to the cellulosome enzyme (dockerin type I) was found in three sequences. Most likely, the leucine-rich repeat protein is part of the dockerin structure associated with the cellulosome enzyme.
3.5.4 Pathway abundance
Ninety-one pathways involved in biosynthesis (75), degradation (5), fermentation (10), and anaerobic respiration (1) in hydrogenotrophic methanogenesis were found in E19. Most biosynthesis pathways in E19 were involved in nucleoside and nucleotide biosynthesis. This class consisted of the synthetic pathways of the nucleoside triphosphates (building blocks of RNA and DNA), such as pyrimidine deoxyribonucleotide phosphorylation (PWY-7197), adenosine ribonucleotide de novo biosynthesis (PWY-7219), adenosine deoxyribonucleotide de novo biosynthesis II (PWY-7220), guanosine ribonucleotide de novo biosynthesis (PWY-7221), and guanosine deoxyribonucleotide de novo biosynthesis II (PWY-7222). The biosynthesis compound of methane production, factor 420 biosynthesis (PWY-5198), a redox-active compound that plays a role in methanogenesis, was found in E19. The multi-step biosynthesis pathway involves enzyme-catalyzed processes and elements such as precursor compounds, catalytic enzymes (which may require coenzymes such as NADH and NADPH), and chemical energy (e.g., ATP). Besides, five pathways of degradation were generated, namely starch degradation V (PWY-6737), D-galactose degradation V (PWY66-422), L-histidine degradation I (HISDEG-PWY), purine nucleobase degradation I (anaerobic: P164-PWY), and adenosine nucleotide degradation IV (PWY-5532). Ten fermentation pathways were found, namely, pathways of homolactic fermentation (ANAEROFRUCAT-PWY), pyruvic acid fermentation to acetic acid and lactic acid II (PWY-5100), pyruvate fermentation to isobutanol (PWY-7111), incomplete reductive TCA cycle (P42-PWY), urea cycling (PWY-4984), and five pathways of glucose fermentation to pyruvate, namely glycolysis I (GLYCOLYSIS), glycolysis II (PWY-5484), glycolysis III (ANAGLYCOLYSIS-PWY), glycolysis IV (PWY-1042), and glycolysis VI (PWY66-400). The respiration pathway METHANOGENESIS-PWY, which is involved in methane production from H2/CO2, was found in HMT.
3.6 Gene functions linked to lignocellulose degradation for methane production
Four main steps of AD for methane production: hydrolysis, acidogenesis, acetogenesis, and methanogenesis, were identified. The dominant enzymes from AF, AB, and MT from the E19 consortium were used to construct the possible pathway of methane production from lignocellulosic biomass, with RS as a substrate.
Hydrolysis is the first step of the AD of RS for methane production. The gene families involved in the degradation of cellulose, hemicellulose, and lignin were characterized. The genes associated with polysaccharide degradation, fermentation, and production of methane are shown in Tables S6 and S7. The gene function of cellulase, hemicellulase, and pectinase in the degradation of RS cell walls were classified into families of glycoside hydrolases (GHs) of CAZy (carbohydrate-active enzymes) database from the reference clusters gene family as shown in Table S8. The glycoside hydrolase (GH) enzymes were divided according to their substrates, EC numbers, abbreviations, and corresponding CAZyme families. Anaerobic bacteria were found in families such as GH2, 3, 5, 15, 28, 29, 43, and 105. Anaerobic fungi were found in three families, GH5, GH11, and GH31. Archaea were found in GH57. The function of these GHs could degrade cellulose, hemicellulose, and pectin (Table 4).
For cellulose degradation, linear cellulose chains of the repeating unit of β-1,4-linked D-glucose in the form of crystalline and amorphous regions were degraded by β-1,4-glucosidase (BGL; EC 3.2.1.21) and β-1,4-endoglucanase (EG; EC 3.2.1.4), from GH3 and GH5, and cellulose 1,4-β-cellobiosidase (nonreducing end) (CBH1; EC 3.2.1.91) from GH5 of P. acetatigenes and AF. For hemicellulose degradation, four structural types of hemicellulose, xylan, galactomannan, xyloglucan, and arabinoxylan, with different main monosaccharide units in their backbones were degraded by GH2, 3, 5, 28, 29, and 43. Xylan was cleaved by 1,4-β-xylosidase (BXL; EC 3.2.1.37). Endo-1,4-β-xylanase (XLN; EC 3.2.1.8) hydrolyzed xylobiose into its monomeric units. Mannose was cleaved by endo-β-1,4-mannosidase (MAN; EC 3.2.1.78) into mannooligosaccharides, and the terminal ends of mannan were cleaved by β-1,4-mannosidase (MND; EC 3.2.1.25), releasing D-mannose. For pectin degradation, enzymes involved in pectin degradation from E19 were GH2, 3, 5, 18, 28, 43, 31, 57, and 105. Xylogalacturonan hydrolase (XGH; EC 3.2.1-) of the glycoside hydrolase family 28 (GH28) could hydrolyze the glycosidic bond of two β-xylose-substituted galacturonic acid residues in pectin. The homogalacturonan backbone of pectin was cleaved by endopolygalacturonase (PGA; EC 3.2.1.15) and exo-polygalacturonase (PGX; EC 3.2.1.67), which act within and at the end of the terminal to release D-galacturonic acid. The backbones of rhamnogalacturonan and xylogalacturonan were cleaved by rhamnogalacturonase (RGH; EC 3.2.1.171) and xylogalacturonan hydrolase (XGH; EC 3.2.1.), respectively. Debranching enzymes cleaved monomers like D-galactose, D-xylose, L-arabinose, L-fucose, D-glucuronic acid, acetate, ferulic acid, and p-coumaric acid and short oligomers such as α-1,2-L-arabinofuranosidase (ARA; EC 3.2.1.-), α-galactosidase (AGL; EC 3.2.1.22), and α-L-arabinofuranosidase (ABF; EC 3.2.1.55) belonging to GH31 from fungi and GH57 from MT.
