Abstract
Background
Malnutrition affects large section of population worldwide. Vitamin A and protein deficiencies have emerged as the major global health-issue. Traditional shrunken2 (sh2)-based sweet corn is deficient in provitamin A (proA), lysine and tryptophan. Natural variant of β-carotene hydroxylase1 (crtRB1) and opaque2 (o2) enhances proA, lysine and tryptophan in maize. So far, no sweet corn hybrid rich in these nutrients has been released elsewhere. Development of biofortified sweet corn hybrids would help in providing the balanced nutrition.
Methods and Results
We targeted three sh2-based sweet corn inbreds (SWT-19, SWT-20 and SWT-21) for introgression of mutant crtRB1 and o2 genes using molecular breeding. The gene-based 3′TE-InDel and simple sequence repeat (SSR) (umc1066) markers specific to crtRB1 and o2, respectively were utilized in foreground selection in BC1F1, BC2F1 and BC2F2. Segregation distortion was observed for crtRB1 and o2 genes in majority of populations. Background selection using 91–100 SSRs revealed recovery of recurrent parent genome (RPG) up to 96%. The introgressed progenies possessed significantly higher proA (13.56 µg/g) as compared to the original versions (proA: 2.70 µg/g). Further, the introgressed progenies had accumulated moderately higher level of lysine (0.336%) and tryptophan (0.082%) over original versions (lysine: 0.154% and tryptophan: 0.038%). Kernel sweetness among introgressed progenies (17.3%) was comparable to original sweet corn (17.4%). The introgressed inbreds exhibited higher resemblance with their recurrent parents for yield and morphological characters.
Conclusion
These newly developed biofortified sweet corn genotypes hold immense promise to alleviate malnutrition.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malnutrition caused by intake of unbalanced food is a major health issue globally [1]. Nearly 2.3 billion people are moderately or severely food insecure, thereby suggesting the poor health status worldwide [2]. Nearly 45% of deaths among children (< 5 years) are linked to malnutrition [3]. Amongst various forms of nutritional disorders, deficiencies caused by vitamin-A and protein have profound effects on human health [4]. Vitamin-A deficiency (VAD) causes growth retardation, mucous membrane damage, reproductive disorders, impaired vision and night blindness [5]. VAD critically affects over seven million pregnant women and 125 million children [6]. Children and infants suffering from VAD are often deficient in other micronutrients further leading to anaemia, impaired mental growth and severe risk of morbidity [7, 8]. Protein deficiency leads to protein energy malnutrition (PEM) and in severe cases may cause ‘marasmus’ and kwashiorkor’ in humans [9]. Highest number of deaths amongst all nutritional disorders have been accounted to PEM [10]. Deficiency of lysine and tryptophan aggravates the occurrence of PEM, and they impair protein biosynthesis and other physiological functions, leading to loss of reproductive tissues, depression, anxiety, anaemia, fatigue and delayed growth [11, 12].
Sweet corn, consumed as fresh and processed vegetable has gained worldwide acceptance in both national and international markets [13]. The global import of sweet corn is US$ 1542 million, and countries like Japan, Britain, USA, Germany, China and Belgium are the leading importers of sweet corn [14]. Among various mutants, recessive shrunken2 (sh2) gene present on chromosome-3L has been abundantly used in sweet corn breeding programmes worldwide [15]. Mutant sh2-based sweet corn possesses six-fold higher sucrose and suitable for prolonged storage and transportation [16]. Large number of sh2-based sweet corn hybrids have been released for commercial cultivation worldwide [17].
Despite of the wide-reaching popularity of sweet corn, the level of provitamin A (proA) (3–4 µg/g), lysine (0.15–0.25%) and tryptophan (0.03–0.04%) is quite low in traditional sweet corn cultivars [18]. Hence, their consumption does not make major contribution to daily requirement in humans [19]. A mutant of β-carotene hydroxylase1 (crtRB1) gene is associated with accumulation of 2–10 fold higher proA in maize endosperm [20]. The recessive o2 gene can enhance lysine and tryptophan by 1.5–2-folds [21]. Biofortification of crops using plant breeding is one of the preferred choices for sustainable solution to malnutrition [4]. The gene-based markers for crtRB1 and o2 can be effectively used for their introgression into elite parental lines through marker-assisted backcross breeding (MABB) approach [22, 23]. So far, no sweet corn hybrid rich in proA, lysine and tryptophan has been released for commercial cultivation elsewhere. Considering the growing demand and wide popularity of sweet corn and significance of nutritional values, development of biofortified sweet corn cultivars with higher nutritional value hold great promise. The present investigation was therefore undertaken to (i) introgress favourable alleles of crtRB1 and o2 into elite sweet corn inbreds using marker-assisted selection (MAS), (ii) assess the MAS-derived sweet corn inbreds for nutritional quality, and (iii) evaluate the introgressed sweet corn inbreds for agronomic performance and yield attributes.
