Abstract
Apple ring rot inflicts severe economic losses in the main apple producing areas of East Asia. The causal agent of the disease has been variously identified as Macrophoma kuwatsukai, Physalospora piricola and Botryosphaeria berengeriana f. sp. piricola, although B. dothidea is currently the most widely accepted pathogen name. The taxonomic uncertainty has delayed research that is needed to manage effectively this destructive disease. In the present study, genealogical concordance phylogenetic species recognition (GCPSR) was applied to pathogenic fungal isolates from apple and pear from several locations in China, along with several reference isolates. Phylogenetic results based on sequences of four nuclear loci (ITS, EF-1α, HIS and HSP) revealed the existence of two species within the examined isolates. One includes an ex-epitype isolate of B. dothidea and the other includes an isolate that was previously designated as B. berengeriana f. sp. piricola. Morphologically, the latter taxon presented an appressed mycelial mat on PDA whereas B. dothidea displayed columns of aerial mycelia reaching the lids, and conidia of the latter species were longer than B. dothidea. Botryosphaeria dothidea had a faster growth rate than the latter taxon under relatively high temperatures. Pathogenicity tests showed that on pear stems the latter taxon caused large-scale cankers along with blisters whereas B. dothidea was non-pathogenic, but on apple shoots the two fungi induced large and small wart-like prominences, respectively. Overall, this cryptic species demonstrated sufficient genetic variations and biological differences from B. dothidea. As a result of taxonomic study, we described here the latter taxon in a new combination, Botryosphaeria kuwatsukai and designate an epitype. Both B. kuwatsukai and B. dothidea are considered to be the main causal agents for apple ring rot in China and Japan.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ring rot has recently become one of the most destructive apple diseases in China, as well as in several neighboring countries, including Japan and South Korea (Ogata et al. 2000; Park 2005; Tang et al. 2012). Symptoms of the disease appear as a soft, light-coloured rot on fruit, especially during storage, and extensive cankers and/or warts on branches and trunks (Chen 1999). Widespread planting of susceptible cultivars (e.g., Fuji) over the past several years and reduced fungicide usage due to fruit bagging have likely resulted in the increased occurrence of ring rot, resulting in serious economic losses to Chinese apple growers (Kang et al. 2009).
Apple ring rot disease was first reported in Japan in 1907. The pathogen was later described as Macrophoma kuwatsukai Hara (Hara 1930). Soon afterwards, the sexual morph of the pathogen was found and named Physalospora piricola (Nose 1933). This taxon had long been known as the causal agent of apple ring rot in other countries. Koganezawa and Sakuma (1980, 1984) reappraised the sexual morph and tried to equate it with the morphologically identical Botryosphaeria berengeriana, a fungus causing apple Botryosphaeria canker (and fruit rot) in Japan. However, distinctly different symptoms, cankers and wart bark, caused by B. berengeriana and by P. piricola, respectively, on branches and trunks resulted in the authors proposing the name Botryosphaeria berengeriana f. sp. piricola for Physalospora piricola (Koganezawa and Sakuma 1984). As Botryosphaeria berengeriana and B. berengeriana f. sp. piricola induced the same apple rot symptom, diseases caused on fruit were referred to both as apple ring rot (Koganezawa and Sakuma 1984). These two pathogen taxons are generally accepted in Japan, but they are rejected by European and American researchers who consider B. berengeriana to be a synonym of B. dothidea (Jones and Aldwinckle 1990; Slippers et al. 2004a). In China, the correct taxonomy of the apple ring rot pathogen is uncertain, in that Physalospora piricola, Botryosphaeria berengeriana, B. berengeriana f. sp. piricola and B. dothidea have been adopted by different researchers (Qu et al. 2007; Peng et al. 2011; Lv et al. 2012; Tang et al. 2012).
The application of molecular techniques to the taxa associated with apple ring rot has shown that genetic heterogeneity exists among these pathogenic fungal isolates. Ogata et al. (2000), using ITS sequences, divided isolates of Botryosphaeria causing ring rot on apple fruit into two groups based on twig symptoms (warts or blight) and size of conidia. Huang and Liu (2001) showed that there was marked a difference between B. berengeriana and B. berengeriana f. sp. piricola by RAPD analysis, in spite of their close genetic relationship. Peng et al. (2011) separated isolates from apple ring rot into two ISSR groups. Lv et al. (2012) proposed that the isolates causing apple ring rot with different variable sites in ITS sequences might behave differently in terms of pathogenicity and some biological characteristics (e.g., the ability to sporulate). Xu et al. (2013) genotyped B. dothidea isolates (including some from apple trees) according to the distribution of ribosomal group I introns. Four genotypes were described and the authors indicated that there may be a correlation between these genotypes and host or geographic origin. The high amount of variation detected by the different approaches suggests that there is a mixture of pathogens rather than a single pathogenic species that causes apple ring rot.
Accurate identification of etiological factors is the foundation of plant disease research. Vague classification and confused appellation of the apple ring rot pathogen has led to the inability to compare research results from different countries, regions and investigators. Therefore, the objectives of this study were to: 1) re-assess the Botryosphaeria isolates associated with symptoms of apple ring rot, including fruit rot, branch canker and wart bark in China, 2) detect the reasons for the high amount of variation among isolates, and 3) clarify whether any cryptic fungal species exist within the hypothetical pathogenic complex. Our approaches to the study included genealogical concordance phylogenetic species recognition (GCPSR) of multi-gene loci (Taylor et al. 2000), as well as analysis of morphology, pathogenicity and growth characteristics.
