Abstract
Sweet corn has gained worldwide popularity. Traditional sweet corn possesses low concentration of essential nutrients such as lysine (0.15–0.25%), tryptophan (0.03–0.04%) and provitamin-A (proA 3–4 ppm), and deficiency leads to serious health problems in humans. Here, stacking of shrunken2 (sh2), opaque2 (o2), lycopene epsilon cyclase (lcyE) and β‐carotene hydroxylase (crtRB1) genes were undertaken in the parents of four hybrids viz., APQH1, APHQ4, APHQ5 and APHQ7 using marker-assisted backcross breeding (MABB). Gene-linked markers (umc2276 and umc1320) for sh2, while gene-based markers for o2 (umc1066 and phi057), lcyE (5′TE-InDel) and crtRB1 (3′TE-InDel), were used for genotyping in BC1F1, BC2F1 and BC2F2. Selected backcross progenies showed high recovery of recurrent parent genome (92.4–97.7%). The reconstituted sweet corn hybrids possessed significantly high lysine (0.390%), tryptophan (0.082%) and proA (21.14 ppm), coupled with high kernel sweetness (brix 18.96%). The improved sweet corn hybrids had high cob yield (12.22–15.33 t/ha) across three environments. These newly developed biofortified sweet corn hybrids possess great significance in providing balanced nutrition. This is the first report of combining sh2, o2, lcyE and crtRB1 genes for enrichment of sweet corn hybrids with multiple essential nutrients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sweet corn possesses economic importance due to its increased worldwide usage as fresh and processed vegetables (Chhabra et al. 2019). Global import of frozen sweet corn was US$433 million, while it was US$957 million for processed sweet corn (FAOSTAT 2017). Due to increasing urbanization, the demand of sweet corn has increased tremendously (Lertrat and Pulam 2007; Hossain et al. 2015). It is usually consumed at immature stages of endosperm development at 20–24 days after pollination (DAP) (Khanduri et al. 2011). The green plants after harvest serve as source of green fodder to cattle.
Recessive shrunken2 (sh2) has been predominantly used for sweet corn cultivars development. Sh2 located on chromosome 3L possess six times more sucrose and reducing sugar than normal maize. The depletion of sugar level in sh2 type is much slower and thus has extended shelf life (Lertrat and Pulam 2007). Due to this reason, sh2-based sweet corn is now more popular worldwide (Hossain et al. 2015). Micronutrient malnutrition is a serious health problem, and nearly two billion people suffer from deficiency of micronutrients (Global Nutrition Report 2018). Deficiency of provitamin-A (proA) leads to measles, diarrhoea, respiratory diseases, night blindness and low resistance to infectious diseases (Bouis and Saltzman 2017). The lack of adequate lysine and tryptophan affects protein biosynthesis inside the body and also leads to fatigue, dizziness, nausea, anaemia, delayed growth, depression, anxiety, loss of appetite and reproductive tissue (Hossain et al. 2018). Therefore, development of nutrition-rich staple crops through plant breeding approach hold great promise to address the problem of malnutrition (Pfeiffer and McClafferty 2007).
Traditional sweet corn possesses low level of lysine (0.15–0.25%), tryptophan (0.03–0.04%) and proA (3–4 ppm). The recessive opaque2 (o2i enhances lysine and tryptophan by 2-fold (Mertz et al. 1964). The mutant version of β-carotene hydroxylase (crtRB1) and lycopene epsilon cyclase (lcyE) increases kernel proA by ~10-fold (Harjes et al. 2008; Yan et al. 2010). The o2, crtRB1 and lcyE genes have been utilized for development of proA rich field corn cultivars worldwide (Prasanna et al. 2020). So far, no sweet corn hybrid enriched with lysine, tryptophan and proA has been commercialized elsewhere in the world. The present investigation was therefore undertaken to (i) combine sh2, o2, crtRB1 and lcyE genes into the parents of elite hybrids using marker-assisted selection (MAS); (ii) evaluate the MAS-derived sweet corn inbreds and hybrids for kernel sweetness, lysine, tryptophan and proA; and (iii) compare the MAS-derived genotypes for agronomic and yield performance with the original versions.