During acidogenesis, P. acetatigenes was involved in glucose fermentation to pyruvate via glycolysis I, II, III, IV, and VI pathways. The α-D-phosphohexomutase superfamily found in fungi was involved in the conversion of D-glucose 1-phosphate to D-glucose 6-phosphate and in the breakdown and synthesis of glucose. In addition, this enzyme could use glucose and mannose from hemicellulose as substrates. The steps of acetogenesis and methane formation are tightly connected. Acetogenesis supplies substrates for (a) acetoclastic/autotrophic methanogens (acetate), (b) hydrogenotrophic methanogens (H2/CO2), and (c) methylotrophic methanogens (methyl group) [55]. Pyruvate from glycolysis is converted to H2, CO2, acetate, and lactate via the PWY-5100 pathway. The Thermotoga sp., which accounted for less than 1% of the total bacteria in E19, was involved in the fermentation of pyruvic acid to acetic acid and lactic acid. The incomplete reductive TCA cycle P42-PWY was also found. Another pathway of acetogenesis found in E19 was PWY-7111, which is involved in pyruvate fermentation to CO2 and then to isobutanol by P. acetatigenes. The syntrophic P. piscolens was not found in the acetogenesis pathway but was involved in L-histidine degradation in L-histidine degradation I (HISDEG-PWY), producing ammonium, which was further degraded for urea production in the urea cycle (PWY-4984).
The methanogenesis pathway METHANOGENESIS-PWY, a hydrogenotrophic (H2/CO2) pathway, was found in M. marisnigri, M. bourgensis, and M. harundinacea. Based on the gene family abundance and pathways, consortium E19 contained specific species of AF, AB, and MT that could hydrolyze RS, ferment soluble monomers to VFAs, generate H2/CO2, and finally, produce methane through hydrogenotrophic methanogenesis pathways.
4 Discussion
The enriched and stabilized AFC was discussed in terms of anaerobic microbial existence, stability of the AD system, enhancement of methane production, and microbial community related to RS degradation and biogas production.
4.1 Existence of anaerobic microorganisms
Rumen microbial communities of AF and AB are well known for lignocellulosic biomass degradation, with a wide range of enzymes and microorganisms involved in the breakdown of lignocellulosic materials [56]. Rumen microorganisms contribute to the digestion of high-fiber feed, significantly promoting biogas yield with lignocellulose as a substrate [44, 57]. In this study, we focused on the enrichment and stabilization of an AF consortium to produce methane from lignocellulosic biomass. Removing AB in the RU inoculum using antibiotics such as penicillin and streptomycin resulted in high FP, MCC, and RS degradation over 7 days. During the screening of the AFC, no effect of antibiotics on AF was found, whereas the AB population was highly affected at the end of the incubation period (Fig. S2). The RU inoculum was used to digest RS by adding antibiotics that could inhibit the growth of AB at the first cycle of the CBS (E1). Thus, the subsequent consecutive batch subcultures with RS (E2 − E19) were no longer supplemented with antibiotics through enrichment and stabilization. One cycle had an incubation period of 14 days and no biogas production occurred. Anaerobic bacteria and AF were detected at the end of the incubation period of 14 days, via analysis by B-ARISA and F-ARISA as shown in Fig. 2b–c. Anaerobic bacteria were not detected in E1 (with antibiotics) and E2 (transition batch) but in E3 − E19 (no antibiotics); the patterns of fragments after several CBS were changed, with stable fragments at E13 − 19 (Fig. 2c). Antibiotics such as penicillin and streptomycin are widely used to remove AB from anaerobic fungal cultures [12, 34, 58,59,60,61,62], although some resistant bacteria may persist [63]. We, therefore, assume that the buffalo rumen sample in this study contained antibiotic-resistant bacteria that can be cultured in the long term with AF and MT for methane production under anaerobic conditions. During E3 − E19 cycles, AB and AF were found in the system, and methane production was high (Fig. 2a). The increase in the anaerobic bacterial population of E3 − E19 did not affect the growth of AF in the culture; thus, methane production in the E13 − E19 CBS cycles remained stable. The fragments of AF and AB in Figs. 2b and c refer only to the quality and not to the quantity of the microbial community. The quantities of AF, AB, and MT in the enriched and stabilized E19 are shown in Table S4.
4.2 Stability of AD system
The efficiency of methane production is strongly dependent on a balanced microbial consortium, whose high-resolution characterization, functional potential, and interactions are pivotal for process optimization [64]. Optimal environmental conditions (temperature, pH, buffering capacity, and fatty acid concentrations) inside the digester are important to promote microbial interactions and boost the conversion of lignocellulosic biomass to methane. Most reactors operate at either mesophilic or thermophilic temperatures, with optimal temperatures at 35 and 55 °C, respectively. The optimum temperature for methanogenesis may not necessarily be the optimum one for other processes in AD, such as hydrolysis or acidification [65]. This experiment with E19 was set at 37 °C, which should not significantly affect microorganisms in the system, being in the mesophilic temperature range. The system pH of 6.9–7.4 should promote the synergy between the anaerobic microbial community, including the hydrolytic/fermentative bacteria (non-MT) and MT, in the consortium. The balance between non-MT and MT in the E19 consortium resulted in low accumulation and a stable neutral pH in the AD system. No acid inhibition was found in this study. The optimum pH for MT is in the range of pH 6.7–7.5. The pH lower than 6.6 could significantly inhibit MT, and pH below 6.0 is toxic to MT [66]. The optimal pH for methanogenesis is a neutral pH, whereas the optimal values for hydrolysis and acidogenesis ranges from 5.5–6.5 [67]. Volatile fatty acids (acetic, propionic, and butyric acids) are key intermediates in AD and can inhibit methanogenesis at high concentrations. The production of fatty acids in anaerobic processes will change the pH, thereby inhibiting methanogenesis [65]. The inhibition of microbial activity caused by VFAs has been reported at concentrations above 2 g/L for cellulolytic activity and concentrations above 4 g/L for methanogenesis in AD [68]. In previous studies, the accumulation of VFAs resulted in a decrease in pH values during fungal growth in an anaerobic reactor [69]. The optimal pH for the growth of AF ranges from 6.0 to 7.0. Also, a slow growth rate of AF can lead to small populations [70].
As expected, the AF stayed alive up to 40 days when inoculated in RS at pH 6.9–7.4 (Fig. 3). During AD (40 d), the pH in E19 was in the neutral range. In this pH range, the concentration of VFAs was less than 2 g/L, and no effect of VFAs on methanogenesis was found. Consortium E19 showed a good balance between acid production by non-MT and acid consumption by MT, resulting in less VFA accumulation and a stable pH.
4.3 Enhancing methane production
The E19 consortium showed high RS degradation and methane production, with 84%VS and 310 mL/g VSadded within 14 days, respectively. Compared to previous studies on RS degradation and methane production at 37 °C in a batch system (Table 5), E19 showed higher RS degradation and methane yield in a shorter incubation time of 14 days. Mesophilic inocula from municipal wastewater treatment systems and sewage sludge showed a methane yield of 200–281 mL/g VSadded and RS removal of 62%VS within 30–36 days [7, 15, 42, 43]. At 40 days, E19 had a methane yield of 336 mL/g VSadded compared to 310 mL/g VSadded for ALMC. The methane yield of E19 at day 14 was similar to that of ALMC at day 40. E19 had slightly higher RS degradation and methane yield than ALMC at 14 days.