Materials and methods
Fungal isolates
Twenty-four isolates (Table 1) were used in this study, 18 of which were collected in the main apple production areas of China (Shaanxi, Henan, Shandong, Liaoning, Jiangsu, Shanxi, Hebei Provinces) during 2008–2011. Of these 18 isolates, 16 were either from warts or cankers on apple branches and trunks or apple fruit with rot symptoms; and two were isolated from the diseased woody tissue of pear exhibiting canker symptoms. The other six were reference isolates, including one isolate each of B. dothidea, B. berengeriana and B. berengeriana f. sp. piricola from the Fruit Tree Research Experiment Station of the Ministry of Agriculture, Forestry and Fisheries (MAFF), Japan, two isolates of B. dothidea from the International Collection of Microorganisms from Plants (ICMP), New Zealand, and one ex-epitype isolate of B. dothidea from the Centraalbureau voor Schimmelcultures (CBS), Netherlands. All the isolates collected in this research were obtained from either pycnidia directly or diseased tissue by cultivating on potato dextrose agar (PDA), and were examined for colony characteristics and microscopic morphology to preliminarily identify them as Botryosphaeria spp. They were then purified by single conidium isolation and stored at −80 °C in College of Plant Protection, Northwest A&F University, China.
DNA extraction, PCR amplification and sequencing
Single-conidial isolates were grown on PDA and incubated for 5 days at 25 °C in the dark. Total genomic DNA was extracted from fungal mycelium following the modified phenol : chloroform DNA extraction method (Smith et al. 2001). Four different gene regions were selected for characterization, including the complete nuclear rDNA internal transcribed spacer (ITS) region (White et al. 1990), partial sequence of translation elongation factor 1 alfa (EF-1α), histone H3 (HIS) and heat shock protein (HSP) genes (Inderbitzin et al. 2010). The polymerase chain reaction (PCR) was performed in a reaction mixture of 25 μl containing approximately 10–30 ng fungal genomic DNA, 10× Taq buffer with (NH4)2SO4, 1.5 mM MgCl2, 0.2 μM of each dNTP, 5 pmol of each primer, 1 U Taq polymerase and sterile ultrapure water. The following thermal protocol for PCR was applied: an initial denaturation at 94 °C for 2 min, followed by 32 amplification cycles of denaturation at 94 °C for 35 s, annealing at their respective dependent temperatures for 30 s, elongating at 72 °C for 3 min and then one final step at 72 °C for 10 min. All PCR products (5 μl) were electrophoresed on 1 % agarose gel and stained with the DNA dye, EZ-Vision One (Amresco, USA), and then visualized under UV illumination. PCR products were purified and sequenced using the same forward and reverse primers at Sangon Biotech (Shanghai, China).
Sequence alignment and phylogenetic analysis
Raw sequences of isolates collected in this study and some reference isolates were obtained from ABI 3730XL DNA Analyzer (Applied Biosystems, USA) and all the other sequences used here were downloaded from GenBank (Table 1) following Blast searches or references to published papers (Liu et al. 2012; Hyde et al. 2014). All nucleotide sequences were initially aligned with the software Clustal X 2.0, and then imported into BioEdit 5.0.9.1 for optimizing manually to analyze their nucleotide polymorphisms (Hall 1999; Larkin et al. 2007). Phylogenetic reconstructions of concatenated and individual gene-trees were performed using Maximum-parsimony (MP) and Bayesian Markov Chain Monte Carlo criteria. In addition, to determine whether the combined sequences can be used to construct phylogenetic analyses, statistical congruence was examined by applying a partition homogeneity test (PHT) (Farris et al. 1994). The PHT was performed in PAUP version 4.0b10 using 1,000 replicates and the heuristic standard search options (Swofford 2003).
PAUP 4.0b10 was used to separately analyze single genes and combined genes to construct parsimony trees (Swofford 2003). Gaps were treated as “missing” and all characters were unordered and of equal weight. Insertions/deletions (indels), irrespective of their size, were each treated as one evolutionary event and weighted as one base substitution. MP trees were found using the heuristic search function with 1,000 random addition replicates and tree bisection and reconstruction (TBR) selected as branch swapping algorithm. Branches of zero length were collapsed and all multiple, equally parsimonious trees were saved. Branch supports were estimated using 1,000 bootstrap replicates (Felsenstein 1985). Tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC) and homoplasy index (HI) were calculated.
Bayesian inferences were performed using MrBayes 3.1.2 for single locus datasets and for the combined dataset of multiple loci (Altekar et al. 2004). The optimal nucleotide substitution models for each gene region and for the combined data were estimated using MrModeltest v2.2 software (Nylander 2004). The option for rates was set to invgamma, whereas all the other parameters of the likelihood model were default. Four simultaneous Markov chains were run starting from a random tree for 5,000,000 generations and trees were sampled every 1,000 generations. The first 1,250 of the 5,000 saved trees were discarded as burn-in, and the consensus tree was based on the remaining 3,750 trees. To determine the confidence of the tree topologies, values of Bayesian posterior probabilities (BPPs) were estimated using MrBayes.
Novel sequence data are deposited in GenBank (Table 1) and the alignment in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2: S15479).
Morphological analysis
Colony traits of single-conidium isolates on potato dextrose agar (PDA) medium were noted after a 5-day incubation at 25 °C in the dark. To induce sporulation, PDA dishes or malt extract agar (MEA) dishes filled with mycelia were incubated at 25 °C under alternating 12 h cycles of light/dark using both fluorescent and UV light for 15 days. In addition to the 18 isolates collected from China’s apple-growing regions that were used for our phylogenetic analyses, 18 additional samples that were collected similarly were used to make conidial measurements (36 total isolates). Conidial measurements were made using conidia collected from pycnidia on the agar surface. Conidia were observed and measured from images taken using an Olympus BX51 microscope with an Olympus DP72 digital camera (Olympus Corp., Japan). Thirty measurements of conidial lengths and widths were taken for each isolate, and the ranges and averages as well as length and width ratio were calculated. SPSS statistical software was used to analyze variability in conidial lengths and widths among the isolates.
