Abstract
Purpose
Endothelial cell (EC) dysfunction underlies the pathology of multiple disease conditions including cardiovascular and pulmonary diseases. Dysfunctional ECs have a distinctive gene expression profile compared to healthy ECs. RNAi therapy is a powerful therapeutic approach that can be used to silence multiple genes of interests simultaneously. However, the delivery of RNAi to ECs in vivo continues to be a major challenge. Here, we optimized a polymer formulation based on poly(β-amino ester)s (pBAEs) to deliver siRNA to vascular ECs.
Methods
We developed a library of bioinspired oligopeptide-modified pBAE nanoparticles (NPs) with different physicochemical proprieties and screened them for cellular uptake and efficacy of RNAi delivery in vitro using ECs, vascular smooth muscle cells, and THP-1 monocytes. From the screening, the lysine-/histidine-oligopeptide modified pBAE (C6-KH) NP was selected and further tested ex vivo using mouse aorta and in mice to determine efficiency of siRNA delivery in vivo.
Results
The in vitro screening study showed that C6-KH was most efficient in delivering siRNA to ECs. Ex vivo study showed that C6-KH nanoparticles containing siRNAs accumulated in the endothelial layer of mouse aortas. In vivo study showed that C6-KH nanoparticles carrying siICAM2 injected via tail-vein in mice significantly reduced ICAM2 level in the artery endothelium (55%), lung (52%), and kidney (31%), but not in the liver, heart, and thymus, indicating a tissue-specific delivery pattern.
Conclusions
We demonstrate that C6-KH pBAE can used for delivery of siRNAs to the artery endothelium and lung, while minimizing potential side or toxic effects in the liver and heart.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
A healthy vascular system is of critical importance in the maintenance of tissue homeostasis and disease. Endothelial cells (ECs) lining the vasculature play critical roles in regulation of vascular permeability, thrombogenicity, immune responses, and vasodilation and vasoconstriction, and endothelial dysfunction is the primary indication of vascular disease.22,23 Endothelial dysfunction is mediated in large part through changes in gene expression,15 which could serve as targets of RNAi therapy.28 RNAi therapies can be easily designed with high specificity with minimum non-specific effects unlike typical pharmacological inhibitors.3,11,15,29,31,32,33 However, delivery of RNAi, such as siRNAs, to ECs in vivo continues to be a major challenge for their therapeutic and research use. In addition, targeted delivery of RNAi to specific ECs, such as healthy vs. diseased regions of vessels in vivo, while avoiding off-target effects and toxicity in other tissues is still a major issue to be solved.30
Non-viral synthetic nanoparticles are a relatively safe and efficacious approach that allows for RNAi stability and increased delivery efficacy. Lipid and polymeric nanoparticles represent a broad class of biomaterials that can deliver RNAi, however, they tend to accumulate in hepatocytes as the lipophilic particles interact with serum lipoproteins.10,12,18,27 Cationic lipids have been used to deliver siRNAs to ECs, however, they often require multiple administrations at high doses to achieve a robust therapeutic effect,13,24,25 raising concerns for potential side effects and tissue toxicity.14,16 7C1 is a low molecular weight lipid-polymer hybrid formulation, which showed an efficient siRNA delivery to ECs with high knockdown efficiency, while avoiding significant effects in hepatocytes and immune cells in mice.2 While 7C1 is one of the first successful nanoparticles that shows endothelial-targeted delivery of siRNAs, it does not discriminate between healthy vs. diseased regions. Therefore, additional nanoparticles need to be developed for improved specificity.
Poly(β-amino ester)s (pBAEs) are attractive candidates as carrier vehicles for siRNAs due to their high biodegradability and biocompatibility.5,17,26 pBAEs are easy to synthetize by the co-polymerization of diacrylate and primary amines, and simple to modify with natural or synthetic amines to improve transfection efficiencies.9,17,26,34 The presence of amines that become protonated in their structure facilitates siRNA endosomal escape through the proton sponge effect in cells.1 We recently developed a large library of stable and bioinspired oligopeptide-modified pBAE nanoparticle (NPs) with various physicochemical proprieties that demonstrate efficient nucleic acid delivery in cancer cells in vitro.4,5,21,26 However, it was unknown whether pBAE NPs can deliver siRNAs to ECs in vivo.
Here, we screened the pBAE nanoparticle libraries and identified a formulation composed of lysine- and histidine- oligopeptide-modified pBAEs (C6-KH), which can efficiently deliver siRNA to ICAM2 (siICAM2) to ECs both in vitro and in vivo. The C6-KH nanoparticle loaded with siICAM2 reduced ICAM2 in the arterial endothelium, lung, but not the liver and heart, indicating a tissue-specific delivery pattern.
Materials and Methods
Materials
Reagents and solvents used for polymer synthesis were purchased from Sigma-Aldrich and Panreac. Oligopeptide moieties used on the polymer modification (H-Cys-Arg-Arg-Arg-NH2, H-Cys-Lys-Lys-Lys-NH2, H-Cys-His-His-His-NH2, and H-Cys-Asp-Asp-Asp-NH2) were obtained from GL Biochem (Shanghai) Ltd. with a purity of at least 98%. Labeled siRNA (AllStars Neg. siRNA AF 555) for uptake experiments was purchased from Qiagen. ON-TARGETplus Non-Targeting Control Pool (D-001810-10) used as siRNA control was obtained from Thermo GE Dharmacon. Polyplus Interferin reagent was purchased from VWR and used according to manufacturer instructions. siRNA against ICAM2 (termed siICAM2) with forward sequence 5′-AGGACGGUCUCAACUUUUC-3′ and reverse sequence 5′-GAAAAGUUGAGACCGUCCU-3′ and siRNA against luciferase (used as a negative control and termed siCON) were used, all obtained from AXOlabs. RNA extraction was carried out using the Direct-zol RNA kit from Genesee Scientific, cDNA reaction was performed using the Applied Biosystems cDNA kit, and cDNA quantification was analyzed using SYBR green from Affymetrix, following the manufacturer’s instructions.