Efforts have been made to accelerate the degradation process using thermal and biological pretreatments, including pre-digestion with hyperthermophilic AB (Caldicellulosiruptor bescii). The pre-digestion of RS with C. bescii at 75 °C in an 1875 L CSTR reactor under anaerobic conditions resulted in 75–85% RS removal and 75% methane [71]. Pretreating RS with physical, chemical, and biological methods in a mesophilic AD bioreactor at 37 °C at ISR of 2–7 for 30–60 days provided methane yield in a range from 258 to 346 mL/g VS [9, 21, 72]. These results showed that AFC performed better than pretreatment, predigestion, and bioaugmentation for methane production and biomass removal in terms of the shorter time for AD, a lower amount of inoculum, and no extra work or equipment needed to prepare feedstock and bioaugmented cells prior to feeding to the AD system. Other factors to be considered include the size and type of reactors as well as the chemical composition and properties of the biomass [1], which may require different conversion technologies and specific microbial consortia [73]. In addition, O’Shea et al. [74] reported on a strategy to maximize the benefits of biogas by balancing the benefits and drawbacks of using a compromised programming approach. This methodology can implement and integrate an AD plant by selecting the optimal blend of by-products to use. It can help decision-makers to design AD projects for decarbonizing accounts with different criteria.
The numbers of AF, AB, and MT in the enriched and stabilized E19, as measured by qPCR, were 9.6 × 104, 3.8 × 108, and 3.2 × 106 copies/mL, respectively. The number of AF in E19 increased by 1–2 log cycle times compared to RU0 and ALMC, whereas that of AB in E19 decreased by 1–2 log cycle times compared to RU0 and ALMC. Consortium E19 could enrich MT and had a higher number, 1 log cycle more, than RU0 (Table S4). The ratio of AF to AB was 1:104 in E19 and 1:108 in ALMC. Also, E19 showed higher RS degradation and methane production than ALMC. The AD of RS by E19 was highly stable, with less VFA accumulation and a stable neutral pH, promoting RS degradation and methane production. In a similar study, Nagler et al. [57] investigated the AD of corn straw (1.6%) using ox rumen fluid at 37 °C within 32 days. They reported an AF to AB ratio of 1:105, high VFAs accumulation, pH of 5.74, MT inhibition, and low methane production from day 7 until the end of the experiment. Most likely, the higher AF to AB ratio helped promote lignocellulosic biomass degradation and work in balance with MT. E19 showed an AF:AB:MT ratio of 1:104:102, mutually converting RS to methane. The ratio of anaerobic microorganisms and the existing key dominant species is important to accelerate lignocelluloses degradation and enhance methane production.
4.4 Microbial community of E19 for rice straw degradation and methane production
The dominant AB in ruminants belongs to the phyla Bacteroidetes, Fibrobacteres, Firmicutes, Proteobacteria, Spirochaetes, Synergistetes, Spirochaetes, and Actinobacteria [44]. In our study, we found three main anaerobic bacterial phyla in the buffalo rumen fluid (RU0), namely Proteobacteria (46%), Bacteroides (31%), and Firmicutes (15%). Following the enrichment (E19), the most dominant was Synergistetes (32%), followed by Bacteroidetes (20%) and Firmicutes (12%). The decrease in Bacteroidetes and Firmicutes as a result of the addition of antibiotics, which allowed syntrophic prokaryotes such as Synergistetes and Spirochaetes to grow. The microbial diversity can vary depending on the conditions of growth and substrates [75].
According to previous studies, Synergistetes like hydrogenotrophic methanogens can degrade acetate by syntrophic acetate oxidation and hydrogen consumption [76]. Spirochaetes are also involved in syntrophic acetate oxidation [44]. Higher abundance of syntrophic acetate oxidizing bacteria (Synergistete and Spirochaetes) and HMT (Methanomethylovorans, Methanofollis, Methanoculleus, Methanobrevibacter, and Methanobacterium) than AMT were found in E19, based on metagenomic shotgun sequencing of E19.
P. piscolens, a syntrophic member of the phylum Synergistetes, is involved in protein degradation and amino acid fermentation to provide H2/CO2 and VFAs for MT. As an anaerobic acid-producing bacterium, it produces acetic, propionic, isobutyric, isovaleric, succinic, and phenylacetic acids as end products of metabolism. This species can grow and oxidize various amino acids in syntropy with HMT [77]. Here, we found the gene function and genes of the peptidase family and an enzyme for lignin degradation from P. piscolens. Thus, the role of P. piscolens in biogas reactors is to provide VFAs and H2 to HMT.
P. acetatigenes, belonging to the phylum Bacteroides, can produce proteolytic enzymes for the production of acetate, propionate, and CO2 [78]. In our study, we found genes encoding cellulolytic enzymes such as glycosidase hydrolysis family proteins (GH2, 3, 5, 15, 26, 28, 29, 43, 92, and 105), involved in peroxiredoxin (Prx) degradation [79], and genes encoding enzymes for degrading starch and glycogen, such as α-amylase [80] in E19. Based on the results, P. acetatigenes can degrade the cellulose of plant cell walls. Genes encoding enzymes and pathways involved in acidogenesis and acetogenesis were found in P. acetatigenes, suggesting that it plays a crucial role in the degradation of complex substrates (cellulose, hemicellulose, lignin, and pectin) to smaller molecules, which are then fermented to acetate, H2, and CO2 and used as substrates. Both P. acetatigenes and P. piscolens are rod-shaped Gram-negative bacteria containing genes encoding enzymes for lignin degradation. According to Wilhelm et al. [81], who studied the diversity of cellulase-producing microbes in coniferous forest soil, fungi showed a higher cellulase activity than other microbes, whereas Gram-negative bacteria were more involved in the decomposition of lignin.