Materials and methods
Genetic materials
Plant materials used in the present study comprised of three sh2-based sweet corn inbreds viz., SWT-19, SWT-20 and SWT-21. These inbreds are the parents of two sweet corn hybrids, viz. PSSC-1 (SWT-19 × SWT-20) and ASKH6 (SWT-21 × SWT-20) developed at ICAR-Indian Agricultural Research Institute (IARI), New Delhi. PSSC-1 is a released sweet corn hybrid during 2018, while ASKH-6 is the promising sweet corn hybrid. These inbreds possess low proA, lysine and tryptophan in the kernels. These parental inbreds were targeted for introgression of favourable alleles of crtRB1 and o2 genes using MAS. PMI-PV5 with favourable allele of crtRB1 and o2 genes was used as a donor parent. PMI-PV5 was developed through the introgression of crtRB1 gene from a HarvestPlus-bred donor inbred (HP704-23) into a subtropically adapted o2-based inbred, HKI161 [24]. PMI-PV5 is a parent of five released biofortified maize hybrids in India.
Development of backcross- and self- progenies
The recurrent parents (SWT-19, SWT-20 and SWT-21) were crossed with donor parent (PMI-PV5) during winter season (December-May) (2015–16) at Winter Nursery Centre (WNC), ICAR-Indian Institute of Maize Research (IIMR), Hyderabad (17◦21′50.39′′N and 78◦29′42.31′′E). The donor was developed in our earlier programme at ICAR-IARI, New Delhi using MAS. F1s were raised at IARI, New Delhi during rainy season (July–October) (2016). F1s were backcrossed to respective recurrent parents to generate BC1F1 generation. BC1F1 seeds were separated for normal (round: sh2sh2Sh2) and shrunken (sh2sh2sh2) phenotypes [25]. BC1F1 seeds with shrunken kernels with segregation of crtRB1 and o2 genes were sown during rainy season (2017) at IARI, New Delhi. BC2F1 and BC2F2 populations (with shrunken seeds) segregating for crtRB1 and o2 genes were further raised during winter season (2017–18) and rainy season (2018) at WNC, IIMR, Hyderabad and IARI, New Delhi, respectively. BC2F3 progenies were advanced for seed increase at WNC, IIMR, Hyderabad during winter season (2018–19). The details of MABB scheme followed for the present study are depicted in Fig. S1.
DNA isolation and marker analysis
Genomic DNA was isolated from the leaves of young plants (3–4 weeks) using the modified standard Cetyl Trimethyl Ammonium Bromide (CTAB) method [26]. Polymerase chain reaction (PCR)-based DNA amplification for crtRB1 and o2 genes was carried out as per Sarika et al. [12] and Muthusamy et al. [27], respectively. The PCR-amplified products of crtRB1 (296 bp and 543 bp) and o2 (140 bp and 160 bp) were resolved in 1.5% and 4% Seakem LE agarose gel (Lonza, Rockland, ME-USA), respectively at 120 V for 2–4 h. The PCR-amplified products were resolved using agarose gel, further visualized under gel documentation system (Alpha-Innotech).
Foreground selection
3′TE-InDel marker specific to crtRB1 and gene-based SSR (umc1066) present within o2 were used for foreground selection [11, 24]. Details of the markers used for foreground selection are given in Table S1. Hybridity testing of F1s was performed using markers specific to crtRB1 and o2 genes. Since, the favourable allele of o2 and crtRB1 were introgressed from donor parent, BC1F1 and BC2F1 populations segregated for unfavourable homozygotes and heterozygotes. Thus, heterozygotes for both o2 and crtRB1 genes were selected in BC1F1 and BC2F1 populations, whereas double homozygotes for favourable allele were selected in BC2F2 populations.
Background selection
Genome-wide SSRs covering 10 chromosomes were used for recovery of recurrent parent genome (RPG) [11]. The SSRs were retrieved from Maize Genome Database (www.maizegdb.org). The selected markers were custom synthesized (Sigma Tech., USA) and used to screen polymorphic markers between recurrent and donor parents. The polymorphic SSRs were further used for RPG recovery among selected foreground positive plants in BC1F1, BC2F1 and BC2F2 generations. The amplification of SSRs was carried out using touch-down PCR procedure [28] and separated in 4% Seakem LE agarose gel for 3–4 h. The resolved PCR products were visualized using gel documentation system (Alpha-Innotech).