Materials and methods
Plant material
Parental lines (PMI-PV5, PMI-PV6, PMI-PV7 and PMI-PV8) of four proA rich quality protein maize (QPM) hybrids, viz., APQH-1, APQH-4, APQH-5 and APQH-7, having recessive alleles of o2, crtRB1 and lcyE developed at ICAR-Indian Agricultural Research Institute (IARI), New Delhi (Zunjare et al. 2018) were targeted for introgression of sh2 allele using marker-assisted backcross breeding (MABB). APQH-1, APQH-4, APQH-5 and APQH-7 are the proA rich version of the released medium-late maturing QPM hybrids (HQPM-1, HQPM-4, HQPM-5 and HQPM-7) in India (Table S1). SWT-147, a sweet corn inbred with recessive sh2 allele, was used as donor line. The pedigree details of the recurrent and donor parents are presented in Table S2.
Development of backcross progenies
Backcross- and self-populations segregating for the alleles of sh2, o2, crtRB1 and lcyE were grown at two locations alternatively viz., Winter Nursery Centre (WNC), ICAR-Indian Institute of Maize Research (IIMR), Hyderabad (17° 21′ 50.39″ N and 78° 29′ 42.31″ E) and ICAR-IARI, New Delhi (29° 41′ 52.13″ N and 77° 0′ 24.95″ E). The detailed MABB scheme followed in the present study is shown in Fig. 1.
Genotyping in segregating generations
Genomic DNA was isolated from the young leaves using the modified standard cetyl trimethyl ammonium bromide (CTAB) procedure (Saghai-Maroof et al. 1984). Foreground selection was carried out in BC1F1, BC2F1 and BC2F2 generations using markers associated with the sh2, o2, lcyE and crtRB1 genes (Table 1). Gene-linked simple sequence repeat (SSR) markers umc1320 and umc2276 were used for sh2 (Hossain et al. 2015). For other three target genes, gene-based markers were used for genotyping backcross populations, viz., umc1066 and phi057 for o2 (Hossain et al. 2018), 5′TE-InDel marker for lcyE (Harjes et al. 2008) and 3′TE-InDel-based marker for crtRB1 (Yan et al. 2010). The polymerase chain reaction (PCR) protocols for all the four genes viz., sh2 (Hossain et al. 2015), o2 (Hossain et al. 2018), lcyE and crtRB1 (Zunjare et al. 2018) standardized at Maize Genetics Unit, ICAR-IARI, New Delhi, were further modified for reducing the time duration. The amplified product was resolved on 4% Seakem LE agarose gel in case of umc1320, umc2276, umc1066 and phi057 and 1.5% Seakem LE agarose gel in case of crtRB1and lcyE genes. The genome-wide SSRs with near uniform coverage across 10 chromosomes of maize were employed for recovering the recurrent parent genome (RPG) in foreground positive plants of BC1F1, BC2F1 and BC2F2 generations. The protocol for amplification of SSRs was as per Hossain et al. (2018).
Evaluation of introgressed progenies and reconstituted hybrids
The selected BC2F4 progenies along with the original inbreds were evaluated in random complete block design (RCBD) with two replications during rainy season (2018) at ICAR-IARI, New Delhi. Twelve reconstituted hybrids (-A, -B and -C versions in each of the four hybrids) along with the four original versions and national check hybrid (Pusa Vivek QPM9 Improved) were evaluated in RCBD with two replications in three dates of sowing (S-I: 8th August, S-II: 28th August and S-III: 17th September) during rainy season at ICAR-IARI, New Delhi (Table 2). Selfed seeds were used for estimation of kernel sweetness (brix), lysine, tryptophan and proA. Plant height, ear height, cob length, cob diameter, number of rows per cob, number of kernels per row and male- and female-flowering were also recorded. The dehusked cob yield and green fodder weight were recorded for hybrids. Newly derived inbreds and hybrids were also characterized for 30 distinctness, uniformity and stability (DUS) characters (PPVFRA 2007).
Estimation of kernel sugar
In each entry, three plants were selfed to avoid any xenia effect. Selfed kernels (60–90 grains per genotype) bulked from the three ears (20–30 grains from each ears) were crushed to measure sweetness (brix) at 20 days after pollination (DAP) using brix meter (Atago, Japan) (Khanduri et al. 2011). The grains were crushed and the milky juice thus produced was used to measure the brix reading.