Cell Lines and Animals
Immortalized mouse aortic endothelial cells (iMAECs) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum, 100 U mL−1 penicillin, 100 µg mL−1 streptomycin, 0.1 mM Non-Essential Amino Acids (NEAA), 2mM L-glutamine, and 50 µg mL−1 ECGS (bovine brain extract 1%) as we reported.20 Smooth Muscle cells (SMC) were obtained from ATCC (CRL-2797) and maintained in DMEM supplemented with 10% fetal bovine serum, 100 U mL−1 penicillin, 100 µg mL−1 streptomycin, and 2 mM L-glutamine. Monocytic cell line (THP-1) was obtained from ATCC (TIB-202) and maintained in RPMI medium supplemented with 10% fetal bovine serum, 100 U mL−1 penicillin, 100 µg mL−1 streptomycin, and 0.05 mM of 2-mercaptoethanol. All cells were cultured at 37 °C, under a 5% CO2/95% air atmosphere until 90% confluence before starting transfections.
All mouse procedures were conducted at Emory University under the protocol approved for this study by the Emory Institutional Animal Care and Use Committee (IACUC). C57BL/6 mice 8 week-old males were used to minimize potential sex-dependent differences for each condition.
Synthesis of Oligopeptide-Modified C6 Polymer
Synthesis of pBAEs was performed via a two-step procedure, as previously described.4 (1) Addition reaction of primary amines to an excess of diacrylates was used to synthesize an acrylate-terminated polymer (termed C6 polymer). Briefly, C6 polymerization was performed by mixing 5-amino-1-pentanol (0.426 g, 4.1 mmol), hexylamine (0.422 g, 4.1 mmol), and 1,4-butanediol diacrylate (2.0 g, 9.1 mmol) under magnetic stirring at 90 °C for 24 h. (2) C6 polymer was end-capped with different thiol-terminated oligopeptides composed of Cys + 3 amino acids (Arg, Lys, His, Asp, or Glu) at a 1:2.1 molar ratio in dimethyl sulfoxide, allowing to react overnight at room temperature. The resulting polymers were collected by precipitation in a mixture of diethyl ether and acetone (7:3). Synthesized structures were confirmed by 1H-NMR, recorded in a 400 MHz Varian (NMR Instruments, Claredon Hills, IL) and methanol-d4 used as a solvent. Molecular weight (MW) relative to polystyrene standard was determined by HPLC (HPLC Elite LaChrom system of VWR-Hitech equipped with a GPC Shodex KF-603 column and THF as mobile phase).
Biophysical Characterization of Oligopeptide End-Modified pBAEs
Nanoparticles were prepared at 75:1 C6 polymer:siRNA ratio, by mixing equal volumes of siRNA at 0.05 mg mL−1 with the polymer at 3.75 mg mL−1 in sodium acetate buffer (AcONa) at 12.5 mM. The siRNA was added over the polymer solution and mixed by pipetting, followed by 10 min incubation at 25 °C. For the formation of discrete nanoparticles, this mixture (volume = Vi) was nanoprecipitated with the same volume (Vi) of PBS 1x. Then, the resulting nanoparticles were characterized by an agarose gel retardation assay and dynamic light scattering (DLS).
To assess RNAi retardation, different siRNA-to-polymer ratios (w:w) between 10:1 and 400:1 were studied. C6 polymer–siRNA nanoparticles were freshly prepared and added to wells of agarose gel (2.5%, containing 1 μg mL−1 ethidium bromide). Samples were run at 80 V for 45 min (Apelex PS 305, France) and visualized by UV illumination.
Hydrodynamic diameter (nm), polydispersity index (PDI), and surface charge of nanoparticles were measured by ZetaSizer Nano ZS (Malvern Instruments Ltd, United Kingdom, 4 mW laser) at 25 °C using 633 nm laser wavelength and 173° signal detector. Nanoparticles were synthesized as previously described and diluted five times with PBS 1x for further analysis.
Endothelial Cell-Specific Polymer Formulation Screening
Polymer screening of end-modified oligopeptide C6 polymers were carried out using fluorescent siRNA-AF555 (siCONF) in iMAECs, SMC, and THP-1. Briefly, cells were seeded in a 12-well plate at 80,000 cells well−1 and incubated overnight to roughly 80% confluence prior to performing the transfection experiments. Nanoparticles were synthesized as described previously at a 75:1 w:w ratio, diluted in complete DMEM medium, and added to the cells at a final siCONF concentration of 50 nM. Cells were incubated for 2 h in 37 °C at 5% CO2 atmosphere. After 2 h of incubation, cells were washed, and nanoparticle internalization was measured by flow cytometry (BD LSRFortessa cell analyzer). siCONF fluorescence between formulations was compared against untreated cells.