Rumen AF of the phylum Neocallimastigomycota are potential lignocellulosic biomass degraders and can potentially be used for biogas production [11]. The phylum Neocallimastigomycota contains one order (Neocallimastigales) and one family (Neocallimastigaceae) with 17 genera of strictly AF, which have recently been described and characterized, namely 13 monocentric rhizoidal genera: Agriosomyces, Aklioshbomyces, Buwchfawromyces, Capellomyces, Feramyces, Ghazallomyces, Joblinomyces, Khoyollomyces, Liebetanzomyces, Neocallimastix, Ontomyces, Piromyces, and Tahromyces, 2 polycentric rhizoidal genera: Orpinomyces and Anaeromyces, and 2 bulbous genera: Caecomyces and Cyllamyces [58,59,60,61,62, 82,83,84]. The AF present in the rumen of herbivores produces hydrolytic enzymes involved in plant fiber digestion [85]. Feramyces, found in E19, can grow on sugar and lignocellulosic biomass [83]. Peng et al. [86] studied the genomic and functional characteristics of AF in goat rumen by metagenomic shotgun sequencing and found genes encoding glycoside hydrolase for cellulose degradation, GH5, GH6, GH9, GH45, and GH48, and for hemicellulose degradation, GH5, GH10, GH11, GH26, and GH43. The assembly of E19 showed genes of eukaryotic metagenomics by MetaEuk Easy Predict tools, with 37 sequences and GH5, 11, and 31. The gene number in our study was lower compared to Peng et al. [86], most likely because of the different ruminants involved and the low ratio of AF to AB in the inoculum.
Methanogenic species in E19 were mostly classified into HMT, such as Methanomethylovorans, Methanofollis, Methanoculleus, Methanobrevibacter, and Methanobacterium. Hydrogenolysis genes and pathways were found in E19. The AMT species, M. harundinacea, was found to contain genes involved in formate degradation to H2 and CO2, based on metagenomic shotgun sequencing, as shown in Table S6. Thus, M. harundinacea in both HMT and AMT, owing to syntrophic acetate oxidation (Synergistetes and Spirochaetes) and an abundance of HMT, methane production from RS, using the enriched and stabilized AFC (E19) as inoculum, was mainly from H2.
4.5 Limitations of the study
Although the ITS gene target analysis could not identify the genera and species, Sanger sequencing of SSU and ITS1 (110 bp) showed four groups of AF (Table S3). The AF genera from the ITS1 gene target and SSU/ITS1 were different; the SSU/ITS1 region presented more species than the ITS1 gene target. Hanafy et al. [87] endeavored to find a barcode marker for anaerobic gut fungi in domain D1/D2 of the large ribosomal subunit. Full-length ITS and the large ribosomal subunit (primer ITS1-NL4) are highly recommended for use in identifying AF [84]. Metagenomic shotgun sequencing was used for functional analysis. This method did not show which specific lignocellulolytic genes are expressed during RS degradation. These key genes can be identified by transcriptomic analysis. Anaerobic fungi in AFC contained fewer lignocellulolytic enzyme genes than AB in ALMC. This higher number of lignocellulolytic enzyme genes in ALMC did not result in higher RS degradation. Other researchers who studied transcriptomic analysis have presented a higher number of cellulase and hemicellulase gene expressions in AF than in AB [86, 88].
5 Conclusions and outlook
Successful CBS with initial supplementation of antibiotics, the stabilized and effective AFC was established and could be used directly as a starter seed in a single AD reactor. The anaerobic lignocellulolytic microbial consortium E19 consisted of AF coexisting with AB and MT under mesophilic and anaerobic conditions. The enrichment and stabilization of AFC boosted lignocellulose degradation and methane production was compared to rumen fluid. The AFC illustrated that the stabilized consortium of anaerobic cellulolytic bacteria, syntrophic acetate-oxidizing bacteria, and HMT worked together in harmony to degrade RS. This AD system could keep pH neutral and lower VFA accumulation. The higher performance of AFC in converting RS to methane, compared with the conventional pretreatment approach, makes it a promising inoculum for lignocellulosic biomass degradation.
The monitoring of AFC in degrading various kinds of lignocellulosic biomass, scaling up the AD system, and shifting the feed pattern of a single AD reactor from batch to semi-continuous/continuous in low/high solid content for methane production can provide pilot data for demonstrating the efficacy of stabilized AFC. Such informative data could be used to conduct a feasibility study of techno-economic analysis with acceptable accuracy. The characterization of isolated AF and their functions, identification of key lignocellulolytic genes by transcriptomic analysis, and qPCR for key genes associated with lignocellulosic biomass conversion to methane are important for process monitoring and enhancement.
Abbreviations
- 16S rRNA:
-
16S ribosomal RNA is the RNA component of the 30S subunit of a prokaryotic ribosome
- AB:
-
Anaerobic bacteria
- AD:
-
Anaerobic digestion
- AF:
-
Anaerobic fungi
- AFC:
-
Efficacy of anaerobic fungal consortia
- ALMC:
-
Anaerobic lignocellulosic microbial consortium
- AMT:
-
Acetoclastic methanogens
- ANOVA:
-
One-way analysis of variance
- AOAC:
-
Association of Official Agricultural Chemists
- APHA:
-
American Public Health Association
- B-ARISA:
-
Automated method of ribosomal intergenic spacer analysis for bacteria
- BMP:
-
Biochemical methane potential
- BUSCO:
-
Benchmarking Universal Single-Copy Orthologs
- CAZy:
-
Carbohydrate-active enzymes
- CBS:
-
Consecutive batch subculturing
- CM:
-
Cow manure
- COD:
-
Chemical oxygen demand
- CSTR:
-
Continuously stirred tank reactor
- E1 − E19:
-
Serial number of subculture during enrichment and stabilization
- F-ARISA:
-
Automated method of ribosomal intergenic spacer analysis for fungi
- FP:
-
Filter paper
- GC:
-
Gas chromatography
- GH:
-
Glycoside hydrolase
- GM:
-
Goat manure
- HMT:
-