Estimation of lysine, tryptophan and proA
Remaining selfed seeds from the three self-pollinated ears left after the estimation of brix at 20-DAP were stored at −80 °C until used for biochemical estimation of lysine, tryptophan and proA. A total of 20–30 selfed seeds from each of the three ears (60–90 grains in total per genotype) were dried by wrapping the seeds in Whatman® blotting paper and incubating in desiccator filled with activated silica gel for 3 days. Deactivated silica gel was removed after 24 h with replacement of fresh activated silica gel for rapid drying. After drying, seeds were ground to fine powder to use them further for estimation of lysine and tryptophan (Hossain et al. 2018), β-carotene and β-cryptoxanthin through UHPLC System (ultra high-performance liquid chromatography; Thermo Scientific, MA, USA) (Muthusamy et al. 2014).
Statistical analysis
The observed segregation pattern of the genes was tested for goodness of fit by χ2 analysis. The recovery of RPG was estimated as per Benchimol et al. (2005). Pooled analysis of variance (ANOVA) with fixed model was carried out using Windostat 10.0 software package. The percentage contribution of sowing dates and genotype × sowing date for each of the traits was calculated as proportion of sum of square over total sum of square expressed in percentage.
Results
Parental polymorphism
All the four recurrent parents viz., PMI-PV5, PMI-PV6, PMI-PV7 and PMI-PV8 possessed favourable allele for o2 (150 bp for umc1066 and 170 bp for phi057), lcyE (650 bp) and crtRB1 (543 bp) genes, while the donor possessed favourable allele for sh2 gene (103 bp for umc1320 and 130 bp for umc2276) and unfavourable allele of o2 (160 bp each for umc1066 and phi057), lcyE (300 bp) and crtRB1 (296 bp) (Fig. 2). For o2, umc1066 was used for PMI-PV5 and PMI-PV6, while phi057 was used for PMI-PV7 and PMI-PV8. For sh2 gene, umc1320 was used in PMI-PV5, while umc2276 in PMI-PV6, PMI-PV7 and PMI-PV8 for genotyping. A set of 258 genome-wide SSRs covering 10 chromosomes was screened between recurrent and donor parents (Table 3). The selected polymorphic SSRs (97–185) were employed for background selection in BC1F1, BC2F1 and BC2F2 generations for recovery of RPG.
Genotyping of backcross progenies
In BC1F1 generation, 112–189 plants were genotyped for sh2, o2, lcyE and crtRB1 (Tables 4 and 5). In the case of PMI-PV5- and PMI-PV7-based progenies, o2 showed segregation distortion (SD), while PMI-PV8 progenies deviated from expected ratio for sh2, lcyE and crtRB1. The RPG among selected seven progenies (heterozygous: sh2, homozygous: o2, lcyE and crtRB1) ranged from 73.5 to 85.3% (Table S3). In BC2F1, 93–146 plants across populations were genotyped. Sh2 followed 1:1 segregation in all populations, barring PMI-PV8-based one (Table 4). The RPG among 12 selected progenies (heterozygous: sh2, homozygous: o2, lcyE and crtRB1) varied from 87.1 to 96.1%. Among BC2F2 seeds (borne on BC2F1 cob), sh2 gene segregated with a ratio of 3 (round): 1 (shrunken) in all the four populations (Table 4). In total, 222, 180, 138 and 140 phenotypically shrunken (homozygous for sh2) seeds were identified from the selected cobs of PMI-PV5-, PMI-PV6-, PMI-PV7- and PMI-PV8-based populations, respectively. A total of 95–150 plants in BC2F2 were genotyped, and progenies with homozygosity at sh2, o2, lcyE and crtRB1 were selected. The percentage of RPG among selected progenies ranged 92.4–97.7%.
Nutritional quality among introgressed progenies
Kernel sweetness
Original parents, PMI-PV5, PMI-PV6, PMI-PV7 and PMI-PV8, had low brix of 8.90%, 9.25%, 8.90% and 8.50%, respectively. Among the introgressed progenies, PMI-PV7-69-21-4 possessed the highest brix (19.25%), followed by PMI-PV5-70-95-13 (18.25%), PMI-PV8-51-71-17 (17.95%) and PMI-PV6-1-32-22 (17.90%) and in each of the four genetic background (Table 6). Thus, the introgressed progenies showed 2-fold enhancement in brix compared to their original inbreds. Though introgressed progenies had higher brix, variation was observed among progenies within each pedigree lineage. For example, PMI-PV7-69-21-4 had 19.25% brix, while PMI-PV7-66-7-15 possessed 17.75% brix.