Evaluation of introgressed progenies
The MAS-derived introgressed progenies along with their original inbreds were evaluated in randomized complete block design (RCBD) with three replications during rainy seasons (2019 and 2020) at ICAR-IARI, New Delhi. Each entry was sown in 3 m row with row-to-row and plant-to-plant spacing of 75 cm and 20 cm, respectively. In each row, all the plants were self-pollinated to avoid any xenia effect. Two–three self-pollinated ears were harvested at 20 days after pollination (DAP) for estimation of kernel sweetness (brix), proA, lysine, and tryptophan [13]. Rest self-pollinated plants were used for recording of plant height (PH), ear height (EH), days to 50% female flowering (FF) and days to 50% male flowering (MF) and grain yield (GY). The introgressed inbreds along with their recurrent parents were further grown in RCBD with three replications at ICAR-IARI, New Delhi during rainy season (2020 and 2021) for recording of 31 DUS (distinctiveness, uniformity and stability) characters as per guidelines of Protection of Plant Varieties and Farmers' Rights Authority (PPVFRA) [29].
Estimation of kernel sweetness
Self-pollinated ears immediately after harvest were used for estimation of kernel sweetness [18]. Manually removed 20–30 grains from each genotype were bulked and crushed for estimation of brix (%) using portable refractometer (Atago, Japan). The crushed grains produced a milky juice which was used for brix reading [13].
Processing of kernels for quality analysis
The remaining grains of the freshly harvested self-pollinated ears were stored at − 80 °C freezer. A total of 50–100 self-pollinated seeds were manually removed from the ears and wrapped in Whatman® blotting paper. The wrapped seeds were incubated in sealed desiccators filled with pre-activated silica gel for 2–3 days [13]. Deactivated silica was removed after every 24 h for 2 days and replaced with freshly activated silica gel for rapid drying. Dried seeds were finely ground to a fine powder for estimation of quality parameters.
Estimation of proA
The dried powder was used to estimate proA through UHPLC system (Ultra-High-Performance Liquid Chromatography, Thermo Scientific, MA, USA. ProA was extracted using protocol given by Kurilich and Juvik [30]. Quantification of β-carotene (BC) and β-cryptoxanthin (BCX) was done using YMC C30 column (5 μm, 4.6 × 250 mm), and individual components were detected at an absorbance of 450 nm wavelength using photodiode array detector (PDA detector). The regression curves were prepared using the standards of BC and BCX (Sigma Aldrich, USA) to determine the final concentration of each carotenoid in each sample. The concentration of proA was estimated as ‘BC + ½(BCX)’ [31].
Estimation of lysine and tryptophan
Lysine and tryptophan from the dried powder was used for estimation of lysine and tryptophan through the same UHPLC system as per the protocol of Sarika et al. [12]. Sample digestion for estimation of lysine was carried out using acid hydrolysis, while basic hydrolysis was used for estimation of tryptophan. Acclaim™ 120 C18 column (5 μm, 120Ao, 4.6 × 150 mm, Thermo Scientific) was used for sample elution. PDA detector with absorbance at 265 and 280 nm wavelength was used for detection of lysine and tryptophan, respectively. The standard regression curve was prepared using external standards of amino acids (AAS 18-5 ML, Sigma Aldrich) for estimating final concentration of lysine and tryptophan in each sample.
Statistical analysis
The goodness of fit test of the observed segregation pattern of sh2, crtRB1 and o2 genes in BC1F1, BC2F1 and BC2F2 generations was performed using chi-square analysis. Recovery of RPG in the selected plants was calculated using the formula: [RPG (in %) = {(A + 1/2H)/ (A + B + H)} × 100], wherein, ‘A’ is the number of SSRs homozygous for the recipient parent, ‘B’ is the number of SSRs homozygous for the donor parent, and ‘H’ is the number of heterozygous SSRs [32]. Graphical representations of mean lysine, tryptophan and proA were determined by Microsoft Excel-2013. Morphological and biochemical data analysis for standard error (SD) and critical difference (CD) was performed using Windostat 10.0 software.
Results
Parental polymorphism
Screening with crtRB1 specific 3’TE-InDel marker produced an amplicon of 296 bp in all three recurrent parents, while donor parent generated an amplicon of 543 bp. Three gene-based SSRs (umc1066, phi057 and phi112) present within o2 were used, of these umc1066 clearly differentiated the recurrent- and donor- parents. The donor parent produced an amplicon of 140 bp, while recurrent parents produced 160 bp amplicon. All three recurrent parents possessed unfavourable alleles, while the donor had the favourable alleles of crtRB1 and o2 genes. Further, parental polymorphism for background selection was conducted using 183 SSRs, of which 100, 95 and 91 markers were polymorphic between recurrent parents (SWT-19, SWT-20 and SWT-21) and donor line, respectively. This accounted 54.64%, 51.91% and 49.73%, polymorphism for (i) SWT-19 × PMI-PV5, (ii) SWT-20 × PMI-PV5, and (iii) SWT-21 × PMI-PV5 populations (Table S2). The polymorphic markers per chromosome ranged from 4 to 15 across crosses. The polymorphic markers hence identified were utilized to recover RPG through background selection in backcross- and self-generations.