Hydrogenotrophic methanogens
- ISR:
-
The inoculum (I) to substrate (S) ratio
- ITS:
-
Internal transcribed spacer
- MCC:
-
Microcrystalline cellulose
- MT:
-
Methanigens
- MS:
-
Microbial sludge from anaerobic wastewater treatment system of a palm oil mill factory
- PCR:
-
Polymerase chain reaction
- PM:
-
Pig manure
- qPCR:
-
Quantitative real-time PCR
- RS:
-
Rice straw
- RU:
-
Rumen fluid
- SSU:
-
Small subunit
- TS:
-
Total solids, defined as mass remaining after drying at 105 °C
- VFAs:
-
Volatile fatty acids
- VS:
-
Volatile solids, determined as weight loss from heating in air at 550 °C
- °C:
-
Degree celsius (temperature unit)
- bp:
-
Base pairs (nucleic acid unit)
- g :
-
Gram (mass unit)
- h :
-
Hour (time unit)
- L :
-
Liter (volume unit)
- m :
-
Meter (length unit)
- μm:
-
Micrometer (length unit)
- mL:
-
Milliliter (volume unit)
- mg:
-
Milligram (mass unit)
- mm:
-
Millimeter (length unit)
- mM:
-
Millimolar (concentration unit)
- min:
-
Minute (time unit)
- M:
-
Molar (concentration unit)
- s:
-
Second (time unit)
References
Tursi A (2019) A review on biomass: importance, chemistry, classification, and conversion. Biofuel Res J 6(2):962–979. https://doi.org/10.18331/brj2019.6.2.3
Theuerl S, Klang J, Prochnow A (2019) Process disturbances in agricultural biogas production-causes, mechanisms, and effects on the biogas microbiome: a review. Energies 12(365):20. https://doi.org/10.3390/en12030365
Office of Agricultural Economics OAE (2020) Information of agricultural production in Thailand http://www.oae.go.th/view/1/Information/EN-US. Accessed 25 Jun 2019
Monlau F, Barakat A, Trably E, Dumas C, Steyer JP, Carrère H (2013) Lignocellulosic materials into biohydrogen and biomethane: impact of structural features and pretreatment. Crit Rev Environ Sci Technol 43(3):260–322. https://doi.org/10.1080/10643389.2011.604258
Sun C, Liu R, Cao W, Yin R, Mei Y, Zhang L (2015) Impacts of alkaline hydrogen peroxide pretreatment on chemical composition and biochemical methane potential of agricultural crop stalks. Energy Fuels 29(8):4966–4975. https://doi.org/10.1021/acs.energyfuels.5b00838
Widjaja T, Noviyanto AA, Gunawan S (2016) The effect of rumen and mixed microorganism (rumen and effective microorganisms) on biogas production from rice straw waste. ARPN JEAS 11(4):2702–2710
Contreras LM, Schelle H, Sebrango CR, Pereda I (2012) Methane potential and biodegradability of rice straw, rice husk and rice residues from the drying process. Water Sci Technol 65(6):1142–1149. https://doi.org/10.2166/wst.2012.951
Dehghani M, Karimi K, Sadeghi M (2015) Pretreatment of rice straw for the improvement of biogas production. Energy Fuels 29(6):3770–3775. https://doi.org/10.1021/acs.energyfuels.5b00718
Mustafa AM, Poulsen TG, Xia Y, Sheng K (2017) Combinations of fungal and milling pretreatments for enhancing rice straw biogas production during solid-state anaerobic digestion. Biores Technol 224:174–182. https://doi.org/10.1016/j.biortech.2016.11.028
Doi RH (2008) Cellulases of mesophilic microorganisms: cellulosome and noncellulosome producers. Ann N Y Acad Sci 1125:267–279. https://doi.org/10.1196/annals.1419.002
Dollhofer V, Podmirseg SM, Callaghan TM, Griffith GW, Fliegerova K (2015) Anaerobic fungi and their potential for biogas production. Adv Biochem Eng Biotechnol 151:41–61. https://doi.org/10.1007/978-3-319-21993-6_2
Youssef N, Couger M, Struchtemeyer C, Liggenstoffer A, Prade R, Najar F, Atiyeh H, Wilkins M, Elshahed M (2013) The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl Environ Microbiol 79(15):4620–4634. https://doi.org/10.1128/AEM.00821-13
Tabatabaei M, Aghbashlo M, Valijanian E, Kazemi Shariat Panahi H, Nizami A-S, Ghanavati H, Sulaiman A, Mirmohamadsadeghi S, Karimi K (2020) A comprehensive review on recent biological innovations to improve biogas production, Part 1: upstream strategies. Renew Energy 146:1204–1220. https://doi.org/10.1016/j.renene.2019.07.037
Tabatabaei M, Aghbashlo M, Valijanian E, Kazemi Shariat Panahi H, Nizami A-S, Ghanavati H, Sulaiman A, Mirmohamadsadeghi S, Karimi K (2020) A comprehensive review on recent biological innovations to improve biogas production, Part 2: mainstream and downstream strategies. Renew Energy 146:1392–1407. https://doi.org/10.1016/j.renene.2019.07.047
Thongbunrod N, Chaiprasert P (2021) Efficacy and metagenomic analysis of the stabilized anaerobic lignocellulolytic microbial consortium from Bubalus bubalis rumen with rice straw enrichment for methane production. BioEnergy Research 14(3):870–890. https://doi.org/10.1007/s12155-020-10167-y
Rouches E, Zhou S, Steyer JP, Carrere H (2016) White-rot fungi pretreatment of lignocellulosic biomass for anaerobic digestion: impact of glucose supplementation. Process Biochem 51(11):1784–1792. https://doi.org/10.1016/j.procbio.2016.02.003
Kainthola J, Kalamdhad AS, Goud VV, Goel R (2019) Fungal pretreatment and associated kinetics of rice straw hydrolysis to accelerate methane yield from anaerobic digestion. Bioresour Technol 286:121368. https://doi.org/10.1016/j.biortech.2019.121368
Kong X, Du J, Ye X, Xi Y, Jin H, Zhang M, Guo D (2018) Enhanced methane production from wheat straw with the assistance of lignocellulolytic microbial consortium TC-5. Bioresour Technol 263:33–39. https://doi.org/10.1016/j.biortech.2018.04.079
Li P, He C, Li G, Ding P, Lan M, Gao Z, Jiao Y (2020) Biological pretreatment of corn straw for enhancing degradation efficiency and biogas production. Bioengineered 11(1):251–260. https://doi.org/10.1080/21655979.2020.1733733
Xu W, Fu S, Yang Z, Lu J, Guo R (2018) Improved methane production from corn straw by microaerobic pretreatment with a pure bacteria system. Bioresour Technol 259:18–23. https://doi.org/10.1016/j.biortech.2018.02.046
Li J, Wu Y, Zhao J, Wang S, Dong Z, Shao T (2022) Bioaugmented degradation of rice straw combining two novel microbial consortia and lactic acid bacteria for enhancing the methane production. Bioresour Technol 344:126148. https://doi.org/10.1016/j.biortech.2021.126148