Ex Vivo Delivery of Labeled siRNA to Vasculature
Mice were sacrificed using a CO2 chamber and perfused with saline containing heparin (10 U/mL) via the vena cava. The abdominal aorta was isolated and carefully cleaned of peri-adventitial fat. Aortas were maintained in PBS 1x at 4 °C until their use. 300 µL of siCONF at 200nM encapsulated within oligopeptide-end modified pBAEs were injected through the abdominal aorta and incubated for 1 h at 37 °C and 5% CO2. After incubation, the abdominal aortic tissue was washed twice using PBS, fixed in 4% paraformaldehyde, and analyzed by confocal microscopy. All the formulations were tested in triplicates.
En Face Staining
The samples were counter-stained using DAPI and mounted en-face on glass slides using fluorescence mounting medium (Dako). Imaging was done using a Zeiss-LSM 510 META confocal microscope (Carl Zeiss).
Histology Staining
PFA-fixed samples were placed in OCT medium and frozen at − 80 °C. Histology sections at 8 µm were carried out using Leica CM1950. Samples were washed twice using PBS 1x and were counter-stained using DAPI. Finally, samples were mounted on coverslips using fluorescence mounting medium (Dako). Imaging was done using a Zeiss-LSM 510 META confocal microscope (Carl Zeiss).
In Vitro Delivery of siICAM2
iMAECs were seeded in a 12-well plate at 80,000 cells well−1 and incubated overnight to roughly 80% confluence prior to performing the siICAM2 delivery. Cells were transfected with C6-KH nanoparticle with siICAM2 at different siRNA concentrations, ranging from 10 to 100 nM. At 48 h post-transfection, the medium was removed, and the cells were washed with PBS and collected for RT-qPCR analysis. Control siRNA (termed siCON) at 100 nM was used as a negative control of ICAM2 knockdown. ICAM2 expression was normalized to siCON and plotted as percentage of gene silencing.
Cell Viability Assay
The influence of C6-KH nanoparticles on cell viability was determined using the MTS assay (CellTiter 96® AQueous One Solution Cell Proliferation Assay, Promega Corporation, USA) at 48 h post-transfection as instructed by the manufacturer. Briefly, transfection assay was performed as earlier described. 48 h later, the medium was removed, cells were washed with PBS 1x and complete medium supplemented with 20% MTS reagent (v:v) was added. Cells were incubated at 37 °C and absorbance was measured every 30 min using a plate reader until appropriate absorbance values were obtained.
Lyophilization of PBAE Nanoparticles for In Vivo Studies
Nanoparticles were generated by mixing an equal volume of siRNA at 0.5 µg/µL with C6-KH polymer at 37.5 µg/µL in AcONa buffer solution (12.5 mM, pH 5.5). The solution was mixed by pipetting for a few seconds. After 10 min of incubation at room temperature, 200 µL of nanoparticles was precipitated in the same volume of DEPC water. Thereafter, the same volume of a HEPES 20 mM containing 4 wt% sucrose solution (added as crio and lioprotectors) was added. The nanoparticles were then freeze-dried, and the lyophilized powder was kept at – 20 °C until use, when it was reconstituted with DEPC water (120 µL).
siICAM2-C6-KH Efficacy Study
Mice (n = 5) were injected intravenously with 50 µg (120 µL) of siICAM2 or siCON with C6-KH polymer. 48 h post-injection, ICAM2 expression was analyzed in the lung, liver, spleen, thymus, heart, and kidneys. Briefly, mice were sacrificed by CO2 inhalation according to Emory University’s IACUC protocol and perfused with saline solution. The organs were collected and snap-frozen in liquid nitrogen. Then, they were crushed using a mortar and pestle device and lysed with QIAzol for RNA extraction and qPCR analysis. ICAM2, PECAM-1, CD45, and 18S (housekeeping) genes were analyzed by RT-qPCR.
Isolation of Endothelial-Enriched RNA
Mice (n = 5) were injected intravenously with 50 µg (120 µL) of siICAM2 or siCON with C6-KH polymer. At 48 h post-injection, endothelial-enriched RNA from the carotids was extracted as we previously described.19 First, mice were sacrificed by CO2 inhalation according to Emory University’s IACUC protocol and perfused with saline containing heparin (10 U/mL) via the left ventricle after severing the inferior abdominal aorta. The left common carotid artery (LCA) and the right common carotid artery (RCA) were then isolated and carefully cleaned of periadventitial fat. Then, the carotid lumen was quickly flushed with 150 µL of QIAzol® lysis reagent using a 29-gauge insulin syringe into an Eppendorf tube. The eluate was snap-frozen in liquid nitrogen for RNA extraction. Additionally, media and adventitia (smooth-muscle cell enriched) were snap-frozen in liquid nitrogen, pulverized by mortar and pestle, and lysed with QIAzol® lysis reagent (300 µL).
RNA Isolation and RT-qPCR Analysis
Total RNA obtained from carotids was extracted and purified using Direct-zol™ RNA Kits according to the manufacturer’s instructions. The amount and purity of RNA was determined by NanoDrop 1000 Spectrophotometer. After RNA isolation, the RNA was reverse-transcribed using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene expression was determined by RT-qPCR using gene-specific primers (sequences listed in Supplementary Table 1). The expressions of genes of interest were normalized against the 18S housekeeping gene. All analyses were performed using the 2−ΔΔCT method.
Statistical Analysis
Statistical analyses were carried out using GraphPad Prism to assess significant differences between experimental groups. Pairwise comparisons were performed using one-way Student t tests. Differences between groups were considered significant at p values below 0.05 (*p < 0.05, **p < 0.01, ***p < 0.001). All data are reported as mean ± SEMs.