Segregation of normal and shrunken seeds in BC1F1 generations
The BC1F1 seeds segregated in round (sh2sh2Sh2) and shrunken (sh2sh2sh2) shapes. Since, the recurrent parents possessed sh2sh2 (recessive homozygous) genotype and F1 hybrids were heterozygous (Sh2sh2), the BC1F1 seeds showed two types of phenotypes. The BC1F1 seeds segregated as per Mendelian ratio of 1 (round): 1 (shrunken) in SWT-19 and SWT-20 -based populations, while it showed segregation distortion in SWT-21 -based population (Table 1).
Marker-assisted selection
F1 generation
The polymorphic markers for crtRB1 and o2 genes confirmed the hybridity of F1 plants in all the three crosses. This indicated the absence of any self-pollinated plants in the F1 generations.
BC1F1 generation
A total number of 112, 127 and 96 plants were genotyped in SWT-19, SWT-20 and SWT-21 -based populations, respectively. Chi square analysis for crtRB1 and o2 revealed that all three populations significantly deviated from Mendelian ratio (1:1) except for o2 in SWT-20 -based population (Table 2). Background selection in heterozygous plants led to a recovery of 69.6–77.7% RPG across the populations (Table S3). Two plants in SWT-19 (SWT-19-3 and SWT-19-62), three plants in SWT-20 (SWT-20-5, SWT-20-54 and SWT-20-96) and four plants in SWT-21 (SWT-21-3, SWT-21-18, SWT-21-49 and SWT-21-90) with higher recovery of RPG were further advanced.
BC2F1 generation
Foreground selection was performed among 147, 241 and 159 plants in SWT-19, SWT-20 and SWT-21-based populations, respectively. Chi-square analysis revealed that two (SWT-19 and SWT-20 based) populations for o2 and one (SWT-20 based) population for crtRB1 significantly deviated from Mendelian ratio (1:1) (Table 2). Recovery of RPG through background selection ranged from 79.2 to 85.9% across populations (Table S3). Two plants each in SWT-19 (SWT-19-3-14 and SWT-19-62-87) and SWT-20 (SWT-20-54-62 and SWT-20-54-84), and three plants in SWT-21 (SWT-21-3-1, SWT-21-3-10 and SWT-21-3-18) derived progenies were selected for further advancement.
BC2F2 generation
Segregating generations with 251, 211 and 213 plant were genotyped in SWT-19, SWT-20 and SWT-21 -based populations, respectively (Fig. S2). Chi-square analysis for o2 revealed that two (SWT-20 and SWT-21 based) populations significantly deviated from expected Mendelian ratio (1:2:1); while segregation distortion for crtRB1 was observed in all three populations (Table 2). RPG recovery ranged from 88.10 to 93.70%, 87.50–95.80% and 89.40–95.00% among in SWT-1-19, SWT-20 and SWT-21-based populations, respectively (Table S3). Three progenies viz., SWT-19-3-14-94, SWT-19-62-87-43 and SWT-19-62-87-49 of SWT19 background were selected, while two progenies each in SWT20 (SWT-20-54-84-4 and SWT-20-54-84-52) and SWT21 (SWT-21-3-1-65 and SWT-21-3-1-70) were advanced. Graphical genotyping (GGT) of recovery among introgressed progenies is given in Fig. S3.
ProA among introgressed inbreds
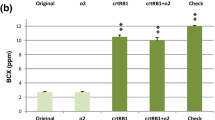
The MAS-derived progenies possessed significantly higher BC (10.10 μg/g), BCX (6.92 μg/g) and proA (13.56 μg/g) over the original inbreds (BC: 1.44 μg/g, BCX: 2.50 μg/g and proA: 2.70 μg/g) (Fig. 1). Among introgressed progenies, SWT-19-62-87-43 (BC: 12.00 μg/g, BCX: 7.61 μg/g, proA: 15.80 μg/g) was the best inbred in SWT-19 background, while SWT-20-54-84-4 (BC: 10.44 μg/g, BCX: 7.46 μg/g, proA: 14.18 μg/g) and SWT-21-3-1-65 (BC: 11.67 μg/g, BCX: 4.93 μg/g, proA: 14.14 μg/g) were the most promising in SWT-20 and SWT-21 background, respectively (Table 3).
Lysine and tryptophan among introgressed inbreds
The introgressed progenies possessed high lysine (0.336%) and tryptophan (0.082%) as compared to original inbreds (lysine: 0.154%, tryptophan: 0.038%). The lysine of introgressed inbreds ranged from 0.306 to 0.373%, while the range for tryptophan was 0.073–0.090% (Figs. 2, 3). Of the introgressed progenies, SWT-19-62-87-43 (lysine: 0.311%, tryptophan: 0.090%) and SWT-20-54-84-4 (lysine: 0.344%, tryptophan: 0.083%) were identified as the most promising inbred for SWT-19 and SWT-20 genetic background, respectively, while SWT-21-3-1-70 (lysine: 0.373%, tryptophan: 0.078%) was the best in SWT-21 background (Table 3).