Ozbayram EG, Akyol C, Ince B, Karakoc C, Ince O (2018) Rumen bacteria at work: bioaugmentation strategies to enhance biogas production from cow manure. J Appl Microbiol 124(2):491–502. https://doi.org/10.1111/jam.13668
Ozbayram EG, Kleinsteuber S, Nikolausz M, Ince B, Ince O (2017) Effect of bioaugmentation by cellulolytic bacteria enriched from sheep rumen on methane production from wheat straw. Anaerobe 46:122–130. https://doi.org/10.1016/j.anaerobe.2017.03.013
Ma Y, Li Y, Li Y, Cheng Y, Zhu W (2020) The enrichment of anaerobic fungi and methanogens showed higher lignocellulose degrading and methane producing ability than that of bacteria and methanogens. World J Microbiol Biotechnol 36(125):1–9. https://doi.org/10.1007/s11274-020-02894-3
Tsapekos P, Kougias PG, Vasileiou SA, Treu L, Campanaro S, Lyberatos G, Angelidaki I (2017) Bioaugmentation with hydrolytic microbes to improve the anaerobic biodegradability of lignocellulosic agricultural residues. Bioresour Technol 234:350–359. https://doi.org/10.1016/j.biortech.2017.03.043
Yıldırım E, Ince O, Aydin S, Ince B (2017) Improvement of biogas potential of anaerobic digesters using rumen fungi. Renew Energy 109:346–353. https://doi.org/10.1016/j.renene.2017.03.021
Akyol C, Ince O, Bozan M, Ozbayram EG, Ince B (2019) Fungal bioaugmentation of anaerobic digesters fed with lignocellulosic biomass: what to expect from anaerobic fungus Orpinomyces sp. Bioresour Technol 277:1–10. https://doi.org/10.1016/j.biortech.2019.01.024
Ferraro A, Dottorini G, Massini G, Mazzurco Miritana V, Signorini A, Lembo G, Fabbricino M (2018) Combined bioaugmentation with anaerobic ruminal fungi and fermentative bacteria to enhance biogas production from wheat straw and mushroom spent straw. Bioresour Technol 260:364–373. https://doi.org/10.1016/j.biortech.2018.03.128
Wei YQ, Long RJ, Yang H, Yang HJ, Shen XH, Shi RF, Wang ZY, Du JG, Qi XJ, Ye QH (2016) Fiber degradation potential of natural co-cultures of Neocallimastix frontalis and Methanobrevibacter ruminantium isolated from yaks (Bos grunniens) grazing on the Qinghai Tibetan Plateau. Anaerobe 39:158–164. https://doi.org/10.1016/j.anaerobe.2016.03.005
Nguyen QH, Le PD, Chim C, Le ND, Fievez V (2019) Potential to mitigate ammonia emission from slurry by increasing dietary fermentable fiber through inclusion of tropical byproducts in practical diets for growing pigs. Asian-Australas J Anim Sci 32(4):574–584. https://doi.org/10.5713/ajas.18.0481
Grenet E, Bernalier A, Jamot J, Fonty G (1993) Degradation of untreated and anhydrous ammonia-treated wheat straw by two strains of rumen anaerobic fungi. Ann Zootech 42(180):1–1
Ha JK, Lee SS, Kim SW, Han IK, Ushida K, Cheng KJ (2001) Degradation of rice straw by rumen fungi and cellulolytic bacteria through mono-, co- or sequential- culture. Asian Australas J Anim Sci 14(6):797–802
APHA (2012) Standard methods for the examination of water and wastewater, 22nd edition out of print. American public health association (APHA), American water works association (AWWA) and water environment federation (WEF), Washington
Bauchop T (1979) Rumen anaerobic fungi of cattle and sheep. Appl Environ Microbiol 38(1):148–158
Mertens DR (2002) Gravimetric determination of amylase-treated neutral detergent fiber in feeds with refluxing in beakers or crucibles: collaborative study. J AOAC Int 85:24
Theodorou M, Williams B, Dhanoa M, McAllan A, France J (1994) A simple gas production method using a pressure transducer to determine the fermentation kinetics of ruminant feeds. Journal of Animal and Feed Sciences 48(3):185–197. https://doi.org/10.1016/0377-8401(94)90171-6
Panichnumsin P, Nopharatana A, Ahring B, Chaiprasert P (2010) Production of methane by co-digestion of cassava pulp with various concentrations of pig manure. Biomass Bioenergy 34(8):1117–1124. https://doi.org/10.1016/j.biombioe.2010.02.018
Ranjard L, Poly F, Lata JC, Mougel C, Thioulouse J, Nazaret S (2001) Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl Environ Microbiol 67(10):4479–4487. https://doi.org/10.1128/aem.67.10.4479-4487.2001
Kittelmann S, Naylor GE, Koolaard JP, Janssen PH (2012) A proposed taxonomy of anaerobic fungi (class neocallimastigomycetes) suitable for large-scale sequence-based community structure analysis. PLoS ONE 7(5):1–3. https://doi.org/10.1371/journal.pone.0036866
Yu Q, Liu R, Li K, Ma R (2019) A review of crop straw pretreatment methods for biogas production by anaerobic digestion in China. Renew Sustain Energy Rev 107:51–58. https://doi.org/10.1016/j.rser.2019.02.020
Belda-Galbis CM, Pina-Perez MC, Espinosa J, Marco-Celdran A, Martinez A, Rodrigo D (2014) Use of the modified Gompertz equation to assess the Stevia rebaudiana Bertoni antilisterial kinetics. Food Microbiol 38:56–61. https://doi.org/10.1016/j.fm.2013.08.009
Li Y, Zhang R, Liu G, Chen C, He Y, Liu X (2013) Comparison of methane production potential, biodegradability, and kinetics of different organic substrates. Bioresour Technol 149:565–569. https://doi.org/10.1016/j.biortech.2013.09.063
Baetge S, Kaltschmitt M (2018) Rice straw and rice husks as energy sources-comparison of direct combustion and biogas production. Biomass Conversion and Biorefinery 8(3):719–737. https://doi.org/10.1007/s13399-018-0321-y
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541. https://doi.org/10.1128/AEM.01541-09
Silva reference files: Release 132 (2018). https://mothur.org/wiki/silva_reference_files/. Accessed 28 Sept 2020
Kessy A, Allan Z, Timo P, Raivo P, Filipp I, Henrik NR, Urmas K (2020) UNITE mothur release for Fungi 2. UNITE Community. https://unite.ut.ee/repository.php. Accessed 25 Sep 2020
Koetschan C, Kittelmann S, Lu J, Al-Halbouni D, Jarvis GN, Muller T, Wolf M, Janssen PH (2014) Internal transcribed spacer 1 secondary structure analysis reveals a common core throughout the anaerobic fungi (Neocallimastigomycota). PLoS ONE 9(3):1–10. https://doi.org/10.1371/journal.pone.0091928