Results
Synthesis and Characterization of a Library of Oligopeptide-Modified pBAE Nanoparticles
Synthesis of end-modified C6 polymer was performed via a two-step procedure, as we described previously.4 First, acrylate-terminated C6 polymer was obtained by conjugate addition of 5-amino-1-pentanol and hexylamine to 1,4-butanediol diacrylate using a slight excess of diacrylate (Fig. 1a). Polymerization was confirmed by HPLC-SEC and the resulting C6 polymer had an apparent average molecular weight of approximately 2500 g/mol (relative to polystyrene standards) and a polydispersity (Mw/Mn) of 1.80, showing a rather broad statistical distribution of polymer chain lengths. The molecular weight was further confirmed by 1H-NMR, and average of 2100 g/mol was obtained. Then, C6 polymer was further modified with cationic and anionic oligopeptide moieties via addition of the thiol group of cysteine-terminated oligopeptides to the acrylate-terminated end-groups of C6 polymer (Fig. 1a). Oligopeptide-terminated pBAEs were characterized in terms of molecular structure by 1H-NMR and FT-IR. The chemical structure of the resultant polymers was confirmed by the disappearance of acrylate signals and the presence of signals typically associated with oligopeptides (Supplementary Information).
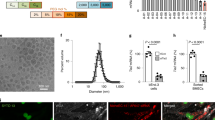
Synthesis and characterization of oligopeptide modified C6 polymer library. (a) Structure and synthetic scheme of oligopeptide-modified pBAEs. R” terminal can be arginine-, lysine-, histidine-, glutamic acid- and aspartic acid- oligopeptides. (b) Polyplexes with different surface proprieties were obtained by combining different oligopeptide moieties. (c) Average hydrodynamic diameter, polydispersity, and zeta potential determination of pBAE:siICAM2 nanoparticles prepared at 75:1 ratio (w:w) with different oligopeptide-modified pBAE polymers were determined by DLS. n = 3, data shown as mean ± SEM.
We created a library of pBAE-based nanoparticles using different oligopeptides or oligopeptide mixtures (Fig. 1b). As we previously demonstrated, oligopeptide end-modification plays an important role in nanoparticle physicochemical properties, where nanoparticles with different size and surface charge can be obtained.5,26 To assess nanoparticle physiochemical properties, freshly prepared nanoparticles were characterized by size (hydrodynamic diameter) and charge (zeta-potential) by Dynamic Light Scattering (DLS). Nanometric size (below 200 nm) and positive surface charge were observed when using cationic or mixtures of different cationic oligopeptides moieties (R, K, RH, KH, and RK). In contrast, bigger nanoparticle sizes were observed when using mixtures of positively- and negatively charged oligopeptides, C6-RD and C6-KD (Fig. 1c). The addition of anionic oligopeptides was used to tune the final nanoparticle surface charge, ranging from negative to positive.
Oligopeptide-Modified pBAE Screening in iMAECs, SMC and THP-1 Cells
To select a polymer formulation that efficiently delivers siRNA to ECs, we screened our oligopeptide-modified pBAE polymers using AF555-labeled siRNA (siCONF) in various cell lines. For this study, iMAECs, SMC, and THP-1 were used and siCONF uptake efficiency was determined by flow cytometry (Fig. 2). Uptake analysis showed differential specificity profiles for each cell line depending on the pBAE formulation. pBAEs end-modified with oligopeptides containing mixtures of lysine-histidine (C6-KH) and lysine-aspartic acid (C6-KD) showed the highest levels of cellular uptake in iMAECs, achieving up to a 2-fold increase when compared to the other formulations (Fig. 2a). In general, SMCs showed a higher cellular uptake compared to iMAECs or THP-1 cells. Arginine-modified pBAEs showed higher uptake in SMCs than formulations containing lysine, and the highest levels of uptake was achieved using arginine-histidine mixtures (C6-RH). Interestingly, C6-KH and C6-KD formulations showed the highest uptake in iMAECs and the lowest uptake in SMCs compared to the other polymer formulations (Fig. 2b). Further, THP-1 cells showed a lower cellular uptake than the iMAECs and SMCs, where the highest siCONF uptake was observed when arginine-modified pBAEs were used (Fig. 2c). Based on these results, we selected to use C6-KH for the next studies.
Cellular uptake of siCONF using different oligopeptide-modified pBAE polymers in (a) iMAEC, (b) SMC and (c) THP-1 cells. Cells were transfected with siCONF at final concentration of 20 nM and fluorescence expression per cell was determined after 2 h post-transfection by flow cytometry. n = 4, data shown as mean ± SEM; *p < 0.05, **p < 0.01, ***p < 0.001.
Ex Vivo siRNA Delivery Using C6-KH Formulation to Mouse Abdominal Aorta
Next, we tested if siCONF can be delivered to mouse aortic ECs by encapsulating it with the C6-KH polymer (siCONF-C6-KH). For this study, mouse abdominal aortas were dissected out and were incubated ex vivo with siCONF-C6-KH or naked siCONF (200nM). Confocal imaging of the cross section of the aorta incubated with siCONF-C6-KH or naked siCONF showed that siCONF-C6-KH signal is predominantly on the endothelial layer, but not in the medial SMCs (Fig. 3a and Fig. S3). Further, en face confocal imaging of the abdominal aorta showed abundant fluorescence signals in the endothelial layer when incubated siCONF-C6-KH, but not naked siCONF (Fig. 3b). In contrast, when arginine was used instead of the lysine (C6-RH nanoparticle), it was much less efficient in the delivery of siRNA compared to C6-KH nanoparticles (Fig. S2). Therefore, the C6-KH formulation, which consists of 50% C6-K and 50% C6-H, was selected as the most appropriate end-oligopeptide modification to efficiently deliver siRNA to endothelial cells in vitro and ex vivo.
siCONF delivery to endothelial layer. Abdominal aortas of C57Bl6 mice were dissected out and incubated ex vivo with siCONF-C6-KH or naked siCONF for 1 h, followed by either frozen cross-section (a) or en face (b) imaging by confocal microscopy. Blue DAPI for nuclear, Green auto-fluorescence for elastic laminas, Red AF555 for siCONF are shown.