Kernel sweetness among introgressed inbreds
The introgressed inbreds displayed similar kernel sweetness (brix: 17.3%) with recurrent parents (brix: 17.4%). The brix among MAS-derived progenies ranged from 16.4 to 18.0%, while the same in original sweet corn inbreds was 17.5% (SWT-19), 16.9% (SWT-20) and 17.8% (SWT-21), respectively (Fig. 4). Among the introgressed progenies, SWT-19-62-87-43 possessed the highest brix (18.0%) in SWT-19 background, while SWT-20-54-84-4 (17.0%) and SWT-21-3-1-70 (17.8%) were the most promising progenies in SWT-20 and SWT-21 background (Table 3).
Phenotypic selection and evaluation of introgressed inbreds
The average dry grain yield (1314 kg/ha) of the introgressed progenies was at par with their original inbreds (1309 kg/ha) (Table S4). Among the progenies, SWT-19-3-14-94 (1436 kg/ha), SWT-20-54-84-4 (1313 kg/ha) and SWT-21-3-1-65 (1220 kg/ha) were the most promising in each of the three genetic backgrounds. Agronomic characters (plant height, ear height and flowering behaviour) of the introgressed inbreds were also similar to their original versions (Table S4). DUS characterization displayed high resemblance between parents and their respective introgressed progenies. However, phenotypic contrast was observed in few cases, for example, attitude and width of blade in leaves was straight and medium in SWT-19, while SWT-19-3-14-94 possessed drooping and broad leaves. Similarly, the angle between main axis and lateral branches in tassel was narrow in SWT-19 and SWT-21, while it was wide in SWT-19-62-87-43 and SWT-21-3-1-65 (Table S5). The density of spikelet in tassel of SWT-19 and SWT-20 was sparse, however SWT-19-62-87-49 and SWT-20-54-84-52 displayed dense spikelets.
Discussion
Globally, the preference and demand of sweet corn has increased steadily in the last decade [17]. Sweet corn consumed as fresh at milky stage possesses nutritional benefits for human health [13]. Traditional sweet corn cultivars lack recommended amount of essential amino acids and proA to meet the daily requirement [19, 33]. Development of biofortified maize with higher lysine, tryptophan and proA in sweet corn possesses great significance to mitigate malnutrition problems, especially in children and pregnant women [13]. Here, we targeted three elite sweet corn inbreds for higher proA, lysine and tryptophan through genomic-assisted breeding for crtRB1 and o2 genes.
Molecular breeding for crtRB1 and o2 genes
Molecular markers for crtRB1 (crtRB1-3′ TE-InDel) and o2 (umc1066) genes were successfully used for the selection of individual plants with favourable alleles of crtRB1 and o2 in the segregating populations [23, 24]. Since gene-based markers were used to identify the target genes, chance of selection of false positive plants due to crossing over between the marker and gene in case of linked markers was not encountered in the study [24]. Segregation distortion (SD) for crtRB1 and o2 gene observed in some of the populations in BC1F1, BC2F1 and BC2F2 could be due to defective kernel mutants, male sterility, specific mutations in embryo and other gametophytic factors [25]. Earlier, SD was also reported by various authors for o2 [11, 13, 33] and crtRB1 [24, 27], thereby necessitating large backcross population size for selecting desirable positive segregants. Further, background selection ensured high recovery of RPG resulting in high resemblance of introgressed progenies with their respective parents in terms of plant, ear and grain characteristics. The introgressed inbreds possessed up to 96% recovery of RPG ensuring higher degree of phenotypic similarity with the original parents [11]. Background selection has been practiced widely in the MABB programme for higher recovery of RPG in maize [11, 12].
Nutritional superiority of introgressed inbreds
The significant amount of proA (five folds) observed among the introgressed inbreds was due to the presence of favourable allele of crtRB1. The mutant allele of crtRB1 located on chromosome-10L reduces the hydroxylation of BC into BCX and further to zeaxanthin, thereby leading to accumulation of higher proA [20]. The introgressed inbreds also possessed significantly higher lysine (2.2-fold) and tryptophan (2.1- fold) compared to the original inbreds due to presence of recessive o2 gene located on the chromosome-7L [21]. The basic leucine zipper (bZIP) transcriptional factor coded by o2 reduces the expression of deficient 22-kDa α-zein genes with concurrent increase in lysine and tryptophan rich non-zein proteins [34, 35]. Besides, reduced activity of lysine keto-reductase enzyme that degrades lysine content and enhancement of various lysine-rich proteins are also responsible for enhancement of lysine [11].