Lysine, tryptophan and provitamin-A
Original inbreds showed variation for lysine (0.312–0.344%), tryptophan (0.077–0.093%) and proA (15.89–18.52 ppm). However, PMI-PV5 had the highest lysine, tryptophan and proA. While PMI-PV8 possessed lowest lysine and tryptophan, PMI-PV6 possessed the lowest proA. MABB-derived progenies were comparable for lysine, tryptophan and proA to their respective original inbreds (Table 6). The lysine among the improved progenies of PMI-PV5 was 0.317–0.354%, while the same for PMI-PV6, PMI-PV7 and PMI-PV8 were 0.306–0.345%, 0.286–0.351% and 0.290–0.331%, respectively. Similarly, tryptophan among the progenies of PMI-PV5 (0.086–0.093%), PMI-PV6 (0.077–0.094%), PMI-PV7 (0.078–0.086%) and PMI-PV8 (0.075–0.086%) were also high. The proA among the MABB-derived introgressed progenies of PMI-PV5, PMI-PV6, PMI-PV7 and PMI-PV8 ranged from 16.82 to 20.19 ppm, 14.87 to 18.12 ppm, 14.25 to 18.40 ppm and 15.48 to 20.06 ppm. The same in the original version was 18.52 ppm, 15.89 ppm, 16.62 ppm and 17.61 ppm, respectively (Table 6). Variation was also observed among the introgressed progenies of the same lineage. For example, PMI-PV5-70-79-22 possessed the highest lysine (0.354%), tryptophan (0.093%) and proA (20.19 ppm) compared to PMI-PV5-70-16-8 and PMI-PV5-70-95-13. Similarly, PMI-PV8-51-71-17 had the highest nutritional values over PMI-PV8-51-29-11 and PMI-PV8-51-93-18.
Agronomic performance of introgressed progenies
The introgressed progenies of PMI-PV5, PMI-PV6, PMI-PV7 and PMI-PV8 showed great degree of similarity with their original versions in relation to plant height, ear height, cob length, cob diameter, number of rows per cob and number of kernels per row. The flowering behaviour of the introgressed progenies was also similar to the original versions (Table S4). DUS characterization also showed high resemblance for majority of the characters (Table S5a and b). However, all the sweet corn progenies lack purple pigmentation in adventitious roots, stem, leaf sheath, silk and glumes. While original inbreds possessed pigmentation in some of the plant parts mentioned above.
Nutritional quality among reconstituted hybrids
Kernel sweetness
Sowing dates and genotype × sowing date had minor effects (<2% of the total variation) on brix (Table S6a). Across sowing dates (S-I, S-II and S-III), the mean brix value of reconstituted hybrids of APQH-1, APQH-4, APQH-5 and APQH-7 was 19.50%, 19.11%, 18.53% and 18.71%, as compared to 9.54%, 9.05%, 8.64% and 8.80% in the original hybrids, respectively, at 20 DAP (Table 7; Fig. 3a). The same in the field corn check hybrid, Pusa Vivek QPM-9 Improved was low (8.73%). Thus, the reconstituted hybrids recorded 2-fold increase in kernel sweetness due to introgression of recessive sh2 gene. However, variation was observed among the reconstituted hybrids of same lineage. For example, APQH-1-SWT-C had the highest brix (20.24%) compared to APQH-1-SWT-B (19.39%) and APQH-1-SWT-A (18.86%).
Lysine, tryptophan and provitamin-A
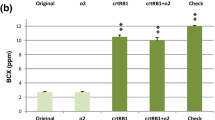
Smaller variation in lysine (6.40%), tryptophan (1.22%) and proA (2.18%) was contributed by sowing dates. However, genotype × sowing date interaction had moderate influence on lysine (34.94%) and tryptophan (23.97%), while it had minor influence on proA (6.82%) (Table S6a). In each of the hybrid versions across sowing time, APQH-5-SWT-B (0.416%) had the highest accumulation of lysine followed by APQH-4-SWT-A and APQH-4-SWT-C (0.401% in each), APQH-7-SWT-A (0.393%) and APQH-1-SWT-A (0.372%). In case of tryptophan, APQH-4-SWT-B (0.092%) possessed the highest accumulation, followed by APQH-5-SWT-A (0.090%), APQH-1-SWT-A (0.079%) and APQH-7-SWT-B (0.075%). Among each of the hybrid versions, the highest accumulation of proA was observed in APQH-7-SWT-B (24.00 ppm), APQH-4-SWT-A (21.59 ppm), APQH-5-SWT-C (20.40 ppm) and APQH-1-SWT-A (19.35 ppm) (Table 7; Fig. 3b, c and d). Variations were also observed among reconstituted hybrids of similar lineage. For example, APQH-1-SWT-A had the highest lysine (0.372%), tryptophan (0.079%) and proA (19.35 ppm) compared to other versions (APQH-1-SWT-B and APQH-1-SWT-C).