Batut B, Hiltemann S, Bagnacani A, Baker D, Bhardwaj V, Blank C, Bretaudeau A, Brillet-Gueguen L, Cech M, Chilton J, Clements D, Doppelt-Azeroual O, Erxleben A, Freeberg MA, Gladman S, Hoogstrate Y, Hotz HR, Houwaart T, Jagtap P, Lariviere D, Le Corguille G, Manke T, Mareuil F, Ramirez F, Ryan D, Sigloch FC, Soranzo N, Wolff J, Videm P, Wolfien M, Wubuli A, Yusuf D, Galaxy Training N, Taylor J, Backofen R, Nekrutenko A, Gruning B (2018) Community-driven data analysis training for biology. Cell Syst 6(6):752–758. https://doi.org/10.1016/j.cels.2018.05.012
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Phan S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, White O, KelleyMethe´SchlossGeversMitrevaHuttenhower TSBPDDMC (2012) Metabolic reconstruction for metagenomic data and its application to the human microbiome. Appl Environ Microbiol 8(6):1–18. https://doi.org/10.1371/journal.pcbi.1002358
Levy Karin E, Mirdita M, Soding J (2020) MetaEuk-sensitive, high-throughput gene discovery, and annotation for large-scale eukaryotic metagenomics. Microbiome 8(48):1–15. https://doi.org/10.1186/s40168-020-00808-x
Keller O, Kollmar M, Stanke M, Waack S (2011) A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27(6):757–763. https://doi.org/10.1093/bioinformatics/btr010
Eko H, Chaiprasert P (2019) Enhancement of methane production from high solid anaerobic digestion of pretreated palm oil decanter cake using a modified solid inclined reactor. J Chem Technol Biotechnol 95(3):781–790. https://doi.org/10.1002/jctb.6266
Liu Y, Whitman WB (2008) Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad Sci 1125:171–189. https://doi.org/10.1196/annals.1419.019
Ozbayram EG, Kleinsteuber S, Nikolausz M (2020) Biotechnological utilization of animal gut microbiota for valorization of lignocellulosic biomass. Appl Microbiol Biotechnol 104(2):489–508. https://doi.org/10.1007/s00253-019-10239-w
Li K, Zhu H, Zhang Y, Zhang H (2017) Characterization of the microbial communities in rumen fluid inoculated reactors for the biogas digestion of wheat straw. Sustainability 9:243. https://doi.org/10.3390/su9020243
Nagler M, Kozjek K, Etemadi M, Insam H, Podmirseg SM (2019) Simple yet effective: microbial and biotechnological benefits of rumen liquid addition to lignocellulose-degrading biogas plants. J Biotechnol 300:1–10. https://doi.org/10.1016/j.jbiotec.2019.05.004
Joblin KN (1981) Isolation, enumeration, and maintenance of rumen anaerobic fungi in roll tubes. Appl Environ Microbiol 42(6):1119–1122
Jin W, Cheng YF, Mao SY, Zhu WY (2011) Isolation of natural cultures of anaerobic fungi and indigenously associated methanogens from herbivores and their bioconversion of lignocellulosic materials to methane. Bioresour Technol 102(17):7925–7931. https://doi.org/10.1016/j.biortech.2011.06.026
Callaghan TM, Podmirseg SM, Hohlweck D, Edwards JE, Puniya AK, Dagar SS, Griffith GW (2015) Buwchfawromyces eastonii gen. nov., sp. nov.: a new anaerobic fungus (Neocallimastigomycota) isolated from buffalo faeces. MycoKeys 9:11–28. https://doi.org/10.3897/mycokeys.9.9032
Dagar SS, Kumar S, Griffith GW, Edwards JE, Callaghan TM, Singh R, Nagpal AK, Puniya AK (2015) A new anaerobic fungus (Oontomyces anksri gen. nov., sp. nov.) from the digestive tract of the Indian camel (Camelus dromedarius). Fungal Biol Rev 119(8):731–737. https://doi.org/10.1016/j.funbio.2015.04.005
Joshi A, Lanjekar VB, Dhakephalkar PK, Callaghan TM, Griffith GW, Dagar SS (2018) Liebetanzomyces polymorphus gen. et sp. nov., a new anaerobic fungus (Neocallimastigomycota) isolated from the rumen of a goat. MycoKeys 40:89–110. https://doi.org/10.3897/mycokeys.40.28337
Jing R, Yan Y (2020) Metagenomic analysis reveals antibiotic resistance genes in the bovine rumen. Microb Pathog 149:1–25. https://doi.org/10.1016/j.micpath.2020.104350
Treu L, Campanaro S, Kougias PG, Sartori C, Bassani I, Angelidaki I (2018) Hydrogen-fueled microbial pathways in biogas upgrading systems revealed by genome-centric metagenomics. Front Microbiol 9:1–16. https://doi.org/10.3389/fmicb.2018.01079
Ward AJ, Hobbs PJ, Holliman PJ, Jones DL (2008) Optimisation of the anaerobic digestion of agricultural resources. Bioresour Technol 99(17):7928–7940. https://doi.org/10.1016/j.biortech.2008.02.044
Chandra R, Vijay VK, Subbarao PMV, Khura TK (2012) Production of methane from anaerobic digestion of jatropha and pongamia oil cakes. Appl Energy 93:148–159. https://doi.org/10.1016/j.apenergy.2010.10.049
Kim M, Gomec CY, Ahn Y, Speece RE (2003) Hydrolysis and acidogenesis of particulate organic material in mesophilic and thermophilic anaerobic digestion. Environ Technol 24(9):1183–1190. https://doi.org/10.1080/09593330309385659
Siegert I, Banks C (2005) The effect of volatile fatty acid additions on the anaerobic digestion of cellulose and glucose in batch reactors. Process Biochem 40(11):3412–3418. https://doi.org/10.1016/j.procbio.2005.01.025
Theodorou MK, Gill M, King-Spooner C, Beever DE (1990) Enumeration of anaerobic chytridiomycetes as thallus-forming units: novel method for quantification of fibrolytic fungal populations from the digestive tract ecosystem. Appl Environ Microbiol 56(4):1073–1078. https://doi.org/10.1128/aem.56.4.1073-1078.1990
Orpin CG (1977) The rumen flagellate Piromonas communis: its life-history and invasion of plant material in the rumen. J Gen Microbiol 99:107–117. https://doi.org/10.1099/00221287-99-1-107
Hansen JC, Aanderud ZT, Reid LE, Bateman C, Hansen CL, Rogers LS, Hansen LD (2021) Enhancing waste degradation and biogas production by pre-digestion with a hyperthermophilic anaerobic bacterium. Biofuel Res J 8(3):1433–1443. https://doi.org/10.18331/brj2021.8.3.3
Mirmohamadsadeghi S, Karimi K, Azarbaijani R, Parsa Yeganeh L, Angelidaki I, Nizami A-S, Bhat R, Dashora K, Vijay VK, Aghbashlo M, Gupta VK, Tabatabaei M (2021) Pretreatment of lignocelluloses for enhanced biogas production: a review on influencing mechanisms and the importance of microbial diversity. Renew Sustain Energy Rev 135:1–18. https://doi.org/10.1016/j.rser.2020.110173