Growth rates testing
To assess colony growth rate, mycelial plugs (5 mm in diam) of all test isolates were taken from the edge of 3-day-old colonies and reversely transferred to the centers of 9 cm PDA dishes. Three replicates for each isolate were used. Colony diameters of each isolate were measured after 5 days of incubation at 25, 35 and 37 °C in the dark, and their average growth rates were calculated and expressed as colony growth rate per 24 h. This experiment was conducted twice. Analysis of variance (ANOVA) of mycelial growth rate was performed using the ANOVA procedure of SPSS statistical software.
Pathogenicity testing
The pathogenicity of tested isolates was evaluated on shoots or stems of apple (Malus × domestica ‘Fuji’) and pear (Pyrus pyrifolia Nakai ‘Suli’). The pathogenicity test was conducted in an orchard (in Yangling, Shaanxi Province) from May to August, 2011, and was repeated in 2012. Scions were grafted on Malus micromalus, and then cultivated in the field with 1.5-m × 4-m spacing. Test isolates were grown on PDA medium at 25 °C in the dark for 3 days before inoculation. The inoculation was performed on the non-wounded bark surface of 2-year-old shoots of 4-year-old apple saplings (3 to 3.5 m high and 15 to 20 branches) and 2-year-old pear (1 to 1.5 m high) stems. In mid-May, mycelial plugs (5 mm in diameter) for each isolate were transferred to the inoculation positions that had been previously disinfected with 70 % ethyl alcohol, and then covered with sterile moist cotton balls and wrapped with Parafilm to maintain high humidity. Five inoculations (10 cm away from each other) were made on a tree as one replicate, and three replicates were used for each isolate; controls were treated in a similar manner with noncolonized PDA plugs. The inoculated trees were arranged in a randomized block design. In mid-August of those two years, the symptoms and sizes of lesions at the inoculated positions were examined, measured and recorded. Measurements on the same tree were averaged, and then data among treatments in each repeated test were compared using ANOVA in SPSS statistical software.
Pathogenicity was tested also on detached non-wounded apple fruit in the laboratory. Mature apple (Fuji) fruit were washed, surface disinfested with 3.5 % NaOCl for 5 min, rinsed twice with sterile-distilled water, and dried in a transfer hood. Three mycelial plugs (5 mm in diameter) cut from the margin of a 5-day-old culture were placed on the surface of each non-wounded fruit with an equal distance between adjacent plugs. Three fruits (replicates) were used for each isolate. Fruit inoculated with PDA plugs without fungus were used as controls. The incidence of lesions at the inoculated positions was determined on the second day after inoculation.
Re-isolation of fungi from diseased parts of inoculated shoots, stems and fruit was performed to complete Koch’s postulates. Warts or edges of the lesions were cleansed with 3.5 % NaOCl for surface disinfestation, washed in sterile-distilled water twice, and then kept on PDA media at 25 °C for 5 days. Fungi were identified based on morphology and the sequence of the rDNA ITS region as described previously.
Intron analysis
The rDNA small-subunit (SSU) regions of all tested isolates were amplified and then sequenced to determine the presence or absence, distribution and primary structure of group I introns with reference to the method of Xu et al. (2013). Sequences of these introns were submitted to GenBank for BLASTN to clarify their attributes.
Results
Phylogenetic placement of tested isolates based on combined ITS and EF1-α sequences
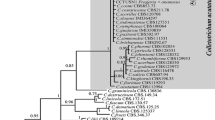
A total of 36 sequences including four randomly selected tested isolates were comprised by the alignment of ITS and EF1-α regions, which contained 760 characters. In the parsimony analysis, 555 characters were constant, 189 characters were parsimony informative while 16 variable characters were parsimony uninformative. The first one (TL = 342, CI = 0.7719, RI = 0.9242, RC = 0.7134, HI = 0.2281) of 100 equally parsimonious trees yielded is presented here (Fig. 1). The Bayesian tree (GTR model) agreed with the topology of the parsimonious tree, so Bayesian posterior probabilities are shown next to bootstrap values at the nodes. This phylogram provided a preliminary guide for identification of the isolates used in the research. In spite of low bootstrap values, isolates PG20 and PG45 cluster together and are evolutionarily relatively far apart from the clade that contains the isolates PG2 and LSP5. In terms of the topology of the tree, PG20 and PG45 are close to B. dothidea, whereas PG2 and LSP5 seemingly belong to an unrecognized taxon under the genus Botryosphaeria.