In Vitro siICAM2 Delivery to iMAECs Using C6-KH Polymer
Next, we tested whether the C6-KH nanoparticle can deliver siRNAs to effectively knockdown a gene of interest in the endothelium. For this, we encapsulated siICAM2 or siCON (non-targeted siRNA as a control) with C6-KH to knockdown ICAM2 in iMAECs. As shown in Fig. 4a, siICAM2-C6-KH nanoparticles reduced ICAM2 level in a concentration dependent manner, reaching a 70% knockdown at 20 nM (Fig. 4a). C6-KH polymer showed no evidence of cytotoxicity as determined by the MTS assay (Fig. 4b). Moreover, treatment with siICAM2-C6-KH did not induce statistically significant induction of inflammatory markers, vascular cell adhesion molecule 1 (VCAM1) and tumor necrosis factor alpha (TNF-α) in comparison to the siCON (Fig. 4c). In addition, we did not observe any noticeable changes in the morphology of iMAECs in this siICAM2-C6-KH study even at 100 nM (data not shown). These results suggest that siICAM2-C6-KH can effectively knockdown ICAM2 in mouse endothelial cells.
Efficient ICAM2 knockdown in iMAECs. iMAECs were transfected with an increasing concentration of siICAM2 encapsulated with C6-KH. (a) ICAM2 expression was determined 48 h post-transfection by RT-qPCR, (b) cell viability of iMAECs were determined by the MTS assay, and (c) VCAM1 and TNF-α expression was analyzed by qPCR. n = 4, mean ± SEM; ***p < 0.001.
siICAM2 Delivery In Vivo Using C6-KH Nanoparticles
Next, we tested whether siICAM2-C6-KH can knockdown ICAM2 in arterial endothelial cells in vivo by injecting mice via tail-vein with siICAM2-C6-KH or siCON-C6-KH (2 mg/kg). At 48 h post-injection, we collected RNAs from endothelial-enriched (intima) and leftover (LO) containing medial layer enriched with SMCs. As we reported previously, the endothelial-enriched fraction showed a high expression of PECAM1 and low level of marker genes of SMCs (SM22a) and leukocytes (CD45).19 In contrast, the leftover samples contained abundant SM22a with low PECAM1 and CD45 (Figs. 5a and 5b). Injection of siICAM2-C6-KH significantly reduced ICAM2 level by 55% compared to the siCON-C6-KH control in the endothelial enriched samples. No significant ICAM2 reduction was observed in SMC-enriched samples, which contained barely detectable level of ICAM2, confirming endothelial-specific expression of ICAM2 (Fig. 5c).
Efficient in vivo ICAM2 knockdown in the endothelium when siICAM2 was delivered using C6-KH polymer. (a) siICAM2-C6-KH and siCON-C6-KH were injected at 2 mg/kg in mice. Two days post-injection, endothelial-enriched (intima) and SMC-enriched fraction (leftover) RNAs were prepared and analyzed by qPCR for gene expression. (b) As a quality control, expression of markers of ECs (PECAM-1), medial SMCs (SM22a), and leukocytes (CD45) were determined, whereas (c) ICAM2 expression was analyzed by RT-qPCR. n = 5, mean ± SEM; ***p < 0.001.
Reduction of ICAM2 Expression in the Lung
Here, we tested whether siICAM2-C6-KH nanoparticles can effectively reduce ICAM2 level in the major organs of the mice using the RT-qPCR method. A single dose of nanoparticles containing siICAM2 were injected at 2 mg/kg by systemic administration and ICAM2 expression levels was evaluated 48 h after delivery in different organs. siCON was used as a negative control (Fig. 6). We found that siICAM2-C6-KH nanoparticles significantly reduced ICAM2 levels in the lung by 50% and in the kidney by 31%, while it was not altered in the spleen, thymus, liver and heart. We also found that ICAM2 expression in the lung, spleen, thymus, and heart was much higher than the kidney and liver.
Tissue-specific effects of siICAM2-C6-KH nanoparticles as a surrogate marker of biodistribution. Following injection of siICAM2-C6-KH or siCON-C6-KH in the mice used in Fig. 5, various organs were prepared and analyzed by RT-qPCR for ICAM2 expression. n = 5, data shown as mean ± SEM; *p < 0.05, **p < 0.01, ns not significant.