Evaluation for yield and agronomic traits
The introgressed progenies displayed high degree of similarities with their original parents with respect to morphology, plant architecture and agronomic characters. The flowering behaviour of the introgressed progenies resembled the original parents. DUS characterization of the introgressed progenies also displayed high degree of similarity for majority of the characters. The grain yield of the introgressed inbreds was similar to their respective parental lines. This is attributed to the background selection where indirect selection for loci associated with agronomic traits and heterosis was performed [11]. Minor differences in the performance are due to interaction of donor parent genome and recurrent parent genome [13].
Acceleration of breeding cycle and cost-effectiveness
In the present study, two-generation based backcross breeding approach led to the development of biofortified sweet corn inbreds through marker-assisted introgression of crtRB1 and o2 genes. These MAS-derived inbreds are very close to their respective recurrent parents for phenotypic, agronomic and yield characters due to high recovery of RPG. Conventional breeding would have taken at least 4–5 backcrosses to achieve comparable results. Due to recessive nature of crtRB1 and o2 genes, each backcross would be have required one generation of selfing after every generation of backcross in traditional backcross breeding approach. Together, conventional breeding would have taken 11–12 seasons for introgression of crtRB1 and o2 genes in the parental sweet corn inbreds compared to 5–6 seasons required in molecular breeding [18]. The present study, thus developed biofortified sweet corn inbreds through accelerating the breeding cycle. In addition, phenotyping of large number of segregating progenies in backcross populations for high proA, lysine and tryptophan using UHPLC is quite expensive (US$ 25–30/trait/sample) compared to the marker-based genotyping which involves US$ 0.5–1/gene/sample. Globally, several researchers have preferred MAS approach to develop nutritionally rich maize with crtRB1 [27], o2 [11] and crtRB1 + o2 [23, 24] over the traditional breeding.
Utilization in breeding programme
The newly developed sweet corn inbreds with crtRB1 and o2 genes possess parental background of two sweet corn hybrids viz., PSSC-1 and ASKH-6. PSSC-1 released in India has wider adaptability in four of the five agroecological zones. ASKH-6 is a promising sweet corn hybrid and has great potential to be released for commercial cultivation. The newly developed biofortified sweet corn inbreds can be used to reconstitute the PSSC-1 and ASKH-6, and their biofortified versions can be tested for their eventual release. So far, no sweet corn hybrid with higher proA, lysine and tryptophan has been released elsewhere. These novel sweet corn hybrids would not only provide higher yield, but also provide better nutrition to the humans. Further, these newly developed inbreds can serve as the valuable donors in the sweet corn breeding programme. Several field corn inbreds with (i) crtRB1, (ii) o2 and (iii) o2 + crtRB1 have been developed by various release groups [11, 23, 24, 27]. However, sh2-based donors with o2 + crtRB1 are quite limited in the sweet corn background. These sweet corn inbreds thus can be effectively used to enrich the sh2-based germplasm for lysine, tryptophan and proA.
Conclusion
Traditional sweet corn lacks desired amount of proA, besides being deficient essential amino acids, lysine and tryptophan. Here, crtRB1 and o2 genes were combined in three elite sh2-based background using MABB strategy. Molecular markers helped in accelerating the breeding cycle, and was found to be cost effective over UHPLC based phenotyping. The introgressed progenies possessed five fold higher proA and two fold higher lysine and tryptophan compared to the parental lines. These nutrient-rich sweet corn inbreds would play vital role in the sweet corn breeding programme to help in eradicating malnutrition in sustainable and cost-effective way.
Data availability
All the data sets supporting the conclusion of this article are included within the article, and its supporting information files are provided as accompanying supplementary materials.
References
von Grebmer K, Bernstein J, Resnick D, Wiemers M, Reiner L, Bachmeier M, Hanano A, Towey O, Chéilleachair RN, Foley C, Gitter S, Larocque G, Fritschel H (2022) Global Hunger Index: Food Systems Transformation and Local Governance. Bonn: Welthungerhilfe; and Dublin: Concern Worldwide.
Global Nutrition Report (2022). Stronger commitments for greater action. Executive summary. www.globalnutritionreport.org.