Phylogram inferred from combined parsimony analysis of ITS and EF1-α sequences from species of the genera Botryosphaeria, Cophinforma, Macrophomina, Neoscytalidium, Dothiorella and Neofusicoccum. The phylogenetic tree resulting from Bayesian inference had a topology identical to the MP tree presented. Bootstrap values/Bayesian posterior probabilities (>50 %) are given at the nodes. The tree was rooted to Neofusicoccum luteum. Clades corresponding to genera and species are highlighted
Phylogeny of individual and combined genes
ITS, EF-1α, HIS and HSP were used to construct phylogeny trees separately based on all 18 tested isolates and eight reference sequences. MrModeltest v2.2 computed appropriate evolutionary models for each of the four loci used in Bayesian analyses as follows: GTR model for ITS, EF-1α and HSP, and HKY model for HIS. For all these genes (except ITS), topologies of their trees are basically identical in the maximum parsimony and Bayesian inference. Only unrooted trees derived from the parsimony analysis are presented with the parsimony bootstrap values and posterior probabilities shown at the branches (Fig. 2). Statistical data for individual trees are summarised in Table 2. In the four gene genealogies, two distinct groups are consistently observed, of which nine tested isolates including PG20 and PG45, B. berengeriana isolate MAFF 645001 and B. dothidea isolates PD 314, CBS 115476, ICMP 13957, MAFF 410826 and ICMP 8019 belong to cluster A while the other nine tested isolates including PG2 and LSP5, B. dothidea isolate PD 313 and B. berengeriana f. sp. piricola isolate MAFF 645002 gather in cluster B. This partition is supported by high bootstrap values and posterior probabilities in EF-1α, HIS and HSP datasets, although slight variations still existed within cluster A according to the three genes (Fig. 2b-d). For the ITS region, however, the two clusters are supported by low bootstrap values and they cannot be distinguished in Bayesian tree. Additionally, for sequences of genes β-tubulin and Actin, too few parsimony-informative characters were found (Table 2), so their phylogeny trees are not presented here.
The datasets including ITS, EF-1α, HIS and HSP were also analyzed collectively. Through the partition homogeneity test (Cunningham 1997), no significant conflicts (P ≥ 0.05) were found among them before catenation. The combined aligned data matrix contained 37 sequences including the outgroup Lasiodiplodia theobromae and 1610 characters, of which 311 characters were parsimony informative, 185 variable and parsimony uninformative, and 1114 constant. The parsimony analysis resulted in a most parsimonious tree (TL = 940, CI = 0.6968, HI = 0.3032, RI = 0.8208, RC = 0.5719) (Fig. 3). Bayesian tree (GTR model) with an identical topology to the MP tree was reconstructed, and its posterior probabilities were thus added next to the bootstrap values. In the phylogenetic reconstruction of combined dataset, two deep clades were recognized and strongly supported with bootstrap values equal to 100 % and posterior probabilities of 1.00, which completely corresponded to the two clusters formed in the individual gene genealogies.
Most parsimonious tree inferred from the combined four locus dataset of ITS, EF-1α, HIS and HSP. The phylogenetic tree resulting from Bayesian inference had a topology identical to the MP tree presented. Bootstrap values/Bayesian posterior probabilities (>50 %) are given at the nodes. The tree was rooted to Lasiodiplodia theobromae. Clades corresponding to genera and species are highlighted
Morphological characteristics
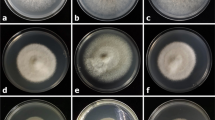
On PDA, colonies of all tested isolates were initially white, gradually becoming greenish brown to grey and then turning dark grey after 5 days of incubation. The reverse side of colonies was at first white, but after 2 or 3 days becoming dark green to olive green from the centre. No pigments were released to the medium by these pure cultures. For the isolates belonging to cluster A, bundles of aerial mycelia were commonly observed and they could even reach the lids of Petri dishes. However, the isolates contained in cluster B generally produced compact and shorter aerial mycelia that can be described as an appressed mycelial mat (Fig. 4a).
a. The colony morphology of cluster A isolates (left) and cluster B isolates (right) from above after 5-day cultivation on PDA at 25 °C. b. The average lengths and widths of 30 conidia measured for each of 36 tested isolates. Seventeen cluster A isolates and 19 cluster B isolates are indicated on the graph as blue triangles and red squares, respectively
Conidial dimensions (lengths and widths) of 17 isolates belonging to cluster A and 19 isolates belonging to cluster B were measured in this study. Conidial lengths and widths of the cluster A isolates ranged from 20.3 to 26.7 μm and 5.3 to 7.0 μm, respectively. Conidial lengths and widths of the cluster B isolates ranged from 18.6 to 24.7 μm and 5.3 to 7.2 μm, respectively (Fig. 4b). Average conidial length (23.3 μm) of cluster A isolates was longer than that (21.9 μm) of cluster B isolates (P < 0.01), whereas there was no significant difference (P > 0.01) in conidial width between these two groups of isolates.
Growth rates
Data from two repeated experiments on mycelial growth were not significantly different (P > 0.05), and therefore were combined to calculate average colony diameter of each tested isolate. Isolates belonging to cluster A and cluster B were treated as two groups and there was no statistical difference (P > 0.05) in growth rate among isolates of the same group when incubated at 25, 35 and 37 °C, respectively. We then compared the average growth rates of these two groups of isolates at the above three incubation temperatures. At 25 °C, the average growth rates of cluster A and cluster B isolates were respectively 12.4 and 11.9 mm per 24 h and there was no difference (P > 0.05) between them. When incubated at 35 °C, however, the growth rate (8.5 mm per 24 h) of cluster A isolates was significantly faster than that (1.7 mm per 24 h) of cluster B isolates (P < 0.01). At 37 °C, no visible colony development was observed for any isolates of cluster B, whereas cluster A isolates could still extend slowly at an average growth rate of 3.3 mm per 24 h (Fig. 5).