Griessmeier V, Gescher J (2018) Influence of the potential carbon sources for field denitrification beds on their microbial diversity and the fate of carbon and nitrate. Front Microbiol 9:1–15. https://doi.org/10.3389/fmicb.2018.01313
O’Shea R, Lin R, Wall DM, Browne JD, Murphy JD (2021) Distillery decarbonization and anaerobic digestion: balancing benefits and drawbacks using a compromise programming approach. Biofuel Res J 8(3):1417–1432. https://doi.org/10.18331/brj2021.8.3.2
Garcia SL, Jangid K, Whitman WB, Das KC (2011) Transition of microbial communities during the adaption to anaerobic digestion of carrot waste. Bioresour Technol 102(15):7249–7256. https://doi.org/10.1016/j.biortech.2011.04.098
Ito T, Yoshiguchi K, Ariesyady HD, Okabe S (2011) Identification of a novel acetate-utilizing bacterium belonging to Synergistes group 4 in anaerobic digester sludge. ISME J 5(12):1844–1856. https://doi.org/10.1038/ismej.2011.59
Downes J, Vartoukian SR, Dewhirst FE, Izard J, Chen T, Yu WH, Sutcliffe IC, Wade WG (2009) Pyramidobacter piscolens gen. nov., sp. nov., a member of the phylum ‘Synergistetes’ isolated from the human oral cavity. Int J Syst Evol Microbiol 59(5):972–980. https://doi.org/10.1099/ijs.0.000364-0
Chen S, Dong X (2005) Proteiniphilum acetatigenes gen. nov., sp. nov., from a UASB reactor treating brewery wastewater. Int J Syst Evol Microbiol 55(6):2257–2261. https://doi.org/10.1099/ijs.0.63807-0
Kumar M, Verma S, Gazara RK, Kumar M, Pandey A, Verma PK, Thakur IS (2018) Genomic and proteomic analysis of lignin degrading and polyhydroxyalkanoate accumulating beta-proteobacterium Pandoraea sp. ISTKB Biotechnol Biofuels 11:154. https://doi.org/10.1186/s13068-018-1148-2
Liu D (2015) Aeromonas. In: Mol. Microbiol. pp 1099–1110.https://doi.org/10.1016/b978-0-12-397169-2.00061-5
Wilhelm RC, Singh R, Eltis LD, Mohn WW (2019) Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J 13(2):413–429. https://doi.org/10.1038/s41396-018-0279-6
Gruninger RJ, Puniya AK, Callaghan TM, Edwards JE, Youssef N, Dagar SS, Fliegerova K, Griffith GW, Forster R, Tsang A, McAllister T, Elshahed MS (2014) Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role, and biotechnological potential. FEMS Microbiol Ecol 90(1):1–17. https://doi.org/10.1111/1574-6941.12383
Hanafy RA, Elshahed MS, Youssef NH (2018) Feramyces austinii, gen. nov., sp. nov., an anaerobic gut fungus from rumen and fecal samples of wild Barbary sheep and fallow deer. Mycologia 110(3):513–525. https://doi.org/10.1080/00275514.2018.1466610
Hanafy RA, Lanjekar VB, Dhakephalkar PK, Callaghan TM, Dagar SS, Griffith GW, Elshahed MS, Youssef NH (2020) Seven new Neocallimastigomycota genera from wild, zoo-housed, and domesticated herbivores greatly expand the taxonomic diversity of the phylum. Mycologia:1–28 https://doi.org/10.1080/00275514.2019.1696619
Kameshwar AKS, Qin W (2018) Genome wide analysis reveals the extrinsic cellulolytic and biohydrogen generating abilities of Neocallimastigomycota fungi. J Genomics 6:74–87. https://doi.org/10.7150/jgen.25648
Peng X, Wilken SE, Lankiewicz TS, Gilmore SP, Brown JL, Henske JK, Swift CL, Salamov A, Barry K, Grigoriev IV, Theodorou MK, Valentine DL, O’Malley MA (2021) Genomic and functional analyses of fungal and bacterial consortia that enable lignocellulose breakdown in goat gut microbiomes. Nat Microbiol 6(4):499–511. https://doi.org/10.1038/s41564-020-00861-0
Hanafy RA, Johnson B, Youssef NH, Elshahed MS (2020) Assessing anaerobic gut fungal diversity in herbivores using D1/D2 large ribosomal subunit sequencing and multi-year isolation. Environ Microbiol.https://doi.org/10.1111/1462-2920.15164
Gruninger RJ, Nguyen TTM, Reid ID, Yanke JL, Wang P, Abbott DW, Tsang A, McAllister T (2018) Application of transcriptomics to compare the carbohydrate active enzymes that are expressed by diverse genera of anaerobic fungi to degrade plant dell wall carbohydrates. Front Microbiol 9:1581. https://doi.org/10.3389/fmicb.2018.01581
Acknowledgements
The authors acknowledge Prof. Dr. Anna Schnürer for the support of barcode preparation and the 16s and ITS1 sequencing at the Department of Molecular Sciences, Swedish University of Agricultural Sciences, Sweden; the Freiburg Galaxy Team: Person X and Prof. Dr. Rolf Backofen for the website of metagenomic data analysis, Bioinformatics, University of Freiburg, Germany, funded by Collaborative Research Centre 992 Medical Epigenetics (DFG grant SFB 992/1 2012) and German Federal Ministry of Education and Research (BMBF grant 031 A538A de. NBI-RBC); the laboratory facility from Excellent Center of Waste Utilization and Management (ECoWaste) at King Mongkut’s University of Technology Thonburi, Thailand.
Funding
The authors gratefully acknowledge the financial support by the Petchra Pra Jom Klao Doctoral Scholarship from King Mongkut’s University of Technology Thonburi (KMUTT) awarded to Ms. Nitiya Thongbunrod. Research funding was supported by the Fundamental Fund (FF) of Thailand Science Research and Innovation (TSRI) and Ministry of Higher Education, Science, Research, and Innovation (MHESI).
Author information
Authors and Affiliations
Contributions
Pawinee Chaiprasert: conceptualization, project administration, funding acquisition, resources, supervision, and writing—reviewing and editing. Nitiya Thongbunrod: methodology, data curation, formal analysis, visualization, investigation, and writing—original draft preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Thongbunrod, N., Chaiprasert, P. Potential of enriched and stabilized anaerobic lignocellulolytic fungi coexisting with bacteria and methanogens for enhanced methane production from rice straw. Biomass Conv. Bioref. 14, 8229–8250 (2024). https://doi.org/10.1007/s13399-022-03129-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13399-022-03129-1