World Health Organization (2021). Malnutrition; Available online: https://www.who.int/en/news-room/fact-sheets/detail/malnutrition Accessed on 8 December 2022
Hossain F, Zunjare RU, Muthusamy V, Kumar A, Madhavan J, Ikkurti G, Katral A, Talukder ZA, Chhabra R, Chand G, Bhatt V, Gul I, Mishra SJ, Duo H, Dutta S, Gain N, Chauhan P, Maman S, Reddappa SB, Kasana R (2023) Genetic improvement of specialty corn for nutritional quality traits. In: Wani SH et al (eds) Boo: maize improvement. Springer International Publishing, Chem, pp 235–257. https://doi.org/10.1007/978-3-031-21640-4_11
Lonzano-Alejo N, Carrillo VG, Pixley K, Rojas NP (2007) Physical properties and carotenoid content of maize kernels and its nixtamalized snacks. Innov Food Sci Emerg Technol 8:385–389. https://doi.org/10.1016/j.ifset.2007.03.015
Giuliano G (2017) Provitamin A biofortification of crop plants: a gold rush with many miners. Curr Opin Biotechnol 44:169–180. https://doi.org/10.1016/j.copbio.2017.02.001
Rice AL, West KP Jr, Black RE (2004) Vitamin A deficiency. Comp Quantification Health Risks 1:0211–0256
Caulfield LE, Richard SA, Rivera JA, Musgrove P, Black RE (2006) Stunting, wasting, and micronutrient deficiency disorders. Disease Control Priorities in Developing Countries. 2nd edition.
Bain LE, Awah PK, Geraldine N, Kindong NP, Siga Y, Bernard N, Tanjeko AT (2013) Malnutrition in sub-Saharan Africa: burden, causes and prospects. Pan Afr Med J 15:1–9. https://doi.org/10.11604/pamj.2013.15.120.2535]
Nyakurwa CS, Gasura E, Mabasa S (2017) Potential for quality protein maize for reducing protein energy undernutrition in maize dependent sub-saharan African countries: a review. Afr Crop Sci J 25:521–537. https://doi.org/10.4314/acsj.v25i4.9
Hossain F, Muthusamy V, Pandey N, Vishwakarma AK, Baveja A, Zunjare RU, Thirunavukkarasu N, Saha S, Manjaiah KM, Prasanna BM, Gupta HS (2018) Marker-assisted introgression of opaque2 allele for rapid conversion of elite hybrids into quality protein maize. J Genet 97:287–298. https://doi.org/10.1007/s12041-018-0914-z
Sarika K, Hossain F, Muthusamy V, Zunjare RU, Baveja A, Goswami R, Bhat JS, Saha S, Gupta HS (2018) Marker-assisted pyramiding of opaque2 and novel opaque16 genes for further enrichment of lysine and tryptophan in sub-tropical maize. Plant Sci J 272:142–152. https://doi.org/10.1016/j.plantsci.2018.04.014
Mehta BK, Muthusamy V, Baveja A, Chauhan HS, Chhabra R, Bhatt V, Chand G, Zunjare RU, Singh AK, Hossain F (2020) Composition analysis of lysine, tryptophan and provitamin-A during different stages of kernel development in biofortified sweet corn. J Food Compos Anal 94:103625. https://doi.org/10.1016/j.jfca.2020.103625
FAOSTAT (2020) https://www.fao.org/faostat/en/#data (data accessed on 12 December, 2022).
Lertrat K, Pulam T (2007) Breeding for increased sweetness in sweet corn. Int J Plant Breed Genet 1:27–30
Feng ZL, Liu J, Fu FL, Li WC (2008) Molecular mechanism of sweet and waxy in maize. Int J Plant Breed Genet 2:93–100. https://doi.org/10.3923/ijpbg.2008.93.100
Chhabra R, Muthusamy V, Baveja A, Katral A, Mehta B, Zunjare RU, Hossain F (2022) Allelic variation in shrunken2 gene affecting kernel sweetness in exotic-and indigenous-maize inbreds. PLoS ONE 17(9):e0274732. https://doi.org/10.1371/journal.pone.0274732
Baveja A, Muthusamy V, Panda KK, Zunjare RU, Das AK, Chhabra R, Mishra SJ, Mehta BK, Saha S, Hossain F (2021) Development of multinutrient-rich biofortified sweet corn hybrids through genomics-assisted selection of shrunken2, opaque2, lcyE and crtRB1 genes. J Appl Genet 62(3):419–429. https://doi.org/10.1007/s13353-021-00633-4
Feng F, Wang Q, Liang C, Yang R, Li X (2015) Enhancement of tocopherols in sweet corn by marker-assisted backcrossing of ZmVTE4. Euphytica 206:513–521. https://doi.org/10.1007/s10681-015-1519-8
Yan J, Kandianis BC, Harjes EC, Bai L, Kim HE, Yang X, Skinner DJ, Fu Z, Mitchell S, Li Q, Fernandez GSM, Zaharoeva M, Babu R, Fu Y, Palacios N, Li J, DallaPanna D, Brutnall T, Buckler SE, Warburton LM, Rocheford T (2010) Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat Genet 42:322–327. https://doi.org/10.1038/ng.551
Mertz ET, Bates LS, Nelson OE (1964) Mutant genes that change protein composition and increase lysine content of maize endosperm. Science 145:279–280. https://doi.org/10.1126/science.145.3629.279
Gupta HS, Vignesh M, Hossain F, Nepolean T (2013) Enrichment of nutritional qualities in maize through marker-assisted selection. National Seminar on Genomics for Crop Improvement, IBAB, Bangalore, pp 69–70.