Pathogenicity
Rounded protuberances were commonly visible approximately 40 to 60 days after inoculating apple shoots with mycelial plugs. They presented distinctly different sizes and denseness between cluster A and cluster B isolates (P < 0.01), and here only measurement data of seven inoculation treatments (six isolates plus one control) are presented (Table 3). Smaller protuberances (0.7–1.0 mm in diameter) induced by the cluster A isolates were distributed densely, and described as a verruculose surface, whereas the cluster B isolates produced bigger but sparser protuberances (3.0–4.1 mm in diameter), described as a verrucose symptom, which occasionally broke through the epidermis (Fig. 6). As the infection progressed, the protuberances usually increased in number, darkened in colour and eventually cracked. When these isolates were inoculated on pear stems, more complex symptoms were observed after about 6 weeks (Table 3, Fig. 6). For the cluster A isolates, just small areas around the inoculation positions turned brown (i.e., local necrosis). In this situation, the above isolates could be considered non-pathogenic on pear trees. For the cluster B isolates, however, the symptoms were extensive and necrosis extended on a large scale into the phloem tissues (cankers 15.0–17.5 mm in diameter). Additionally, blisters (2.5–3.0 mm in diameter) were noticed to form over the cankers and occasionally cause splitting of the periderm in the necrotic areas. No symptoms were observed on apple and pear trees that belonged to the control group
Symptoms on non-wounded apple and pear shoots or stems one or two months after inoculation (via mycelial plugs) with different isolates from cluster A and cluster B. a. Small protuberances on apple shoots. b. Large protuberances on apple shoots. c. Localized necrotic spots on pear stems. d. Cankers with blisters on pear stems. a-1 and c-1: PG 45. b-1 and d-1: PG 2. a-2 and c-2: PG 293. b-2 and d-2: PG330. a-3 and c-3: PG327. b-3 and d-3: LSP 20. 4: Control
On apple fruit, all tested isolates caused brown soft rot symptoms on non-wounded fruit, and exudation could be observed in the lesion areas. There was no marked difference between cluster A and cluster B isolates in pathogenicity. Fruit that were inoculated with PDA plugs without fungi remained healthy.
Analysis of introns
The rDNA SSU of 18 tested isolates used in this study (not published) were scanned for group I introns that related to the genetic diversity (four genotypes) within B. dothidea discussed in our previous research (Xu et al. 2013). As a consequence, all cluster A isolates possessed a 1349-bp intron (Bdo.S1199-A) at position 1199, which agreed with the genotype I of B. dothidea. The cluster B isolates were detected to have the same primary nucleotide structure of rDNA SSU as genotype II of B. dothidea that was characterized by two group I introns (Bdo.S943 and Bdo.S1506) inserted at positions 943 and 1506, respectively (Fig. 7).
Primary structures of the rDNA SSU in cluster A isolates and cluster B isolates. The entire SSU rDNA is shown by the blue rectangle. The triangles in purple, green and yellow indicate three different group I introns. The position, length and name of each intron are given separately above, in and below the triangles. The insertion sites correspond to the 16S rRNA of E. coli J01859
Taxonomy
Based on these individual gene genealogies (ITS, EF-1α, HIS and HSP), multi-locus phylogeny inferred by their combined alignment and biological characteristics, the 18 tested isolates associated with branch or fruit ring rot of Malus × domestica and Pyrus pyrifolia were shown to belong to cluster A and cluster B, respectively, which should be identified as two species of Botryosphaeria, i.e., a previously described species B. dothidea and one new taxon.
Botryosphaeria kuwatsukai (Hara) G.Y. Sun and E. Tanaka, comb. nov. et. emend (Fig. 8)
Asexual structures of Botryosphaeria kuwatsukai (from ex-epitype CBS 135219). a. Infected fruit of apple (black conidiomata scattered on the lesion) b. Culture on PDA (5 days old) c. Conidiomata on PDA d. Conidia and microconidia e-g. Conidia before germination with 1–3 septa h. Microconidiomata i. Microconidiophores j. Conidia k-l. Conidiophores and conidiogenous cells. Scale bars: c = 500 μm d, f, h and i = 10 μm e, g = 5 μm i, j and l = 20 μm
MycoBank: MB 808073.
Facesoffungi Number: FoF00170.
Basionym: Macrophoma kuwatsukai Hara, Pathologia Agriculturalis Plantarum 482 (1930)
= Physalospora pyricola Nose, Ann Agr. Exper. Stat. Gov.-Gen. Chosen 7. 161(1933). (as “piricola”)
≡ Guignardia pyricola (Nose) W. Yamamoto, Sci. Rep. Hyogo Univer. Agric. 5. 11 (1961). (as “piricola”)
= Botryosphaeria berengeriana De Notaris f. sp. pyricola Koganezawa & Sakuma, Bull. Fruit Tree Res. Stat., C11: 58, 1984. (as “piricola”)
Lectotype designated here (MB 808072): illustration of Macrophoma kuwatsukai Hara, Pathologia Agriculturalis Plantarum 482: figure 191 (1930)
Epitype: CHINA, Shaanxi Province, Yangling, from fruit of M. ×domestica, 7 September 2011, CS Wang, dried culture, HMAS 245112 (PG 2); living cultures derived from ex-epitype — CBS 135219 = CGMCC 3.15244.