Agronomic performance of the reconstituted hybrids
Sowing date and genotype × sowing date interaction had the minor influence on cob yield (1.50% and 16.56%) and fodder yield (3.40% and 3.59%) (Table S6b). Across dates of sowing, the reconstituted hybrids showed high degree of resemblance for various agronomic characters with their original versions (Table S7a and b). The cob yield among the original hybrids was 11.56–14.67 t/ha, while the same in the reconstituted sweet corn hybrids was 12.22–15.33 t/ha. The reconstituted hybrids also showed similar plant and cob characteristics. The DUS characterization also showed high degree of similarity among the reconstituted and original hybrids (Table S8a and b). However, all the original hybrids had purple pigmentation in various plant parts like, glume, silk, leaf sheath, stem base and adventitious roots, while they were absent in the reconstituted versions.
Discussion
Segregation of genes and recovery of RPG
In BC1F1 and BC2F1, SD was observed for sh2, o2, crtRB1 and lcyE in some of the progenies. This could be due to various gametophytic factors, defective kernel mutants, male sterility and embryo-specific mutation (Neuffer et al. 1997). Earlier, Muthusamy et al. (2014) and Zunjare et al. (2018) reported SD for crtRB1, while Hossain et al. (2018) observed SD for o2. SD thus necessitates raising of the large backcross populations in order to select desirable number of the positive segregants. Background selection was successfully used for high recovery of RPG. The average recovery of RPG after two backcrosses is 87.13%; however, the SSRs distributed throughout the genome helped achieving higher RPG. Various researchers have reported high RPG in the two-generation based MABB programme (Muthusamy et al. 2014; Hossain et al. 2018; Zunjare et al. 2018). The high recovery also led to the high degree of resemblance with their corresponding recurrent parents with respect to plant, ear and grain characteristics.
Enhancement of kernel sweetness
The introgressed progenies and the reconstituted hybrids recorded 2-fold increase in brix. This is due to the use of closely linked markers which led to the indirect selection of the sh2 gene (Hossain et al. 2015). The sh2 gene, located on the chromosome 3, codes for a large subunit of ADP-glucose pyrophosphorylase (AGPase) and is involved in the synthesis of ADP-glucose from glucose-1-phosphate and adenosine triphosphate. Since the recessive version of the sh2 blocks the conversion, starch biosynthesis such as amylose and amylopectin is restricted, leading to the higher accumulation of sugars in the kernels at 20–24 DAP (Chhabra et al. 2019). Though all the MAS-derived genotypes possessed same sh2 allele, they reported moderate level of variation in brix (17.30–19.25%). This variation of kernel brix is attributed to various modifier loci influencing sh2 gene (Mehta et al. 2020).
Accumulation of high lysine, tryptophan and proA
In the present study, the sweet corn hybrids possessed significantly high mean lysine (0.390%) and tryptophan (0.082%) due to the presence of recessive o2 gene. The recessive o2 mutant located on chromosome 7 enhances lysine and tryptophan by double (Mertz et al. 1964). The o2 codes for basic leucine-zipper (bZIP) transcriptional factor that regulates the expression of 22-kDa α-zein genes. The enhancement is due to (i) less affinity of binding to α-zein genes, (ii) reduced transcription of lysine keto-reductase (LKR), and (iii) synthesis of various lysine-rich proteins and enzymes (Hossain et al. 2018). The high proA (21.14 ppm) observed in the sweet corn was due to lcyE (chromosome 8) that converts the flux of lycopene more towards β-branch, thereby leading to the formation of more β-carotene (Harjes et al. 2008). A transposable element (TE) in the 5′UTR of the lcyE is responsible for the polymorphism. On the other hand, crtRB1 (chromosome 10) causes hydroxylation of β-carotene and β-cryptoxanthin into non-proA carotenoid viz., zeaxanthin. The natural variant of the crtRB1 with no TE in the 3′UTR region drastically slows down the conversion, leading to more accumulation of proA carotenoids (Yan et al. 2010).