Chandrasekharan N, Ramanathan N, Pukalenthy B, Chandran S, Manickam D, Adhimoolam K, Natesan (2022) Development of β-carotene, lysine, and tryptophan-rich maize (Zea mays) inbreds through marker-assisted gene pyramiding. Sci Rep 12(1):8551
Zunjare RU, Hossain F, Muthusamy V, Baveja A, Chauhan HS, Bhat JS, Thirunavukkarasu N, Saha S, Gupta HS (2018) Development of biofortified maize hybrids through marker-assisted stacking of β-carotene hydroxylase, lycopene-ε-cyclase and opaque2 genes. Front Plant Sci 9:178. https://doi.org/10.1371/journal.pone.0113583
Neuffer MG, Coe EH, Wessler SR (1997) Mutants of maize. Cold Spring Harbor Laboratory Press, New York
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325. https://doi.org/10.1093/nar/8.19.4321
Muthusamy V, Hossain F, Thirunavakkarasu N, Choudhary M, Saha S, Bhat JS, Prasanna BM, Gupta HS (2014) Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase Allele. PLoS ONE 9(12):e113583
Duo H, Hossain F, Muthusamy V, Zunjare RU, Goswami R, Chand G, Mishra SJ, Chhabra R, Gowda MM, Pal S, Baveja A, Bhat JS, Kamboj MC, Kumar B, Amalraj JJ, Khulbe R, Prakash B, Neeraja CN, Rakshit RS, Yadav OP (2021) Development of sub-tropically adapted diverse provitamin-A rich maize inbreds through marker assisted pedigree selection, their characterization and utilization in hybrid breeding. PLoS ONE 16(2):e0245497. https://doi.org/10.3382/ps.2011-01719
PPVFRA (2007) Guidelines for the conduct of test for distinctiveness, uniformity and stability on maize (Zea mays L.). Plant Var J India 1(1):1–13
Kurilich AC, Juvik JA (1999) Quantification of carotenoid and tocopherol antioxidants in Zea mays. J Agric Food Chem 47:1948–1955. https://doi.org/10.1021/jf981029d
Babu R, Rojas NP, Gao S, Yan J, Pixley K (2013) Validation of the effects of molecular marker polymorphisms in LcyE and CrtRB1 on provitamin A concentrations for 26 tropical maize populations. Theor Appl Genet 126:389–399. https://doi.org/10.1007/s00122-012-1987-3
Chand G, Muthusamy V, Allen T, Zunjare RU, Mishra SJ, Singh B, Mehta BK, Talukder ZA, Ismail MR, Sarika K, Kamboj MC (2022) Composition of lysine and tryptophan among biofortified-maize possessing novel combination of opaque2 and opaque16 genes. J Food Compos Anal 107:104376. https://doi.org/10.1016/j.jfca.2021.104376
Chauhan HS, Muthusamy V, Rashmi T, Basu S, Anand A, Mehta BK, Gain N, Zunjare RU, Singh AK, Gupta HS, Hossain F (2022) Characterization of crtRB1-and vte4-based biofortified sweet corn inbreds for seed vigour and physico-biochemical traits. J Appl Genet 63(4):651–662. https://doi.org/10.1007/s13353-022-00715-x
Kodrzycki R, Boston RS, Larkins BA (1989) The opaque-2 mutation of maize differentially reduces zein gene transcription. Plant Cell 1(1):105–114. https://doi.org/10.1105/tpc.1.1.105
Ueda T, Waverczak W, Ward K, Sher N, Ketudat M, Schmidt RJ, Messing J (1992) Mutations of the 22-and 27-kD zein promoters affect transactivation by the opaque-2 protein. Plant Cell 4(6):701–709. https://doi.org/10.1105/tpc.4.6.701
Acknowledgements
We thank WNC, IIMR, Hyderabad for providing the off-season nursery.
Funding
We are thankful to the ICAR-IARI, New Delhi for financial support.
Author information
Authors and Affiliations
Contributions
Conduct of the experiment: BS, Generation of backcross populations: RUZ, Recording of phenotypic data: GC and VB, Estimation of quality parameters: NG, Statistical analysis: VM, Writing of manuscript: BS, GC and FH, Designing of experiment: FH and SS.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that no conflict of interest exits.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, B., Zunjare, R.U., Shrivastava, S. et al. Provitamin A, lysine and tryptophan enrichment in shrunken2-based sweet corn genotypes through genomics-assisted breeding for crtRB1 and opaque2 genes. Mol Biol Rep 50, 4965–4974 (2023). https://doi.org/10.1007/s11033-023-08446-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08446-w