Colonies on PDA attaining 52 mm diam. after 4 days at 25 °C in the dark, initially white with moderately dense, appressed mycelial mat and aerial mycelium without columns, gradually becoming grey to dark grey. The reverse side of the colonies at first white, but after 2–3 days becoming dark green to olive-green from the centre. This colouration gradually spreads to the edge and becomes darker from the centre until the entire underside of the colony is black. Conidiomata absent in culture on PDA or on MEA in the dark, formed under the 12-h interactive treatment with black light and fluorescent lamp within 15–20 d, superficial, dark brown to black, globose, mostly solitary and covered by mycelium. Conidiogenous cells holoblastic, hyaline, sub-cylindrical, 7–18 × 2–4 μm, proliferating percurrently with one or more proliferations and occasionally resulting in periclinical thickening. Conidia produced in culture similar to those formed in nature, narrowly fusiform, or irregularly fusiform, base subtruncate to bluntly rounded, smooth with granular contents, widest in the middle to upper third, (18.5–)20–24.5(−26) × 5–7(−8) μm (mean ± SD = 22.3 ± 2.1 × 6.2 ± 0.9 μm, n = 60, L/W ratio = 3.6), forming 1–3 septa before germination. Microconidiomata globose, dark brown to black. Microconidiophores hyaline, cylindrical to sub-cylindrical, 3–10 × 1–2 μm. Microconidia unicellular, hyaline, allantoid to rod-shaped, 3–8 × 1–2 μm. Sexual state not observed in culture.
Known hosts: Malus × domestica and Pyrus pyrifolia
Known distribution: China, Japan and North America
Additional material examined: CHINA, Shaanxi Province, on branch of M. ×domestica, 2011, CS Wang, culture PG 55; Henan Province, on fruit of M. ×domestica, 2011, CS Wang, culture PG 259; Shandong Province, on fruit of M. ×domestica, 2011, CS Wang, culture PG 297; Hebei Province, on fruit of M. ×domestica, ZQ Zhou, culture PG 328; Jiangsu Province, on fruit of M. ×domestica, ZQ Zhou, culture PG 330; Shanxi Province, on fruit of M. ×domestica, ZQ Zhou, culture PG 332; Shaanxi Province, on branch of Pyrus pyrifolia, 2012, CS Wang culture LSP 5; Shaanxi Province, on fruit of P. pyrifolia, 2012, CS Wang, culture LSP 20; JAPAN, Iwate, on twigs of M. pumila Mill. var. domestica, 1980, H. Koganezawa, culture MAFF 645002.
Notes: Miura (1917) found ring rot disease on pears and named the fungus as “Coniothecium”. Kuwatsuka (1921) said that the fungus belonged to “Macrophoma”, and Hara (1930) described it as Macrophoma kuwatsukai Hara. The original description was only in Japanese. Nose (1933) found the sexual stage, which he described as Physalospora piricola Nose, also with only a Japanese description. Yamamoto (1961) transferred the fungus to Guignardia piricola (Nose) W. Yamamoto, but this was not accepted by others. Koganezawa and Sakuma (1984) proposed the name, Botryosphaeria berengeriana De Notaris f. sp. piricola Koganezawa & Sakuma for the fungus causing apple wart bark to separate typical B. berengeriana (=B. dothidea) causing cankers on apple and pear. Even though there was no Latin description or diagnosis for both Macrophoma kuwatsukai and Physalospora piricola, they are validly published (Art.39.1). Macrophoma kuwatsukai has priority over Physalospora piricola, even though it is an asexual stage name (Art.59.1), and should be treated as the basionym.
Botryosphaeria kuwatsukai, which had been always confounded with B. dothidea, was described and identified in this study based on multiple locus genealogies. Biological characteristics including aerial mycelia growth, mycelial growth rate and pathogenicity also supported the segregation of these two species. Morphologically, however, it is hard to discriminate B. kuwatsukai from B. dothidea as they produce similar conidia. In the original description of Macrophoma kuwatsukai Hara (1930), the holotype was not designated. The illustration should be treated as the lectotype. Based on the lectotype, it is hard to discriminate it from other morphological similar species, thus we designate an epitype.
Botryosphaeria dothidea (Moug. ex Fr.) Ces. & De Not., Commentario della Società Crittogamologica Italiana 1 (4): 212 (1863)
Material examined: CHINA, Shaanxi Province, on fruit of M. ×domestica, 2011, CS Wang, culture PG 20; Shaanxi Province, on trunk of M. ×domestica, 2011, CS Wang, culture PG 45; Shaanxi Province, on branch of M. ×domestica, 2011, CS Wang, culture PG 77; Henan Province, on fruit of M. ×domestica, 2011, CS Wang, culture PG 267; Shandong Province, on fruit of M. ×domestica, 2011, CS Wang, culture PG 293; Liaoning Province, on fruit of M. ×domestica, ZQ Zhou, culture PG 320; Hebei Province, on branch of M. ×domestica, ZQ Zhou, culture PG 327; Jiangsu Province, on fruit of M. ×domestica, ZQ Zhou, culture PG 329; Shanxi Province, on fruit of M. ×domestica, 2012, CS Wang, culture PG 331; SWITZERLAND, Crocifisso, isolated from Prunus sp., October 2000, B. Slippers, culture CBS 115476; NEW ZEALAND, Bay of Plenty, Te Puke, from bark of dead twig of Populus nigra, 1 December 1981, G.J. Samuels, culture ICMP 8019; NEW ZEALAND, Waikato, on fruit of M. ×domestica, 1 April 1999, M.A. Manning, culture ICMP 13957; JAPAN, IBARAKI, isolated from Prunus sp., May 1993, T. Yamada, culture MAFF 410826; JAPAN, Iwate, on twigs of M. pumila Mill. var. domestica, 1978, H. Koganezawa, culture MAFF 645001.
Notes: Botryosphaeria dothidea was isolated as the pathogen causing apple ring rot from fruit and branches of Malus × domestica in this study. The colony characteristics and microscopic morphology is exactly similar to the ex-epitype of B. dothidea (Slippers et al. 2004a; Liu et al. 2012; Hyde et al. 2014). In the phylograms, our nine isolates confidently clustered together with the ex-epitype isolate of B. dothidea (CBS 115476) and other referential B. dothidea isolates including ICMP 8019, ICMP 13957, MAFF 410826 and MAFF 645001 (Figs. 2, and 3).