Discussion
We demonstrated that C6-KH nanoparticles can deliver siRNA to the artery endothelium, lung, and kidney, while avoiding some key tissues such as the liver, heart and thymus. This tissue-specific delivery pattern of siRNAs may be of significant advantage in vascular and pulmonary therapeutics while minimizing potential side effects or toxicity in the liver and heart.
pBAEs have been used to deliver different types of nucleic acids, from small RNAi’s to large plasmids.5,21,26 Nucleic acids are electrostatically entrapped in pBAEs forming discrete nanoparticles and protecting them from non-specific degradation. pBAE NPs display excellent endosomal escape capability once they are internalized by cells due to the presence of ternary amines in their structure.1,17 Previously, we synthetized a library of pBAE polymers using various natural oligopeptides (K, R, H, D, and E) displaying a large array of physicochemical proprieties.5,26 It demonstrated that small changes in the structure of these polymers have a remarkable impact on the design of cell-specific nanoparticles. In this study, in vitro screening results showed that the C6-KH formulation was the most efficient formulation for delivering siRNA to ECs (Fig. 2). The underlying mechanisms for endothelial delivery of siRNAs using the C6-KH nanoparticles is unknown. Nevertheless, lysine (K) may facilitate nanoparticle interactions with the cell surface through its protonated primary amine, whereas histidine (H) may stimulate endosomal escape of siRNAs because of its ternary amine.1,26 In addition, nanoparticles with a positive surface charge interact with the proteoglycans on the cell membrane, facilitating their preferential delivery to endothelial cells or SMCs.8 In contrast, negatively charged particles are easily recognized by phagocytic cells. The surface charges of oligopeptide-modified pBAEs range from 10 to 20 mV, making pBAEs less efficiently internalized by THP-1 cells compared to iMAECs or SMCs in our study (Fig. 2). Although we did not test the stability of the nanoparticles in vivo, we previously showed that the C6-KH nanoparticles are stable in the medium containing 10% fetal bovine serum for at least 24 h.4
We found that siICAM2-C6-KH nanoparticle reduced ICAM2 in the lung and kidney, but not in the liver, thymus, and heart (Fig. 6). While the specific underlying mechanism for the tissue-specific effects of siICAM2-C6-KH nanoparticles is not known, its inability to knockdown ICAM2 in the liver and heart could be a crucial benefit to avoid potential side effects or toxicity in those tissues. Recently, we demonstrated that C6-KH nanoparticles show reduced liver accumulation.6 We further demonstrated that biodistribution of pBAE nanoparticles can be tuned using different targeting moieties. When a liver targeting moiety, such as retinol, was added, the pBAE nanoparticles preferentially accumulated in the liver due in part to the protein corona formed in a retinol-dependent manner.7 Given the prevalence of liver and heart toxicities for most drugs and gene therapeutics, the ability of C6-KH nanoparticles to deliver siRNAs could provide an important tool to deliver therapeutics safely.
Conclusion
In summary, we have developed a pBAE nanoparticle that can deliver siRNAs to certain tissues such as the artery endothelium, lung, and kidney, but not in the liver and heart. Interestingly, C6-KH NPs accumulate in the arterial endothelial layer, which may be due to the internal elastic laminar preventing its penetration to the medial layer. This could be used to target endothelial genes in the artery, lung, and kidney, while avoiding other critical major organs such as the liver and heart. This tissue-specific delivery pattern of siRNAs by C6-KH could be a significant advantage in RNAi therapeutics for vascular and pulmonary diseases, while minimizing the potential side-effects or toxicity in the liver and heart.
References
Behr, J. The proton sponge: a trick to enter cells the viruses did not exploit. Chimia 2:34–36, 1997.
Dahlman, J. E., C. Barnes, O. F. Khan, A. Thiriot, S. Jhunjunwala, T. E. Shaw, Y. Xing, H. B. Sager, G. Sahay, L. Speciner, A. Bader, R. L. Bogorad, H. Yin, T. Racie, Y. Dong, S. Jiang, D. Seedorf, A. Dave, K. Singh Sandhu, M. J. Webber, T. Novobrantseva, V. M. Ruda, A. K. R. R. Lytton-Jean, C. G. Levins, B. Kalish, D. K. Mudge, M. Perez, L. Abezgauz, P. Dutta, L. Smith, K. Charisse, M. W. Kieran, K. Fitzgerald, M. Nahrendorf, D. Danino, R. M. Tuder, U. H. Von Andrian, A. Akinc, D. Panigrahy, A. Schroeder, V. Koteliansky, R. Langer, and D. G. Anderson. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol. 9:648–655, 2014. https://doi.org/10.1038/nnano.2014.84.
Dai, G., M. R. Kaazempur-Mofrad, S. Natarajan, Y. Zhang, S. Vaughn, B. R. Blackman, R. D. Kamm, G. Garcia-Cardena, and M. A. Gimbrone. Distinct endothelial phenotypes evoked by arterial waveforms derived from atherosclerosis-susceptible and -resistant regions of human vasculature. Proc. Natl. Acad. Sci. 101:14871–14876, 2004. https://doi.org/10.1073/pnas.0406073101.
Dosta, P., V. Ramos, and S. Borrós. Stable and efficient generation of poly(β-amino ester)s for RNAi delivery. Mol. Syst. Des. Eng. 3:677–689, 2018. https://doi.org/10.1039/C8ME00006A.
Dosta, P., N. Segovia, A. Cascante, V. Ramos, and S. Borrós. Surface charge tunability as a powerful strategy to control electrostatic interaction for high efficiency silencing, using tailored oligopeptide-modified poly(beta-amino ester)s (PBAEs). Acta Biomater. 20:82–93, 2015. https://doi.org/10.1016/j.actbio.2015.03.029.
Fornaguera, C., M. Guerra-Rebollo, M. Ángel Lázaro, C. Castells-Sala, O. Meca-Cortés, V. Ramos-Pérez, A. Cascante, N. Rubio, J. Blanco, and S. Borrós. mRNA delivery system for targeting antigen-presenting cells in vivo. Adv. Healthc. Mater. 7:1800335, 2018. https://doi.org/10.1002/adhm.201800335.