Though several lysine-, tryptophan- and proA-rich maize hybrids have been developed and commercialized worldwide, all these hybrids are field corn type where dried grains are used as food (Prasanna et al. 2020). However, no sweet corn hybrid with high lysine, tryptophan and proA has been developed elsewhere. In China, Yang et al. (2018) introgressed lcyE into four sh2-based sweet corn inbreds and reported low increase in proA (2.07–3.86 ppm) over the original inbreds (0.004–3.09 ppm). However, in the present study, we have achieved very high increase in proA (>18 ppm) among the newly developed sweet corn hybrids. This high value of proA was possible due to use of crtRB1 gene coupled with lcyE (Zunjare et al. 2018). However, the variation for lysine, tryptophan and proA observed among the introgressed inbreds and reconstituted hybrids is due to various modifier loci influencing o2, crtRB1 and lcyE genes (Hossain et al. 2018, Zunjare et al. 2018). Thus, original inbreds and their improved versions were at par with lysine, tryptophan and proA due to the presence of recessive o2 and mutant crtRB1 genes in the recurrent parent. However, the introgressed progenies and reconstituted hybrids possessed high kernel sweetness in comparison to their original versions due to the introgression of recessive sh2 gene. The cob and fodder yield of the reconstituted hybrids was also at par with the original versions. Thus, newly derived nutritionally rich sweet corn hybrids would provide livelihood to large number of farmers, besides providing balanced nutrition to the consumers. This is the first report of marker-assisted stacking of sh2, o2, lcyE and crtRB1 genes for enhancement of essential amino acids and proA in sweet corn genotypes.
Conclusion
Traditional sweet corn lacks the desired concentration of lysine, tryptophan and proA, deficiency of which causes serious health problems. Here we combined sh2, o2, lcyE and crtRB1 genes in a single genetic background using genomic-assisted breeding. The application of gene-specific molecular markers helped to precisely identify the favourable genotypes in the segregating populations. Further, stringent background selection through SSRs led to the high recovery of recurrent parent genome and similar phenotypic characters of the recurrent parents in just two backcrosses. The reconstituted hybrids possess 2-fold more lysine and tryptophan and 4-fold more proA. The molecular breeding thus helped in rapidly developing nutritious sweet corn hybrids which would have taken more than double the time in case of conventional breeding. This is the first study to combine sh2, o2, lcyE and crtRB1 genes for the development of biofortified sweet corn hybrids. These multi-nutrient sweet corn hybrids would help in providing balanced food and address the malnutrition in a sustainable and cost-effective way.
Data availability
All supporting data are included within the article and its additional files.
References
Benchimol LL, Souza CLD Jr, Souza APD (2005) Microsatellite-assisted backcross selection in maize. Genet Mol Biol 28(4):789–797
Bouis HE, Saltzman A (2017) Improving nutrition through biofortification: a review of evidence from HarvestPlus, 2003 through 2016. Glob Food Sec 12:49–58
Chhabra R, Hossain F, Muthusamy V, Baveja A, Mehta B, Zunjare RU (2019) Mapping and validation of plant anthocyanin-1 pigmentation gene for its effectiveness in early selection of shrunken-2 gene influencing kernel sweetness in maize. J Cereal Sci. https://doi.org/10.1016/j.jcs.2019.04.012
FAOSTAT (2017) http://faostat.fao.org
Global Nutrition Report (2018) www.globalnutritionreport.org
Harjes CE, Rocheford TR, Bai L, Brutnell TP, Kandianis CB, Sowinski SG, Stapleton AE, Vallabhaneni R, Williams M, Wurtzel ET, Yan J, Buckler ES (2008) Natural genetic variation in Lycopene Epsilon Cyclase tapped for maize biofortification. Science 319:330–333
Hossain F, Muthusamy V, Pandey N, Vishwakarma AK, Baveja A, Zunjare R, Thirunavukkarasu N, Saha S, Manjaiah KM, Prasanna BM, Gupta HS (2018) Marker-assisted introgression of opaque2 allele for rapid conversion of elite hybrids into quality protein maize. J Genet 97:287–298