Discussion
The theory of multiple gene genealogies has been increasingly applied in studies of species boundaries in both human and plant pathogenic fungi, revealing vast numbers of cryptic species and species complexes in fungal taxa previously identified as one morphospecies (Pringle et al. 2005; Hyde et al. 2010, 2014; Liu et al. 2012; Maharachchikumbura et al. 2012; Udayanga et al. 2012; Morgado et al. 2013). In Botryosphaeriaceae, for example, Slippers et al. (2004a) redefined the circumscription of B. dothidea sensu stricto and separated out B. parva and B. ribis that had previously served as synonyms of B. dothidea. Pavlic et al. (2009) revealed three cryptic species within the Neofusicoccum parvum/N. ribis complex isolated from Syzygium cordatum trees in South Africa. Furthermore, Diplodia scrobiculata was described as a sister species of D. pinea (De Wet et al. 2003) and N. eucalypticola and N. australe were perceived as sister species of N. eucalyptorum and N. luteum, respectively (Slippers et al. 2004b, c). In this study, Botryosphaeria kuwatsukai is described and identified from pathogenic isolates causing ring rot of apple and pear trees in China, thus proving our hypothesis that the previous ring rot pathogen, B. dothidea, is a species complex. This cryptic species was recognized primarily based on DNA sequence data of four nuclear genes combined with some phenotypic characters as supplementary evidence.
According to the phylogram of combined ITS and EF1-α sequences (Fig. 1), we can estimate that the four randomly selected tested isolates belong to two different taxa (one probably equals to B. dothidea) in Botryosphaeria. Further analysis of the genealogies of EF-1α, HIS and HSP genes and their combination (Figs. 2, and 3) demonstrated that all tested isolates were subsumed in two genetically well-separated clusters (cluster A and cluster B), which correspond to the above two taxa. In cluster A, despite some slight variations, nine tested isolates gathered well with five reference B. dothidea isolates including the ex-epitype CBS 115476, indicating that they truly belong to B. dothidea. In cluster B, the remaining nine tested isolates and two reference B. dothidea isolates were re-identified and named as B. kuwatsukai, thus suggesting that previous B. dothidea identified from apple ring rot was a species complex. Two other genes, β-tubulin and Actin, widely used in phylogenetics, were also analyzed here. They were more inclined to make all tested isolates fall together in the B. dothidea clade. This is consistent with the result of Tang et al. (2012), who considered according to the multi-gene sequence data of ITS, β-tubulin and Actin that isolates associated with symptoms of apple and pear ring rot (warts, cankers and fruit rot) all belong to the same pathogen, B. dothidea.
In microscopic morphology, B. kuwatsukai and B. dothidea exhibited a statistically significant difference in conidial length. However, this variation represented a continuum among isolates of the B. dothidea/B. kuwatsukai complex and, therefore, it was too difficult for us to use this character to set a clear delimitation of groups, and then to screen B. kuwatsukai out of B. dothidea prior to the application of some other effective methods. This indicates that genetically isolated species do not necessarily show divergence in some morphological characters such as conidial morphology, which is consistent with conclusions of several previous studies (Pavlic et al. 2009; Maharachchikumbura et al. 2012; Udayanga et al. 2012; Muggia et al. 2014). Colony morphology and different growth patterns of aerial mycelia (i.e., appressed mycelial mat from B. kuwatsukai and columns of aerial mycelia from B. dothidea) can be used to preliminarily distinguish these two species.
When cultured below their optimal temperatures (generally 22 to 28 °C) in darkness, both B. kuwatsukai and B. dothidea grew faster on PDA as temperature increased and there was no significant difference between their growth rates (unpublished data). When cultured above their optimal temperatures, both species grew more slowly as temperature increased; however, the decline in growth rate was more pronounced with B. kuwatsukai. Compared to the demanding requirement for techniques and equipment used in molecular identification, it is undoubtedly a more expeditious approach to differentiate the two species by simultaneously cultivating them at relatively high temperatures (e.g., 35 °C) and then assessing their colony diameters after 3–5 days.
Although the host affiliations of isolates collected in this study were limited to apple and pear, host ranges of the two fungi could be inferred according to previous research (Inderbitzin et al. 2010; Marques et al. 2013; Xu et al. 2013). As mentioned above, the isolates of B. kuwatsukai actually correspond to the genotype II of previous B. dothidea, whereas the other three genotypes (III and IV were not isolated here) are still considered to be genetically different populations within current B. dothidea. Through investigation and statistics, Xu et al. (2013) found that genotype II isolates of B. dothidea could be isolated only from apple and pear, and was the only population detected on pear, whereas the other three genotypes of B. dothidea collectively infected dozens of shrubs and trees, except pear. From the above, it is conjectured that B. kuwatsukai may have host specificity for apple and pear.
The phylogenetic analyses showed that both species causing apple ring rot belong to the genus Botryosphaeria (Liu et al. 2012; Hyde et al. 2014). The Japanese B. berengeriana isolate MAFF645001 belonged to B. dothidea, which agrees with the conclusion of Slippers et al. (2004a), whereas another Japanese isolate B. berengeriana f. sp. piricola MAFF645002 and B. dothidea isolate PD313 from the USA were both re-identified as B. kuwatsukai, which suggests that this species is a cosmopolitan fungus, not unique to China. According to the morphological and biological description of B. berengeriana f. sp. piricola by Koganezawa and Sakuma (1984), this fungus is morphologically identical with B. dothidea, grows slower than B. dothidea and mainly causes ring rot of apple and pear. All these characters are shared by B. kuwatsukai, supporting our phylogenetic results. Reference isolates PD313 and PD314 (Inderbitzin et al. 2010), which were isolated from fruit of M. domestica in USA, were once both described as B. dothidea and caused a prevalent disease called apple white rot (Jones and Aldwinckle 1990; Inderbitzin et al. 2010). However, in this study the two isolates were classified as different species, indicating that apple white rot, which is widely distributed in the North America, is probably able to be induced by two pathogens, B. dothidea and/or B. kuwatsukai.