Fornaguera, C., M. Guerra-Rebollo, M. Á. Lázaro, A. Cascante, N. Rubio, J. Blanco, and S. Borrós. In vivo retargeting of poly(beta aminoester) (OM-PBAE) nanoparticles is influenced by protein Corona. Adv. Healthc. Mater. 8:1900849, 2019. https://doi.org/10.1002/adhm.201900849.
Fröhlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 7:5577, 2012. https://doi.org/10.2147/IJN.S36111.
Green, J. J., R. Langer, and D. G. Anderson. A combinatorial polymer library approach yields insight into nonviral gene delivery. Acc. Chem. Res. 41:749–759, 2008. https://doi.org/10.1021/ar7002336.
Hajj, K. A., J. R. Melamed, N. Chaudhary, N. G. Lamson, R. L. Ball, S. S. Yerneni, and K. A. Whitehead. A potent branched-tail lipid nanoparticle enables multiplexed mRNA delivery and gene editing in vivo. Nano Lett. 2020. https://doi.org/10.1021/acs.nanolett.0c00596.
Heath, J. M., J. Fernandez Esmerats, L. Khambouneheuang, S. Kumar, R. Simmons, and H. Jo. Mechanosensitive microRNA-181b regulates aortic valve endothelial matrix degradation by targeting TIMP3. Cardiovasc. Eng. Technol. 9:141–150, 2018. https://doi.org/10.1007/s13239-017-0296-z.
Kanasty, R., J. R. Dorkin, A. Vegas, and D. Anderson. Delivery materials for siRNA therapeutics. Nat. Mater. 12:967–977, 2013. https://doi.org/10.1038/nmat3765.
Kaufmann, J., K. Ahrens, and A. Santel. RNA interference for therapy in the vascular endothelium. Microvasc. Res. 80:286–293, 2010. https://doi.org/10.1016/j.mvr.2010.02.002.
Knudsen, K. B., H. Northeved, P. E. K. Kumar, A. Permin, T. Gjetting, T. L. Andresen, S. Larsen, K. M. Wegener, J. Lykkesfeldt, K. Jantzen, S. Loft, P. Møller, and M. Roursgaard. In vivo toxicity of cationic micelles and liposomes. Nanomed. Nanotechnol. Biol. Med. 11:467–477, 2015. https://doi.org/10.1016/j.nano.2014.08.004.
Kumar, S., D. Williams, S. Sur, J.-Y. Wang, and H. Jo. Role of flow-sensitive microRNAs and long noncoding RNAs in vascular dysfunction and atherosclerosis. Vascul. Pharmacol. 114:76–92, 2019. https://doi.org/10.1016/j.vph.2018.10.001.
Lv, H., S. Zhang, B. Wang, S. Cui, and J. Yan. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release. 114:100–109, 2006. https://doi.org/10.1016/j.jconrel.2006.04.014.
Lynn, D. M., and R. Langer. Degradable poly(β-amino esters): synthesis, characterization, and self-assembly with plasmid DNA. J. Am. Chem. Soc. 122:10761–10768, 2000. https://doi.org/10.1021/ja0015388.
Miao, L., J. Lin, Y. Huang, L. Li, D. Delcassian, Y. Ge, Y. Shi, and D. G. Anderson. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 11:2424, 2020. https://doi.org/10.1038/s41467-020-16248-y.
Nam, D., C.-W. Ni, A. Rezvan, J. Suo, K. Budzyn, A. Llanos, D. Harrison, D. Giddens, and H. Jo. Partial carotid ligation is a model of acutely induced disturbed flow, leading to rapid endothelial dysfunction and atherosclerosis. Am. J. Physiol. Circ. Physiol. 297:H1535–H1543, 2009. https://doi.org/10.1152/ajpheart.00510.2009.
Ni, C.-W., S. Kumar, C. J. Ankeny, and H. Jo. Development of immortalized mouse aortic endothelial cell lines. Vasc. Cell 6:7, 2014. https://doi.org/10.1186/2045-824X-6-7.
Núñez-Toldrà, R., P. Dosta, S. Montori, V. Ramos, M. Atari, and S. Borrós. Improvement of osteogenesis in dental pulp pluripotent-like stem cells by oligopeptide-modified poly(β-amino ester)s. Acta Biomater. 53:152–164, 2017. https://doi.org/10.1016/j.actbio.2017.01.077.
Pober, J. S., and W. C. Sessa. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 7:803–815, 2007. https://doi.org/10.1038/nri2171.
Pober, J. S., and W. C. Sessa. Inflammation and the blood microvascular system. Cold Spring Harb. Perspect. Biol. 7:a016345, 2015. https://doi.org/10.1101/cshperspect.a016345.
Santel, A., M. Aleku, O. Keil, J. Endruschat, V. Esche, B. Durieux, K. Löffler, M. Fechtner, T. Röhl, G. Fisch, S. Dames, W. Arnold, K. Giese, A. Klippel, and J. Kaufmann. RNA interference in the mouse vascular endothelium by systemic administration of siRNA-lipoplexes for cancer therapy. Gene Ther. 13:1360–1370, 2006. https://doi.org/10.1038/sj.gt.3302778.
Santel, A., M. Aleku, O. Keil, J. Endruschat, V. Esche, G. Fisch, S. Dames, K. Löffler, M. Fechtner, W. Arnold, K. Giese, A. Klippel, and J. Kaufmann. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 13:1222–1234, 2006. https://doi.org/10.1038/sj.gt.3302777.