Hossain F, Nepolean T, Vishwakarma AK, Pandey N, Prasanna BM, Gupta HS (2015) Mapping and validation of microsatellite markers linked to sugary1 and shrunken2 genes in maize. J Plant Biochem Biotechnol. https://doi.org/10.1007/s13562-013-0245-3
Khanduri A, Hossain F, Lakhera PC, Prasanna BM (2011) Effect of harvest time on kernel sugar concentration in sweet corn. Indian J Genet 71:231–234
Lertrat K, Pulam T (2007) Breeding for increased sweetness in sweet corn. Intl J Plant Breed 1:27–30
Mehta BK, Muthusamy V, Baveja A, Chauhan HS, Chhabra R, Bhatt V, Chand G, Zunjare RU, Singh AK, Hossain F (2020) Composition analysis of lysine, tryptophan and provitamin-A during different stages of kernel development in biofortified sweet corn. J Food Compos Anal 94:103625
Mertz ET, Bates LS, Nelson OE (1964) Mutant genes that change protein composition and increase lysine content of maize endosperm. Science 145:279–280
Muthusamy V, Hossain F, Nepolean T, Choudhary M, Saha S, Bhat JS, Prasanna BM, Gupta HS (2014) Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS ONE 9(12):e113583
Neuffer MG, Coe EH, Wessler SR (1997) Mutants of maize. Cold Spring Harbor Laboratory Press, New York
Pfeiffer WH, McClafferty B (2007) HarvestPlus: breeding crops for better nutrition. Crop Sci 47:S89–S105. https://doi.org/10.2135/cropsci2007.09.0020IPBS
PPVFRA (2007). Guidelines for the conduct of test for distinctiveness, uniformity and stability on maize (Zea mays L.). pp. 13
Prasanna BM, Palacios-Rojas N, Hossain F, Muthusamy V, Menkir A, Dhliwayo T, Ndhlela T, Vicente FS, Nair SK, Vivek BS, Zhang X, Olsen M, Xingming F (2020) Molecular breeding for nutritionally enriched maize: status and prospects. Front Genet. https://doi.org/10.3389/fgene.2019.01392
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A 81:8014–8018
Yan J, Kandianis BC, Harjes EC, Bai L, Kim HE, Yang X, Skinner DJ, Fu Z, Mitchell S, Li Q, Fernandez GSM, Zaharoeva M, Babu R, Fu Y, Palacios N, Li J, DellaPenna D, Brutnell T, Buckler SE, Warburton LM, Rocheford T (2010) Rare genetic variation at Zea mays crtRB1 increases beta carotene in maize grain. Nat Genet 42:322–327
Yang W, Zheng Y, Ni S, Wu J (2004) Recessive allelic variations of three microsatellite sites within the o2 gene in maize. Plant Mol Biol Rep 22(4):361–374
Yang R, Yan Z, Wang Q, Li X, Feng F (2018) Marker-assisted backcrossing of lcyE for enhancement of proA in sweet corn. Euphytica 214:130
Zunjare RU, Hossain F, Muthusamy M, Baveja A, Chauhan HS, Bhat JS, Saha S, Gupta HS (2018) Development of biofortified maize hybrids through marker-assisted stacking of β-carotene hydroxylase, lycopene-ε-cyclase and opaque2 genes. Front Plant Sci. https://doi.org/10.3389/fpls.2018.00178
Acknowledgements
Thanks are due to ICAR-IIMR, Ludhiana, for providing the off-season nursery at Hyderabad. The help of Mr. Manish Kapasia, technical assistant, for management of field activities and pollination programme is thankfully acknowledged.
Code availability
Not applicable
Funding
Financial support was from ICAR-IARI, New Delhi.
Author information
Authors and Affiliations
Contributions
Conduct of the experiments, AB; generation of backcross populations, VM and AKD; phenotypic evaluation, BKM and RUZ; statistical analysis, RUZ; biochemical analysis, AB, SJM and SS; genotyping, AB and RC; drafting of manuscript, AB, VM and FH; design of experiment, FH and KKP.
Corresponding author
Ethics declarations
Ethics approval
Not applicable
Consent to participate
Not applicable
Consent for publication
All the authors have read the content and consented to submit the manuscript
Competing interests
The authors declare no competing interests.
Additional information
Communicated by: Izabela Pawłowicz
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOC 361 kb)
Rights and permissions
About this article
Cite this article
Baveja, A., Muthusamy, V., Panda, K.K. et al. Development of multinutrient-rich biofortified sweet corn hybrids through genomics-assisted selection of shrunken2, opaque2, lcyE and crtRB1 genes. J Appl Genetics 62, 419–429 (2021). https://doi.org/10.1007/s13353-021-00633-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-021-00633-4