Segovia, N., P. Dosta, A. Cascante, V. Ramos, and S. Borrós. Oligopeptide-terminated poly(β-amino ester)s for highly efficient gene delivery and intracellular localization. Acta Biomater. 10:2147–2158, 2014. https://doi.org/10.1016/j.actbio.2013.12.054.
Semple, S. C., A. Akinc, J. Chen, A. P. Sandhu, B. L. Mui, C. K. Cho, D. W. Y. Sah, D. Stebbing, E. J. Crosley, E. Yaworski, I. M. Hafez, J. R. Dorkin, J. Qin, K. Lam, K. G. Rajeev, K. F. Wong, L. B. Jeffs, L. Nechev, M. L. Eisenhardt, M. Jayaraman, M. Kazem, M. A. Maier, M. Srinivasulu, M. J. Weinstein, Q. Chen, R. Alvarez, S. A. Barros, S. De, S. K. Klimuk, T. Borland, V. Kosovrasti, W. L. Cantley, Y. K. Tam, M. Manoharan, M. A. Ciufolini, M. A. Tracy, A. de Fougerolles, I. MacLachlan, P. R. Cullis, T. D. Madden, and M. J. Hope. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 28:172–176, 2010. https://doi.org/10.1038/nbt.1602.
Setten, R. L., J. J. Rossi, and S. Han. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 18:421–446, 2019. https://doi.org/10.1038/s41573-019-0017-4.
Son, D. J., S. Kumar, W. Takabe, C. WooKim, C.-W. Ni, N. Alberts-Grill, I.-H. Jang, S. Kim, W. Kim, S. WonKang, A. H. Baker, J. WoongSeo, K. W. Ferrara, and H. Jo. The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nat. Commun. 4:3000, 2013. https://doi.org/10.1038/ncomms4000.
Soutschek, J., A. Akinc, B. Bramlage, K. Charisse, R. Constien, M. Donoghue, S. Elbashir, A. Geick, P. Hadwiger, J. Harborth, M. John, V. Kesavan, G. Lavine, R. K. Pandey, T. Racie, K. G. Rajeev, I. Röhl, I. Toudjarska, G. Wang, S. Wuschko, D. Bumcrot, V. Koteliansky, S. Limmer, M. Manoharan, and H.-P. Vornlocher. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 432:173–178, 2004. https://doi.org/10.1038/nature03121.
Tang, F., and T.-L. Yang. MicroRNA-126 alleviates endothelial cells injury in atherosclerosis by restoring autophagic flux via inhibiting of PI3K/Akt/mTOR pathway. Biochem. Biophys. Res. Commun. 495:1482–1489, 2018. https://doi.org/10.1016/j.bbrc.2017.12.001.
Vaishnaw, A. K., J. Gollob, C. Gamba-Vitalo, R. Hutabarat, D. Sah, R. Meyers, T. de Fougerolles, and J. Maraganore. A status report on RNAi therapeutics. Silence. 1:14, 2010. https://doi.org/10.1186/1758-907X-1-14.
Zampetaki, A., S. Kiechl, I. Drozdov, P. Willeit, U. Mayr, M. Prokopi, A. Mayr, S. Weger, F. Oberhollenzer, E. Bonora, A. Shah, J. Willeit, and M. Mayr. Plasma MicroRNA profiling reveals loss of endothelial MiR-126 and other MicroRNAs in Type 2 diabetes. Circ. Res. 107:810–817, 2010. https://doi.org/10.1161/CIRCRESAHA.110.226357.
Zugates, G. T., N. C. Tedford, A. Zumbuehl, S. Jhunjhunwala, C. S. Kang, L. G. Griffith, D. Lauffenburger, R. Langer, and D. G. Anderson. Gene Delivery properties of end-modified poly(β-amino ester)s. Bioconjug. Chem. 18:1887–1896, 2007. https://doi.org/10.1021/bc7002082.
Author Contributions
PD: Conception and design, Collection and assembly of data, Data analysis and interpretation, Manuscript writing. CD: Data analysis and interpretation, Manuscript writing. VR: Conception and design Data analysis and interpretation, Manuscript writing, Final approval of manuscript. SK: Collection of data. DWK: Collection of data. HJ: Conception and design, Data analysis and interpretation, Manuscript writing, Final approval of manuscript. SB: Conception and design, Data analysis and interpretation, Manuscript writing, Final approval of manuscript.
Funding
This work was supported by funding from the National Institutes of Health grants HL119798 and HL095070 to HJ. It was also supported by funding from the Spanish Ministerio de Ciencia, Innovación y Universidades for the Grant RTI2018-094734-B-C22 to SB. PD received the financial support from AGAUR (Generalitat de Catalunya) 2017FI_B2 00141. HJ was supported by John and Jan Portman Professorship and Wallace H. Coulter Distinguished Faculty Professorship. This study was also funded by Grup d’Enginyeria dels Materials (GEMAT). GEMAT would like to acknowledge Agència de Gestió d’ajuts Universitaris i de Recerca, Generalitat de Catalunya (SGR 2017) nº 1559.
Conflict of interest
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Associate Editor Keefe B. Manning oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dosta, P., Demos, C., Ramos, V. et al. Delivery of siRNA to Endothelial Cells In Vivo Using Lysine/Histidine Oligopeptide-Modified Poly(β-amino ester) Nanoparticles. Cardiovasc Eng Tech 12, 114–125 (2021). https://doi.org/10.1007/s13239-021-00518-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13239-021-00518-x