Pathogenicity tests revealed that both B. kuwatsukai and B. dothidea could induce protuberances on the surfaces of apple shoots, with the two species associated with protuberances of different size and density (described as verrucose and verruculose, respectively). Protuberances caused by B. kuwatsukai were nearly four times as larger than those by B. dothidea. Previously, this phenomenon was often attributed to differences in virulence of different B. dothidea isolates and was applied to the establishment of evaluation criteria for host resistance (Zhou et al. 2010; Lin et al. 2011). Our results show that size of protuberances is not related to virulence differences and this character should not be applied when screening for resistance.
The common canker symptom of apple ring rot on shoots was not observed in the inoculation tests. Koganezawa and Sakuma (1984) inoculated mycelial plugs of B. berengeriana (B. dothidea) isolates and B. berengeriana f. sp. piricola (=B. kuwatsukai) isolates on wounded apple trunks, and reported the former produced typical cankers while the latter formed rough callus bark on inoculation sites. They also inoculated spore suspension of the two pathogens on non-wounded apple trunks, and consequently B. berengeriana caused no symptoms while B. berengeriana f. sp. piricola produced typical wart-like protrusions. Tang et al. (2012) performed similar pathogenicity tests with several B. dothidea isolates, in which warts formed only on non-wounded apple shoots whereas cankers often appeared on wounded apple shoots. Therefore, we speculate that different inoculation methods (non-wounded or wounded and mycelium plugs or spore suspension) lead to different symptoms on apple trunks. B. kuwatsukai tends to infect hosts from the lenticels and causes wart-like protuberances, whereas B. dothidea tends to infect hosts from wounds and causes cankers and, in fact, when the inoculum dose is enough (e.g., mycelium plugs), B. dothidea can cause similar symptoms as B. kuwatsukai (Koganezawa and Sakuma 1984; Zhang et al. 2011). On pear stems, large-scale cankers along with blisters were produced by B. kuwatsukai, whereas B. dothidea just induced localized necrotic spots, which could be regarded as non-infectious. This agrees with the view of Koganezawa and Sakuma (1984) that B. berengeriana f. sp. piricola (=B. kuwatsukai) is the true and only pathogen of pear ring rot.
Our research provided sufficient evidence to prove that apple ring rot disease is caused by two different pathogens, B. dothidea and B. kuwatsukai, whereas B. kuwatsukai alone is the pathogen responsible for pear ring rot. With increased understanding of the etiology of apple ring rot, we can begin to develop targeted management strategies based on sanitation, cultural methods, chemical methods and resistance breeding.
References
Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F (2004) Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20:407–415
Chen C (1999) Advances in the research of apple ring rot. Acta Phytopathol Sinica 29(3):1–7 (in Chinese)
Cunningham CW (1997) Can three incongruence tests predict when data should be combined? Mol Biol Evol 14:733–740
De Wet J, Burgess T, Slippers B, Preisig O, Wingfield BD, Wingfield MJ (2003) Multiple gene genealogies and microsatellite markers reflect relationships between morphotypes of Sphaeropsis sapinea and distinguish a new species of Diplodia. Mycol Res 107:557–566
Farris JS, Källersjö M, Kluge AG, Bult C (1994) Testing significance of incongruence. Cladistics 10:315–319
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hara K (1930) Pathologia Agriculturalis Plantarum. Yokendo, Tokyo, pp 481–483 (in Japanese)
Huang C, Liu K (2001) RAPD analysis of the pathogenic fungi of apple ring rot and other major related diseases. Acta Phytopathol Sinica 31(2):69–74
Hyde KD, Chomnunti P, Crous PW, Groenewald JZ, Damm U, Ko TWK, Shivas RG, Summerell BA, Tan YP (2010) A case for re-inventory of Australia’s plant pathogens. Persoonia 25:50–60
Hyde KD, Nilsson RH, Alias SA, Ariyawansa HA, Blair JE, Cai L, de Cock AWAM, Dissanayake AJ, Glockling SL, Goonasekara ID, Gorczak M, Hahn M, Jayawardena RS, van Kan JAL, Laurence MH, Lévesque CA, Li X, Liu JK, Maharachchikumbura SSN, Manamgoda DS, Martin FN, McKenzie EHC, McTaggart AR, Mortimer PE, Nair PVR, Pawłowska J, Rintoul TL, Shivas RG, Spies CFJ, Summerell BA, Taylor PWJ, Terhem RB, Udayanga D, Vaghefi N, Walther G, Wilk M, Wrzosek M, Xu JC, Yan JY, Zhou N (2014) One stop shop: backbones trees for important phytopathogenic genera: I. Fungal Divers 67:21–125. doi:10.1007/s13225-014-0298-1
Inderbitzin P, Bostock RM, Trouillas FP, Michailides TJ (2010) A six locus phylogeny reveals high species diversity in Botryosphaeriaceae from California almond. Mycologia 102:1350–1368
Jones AL, Aldwinckle HS (1990) Compendium of apple and pear diseases. American Phytopathological Society, St. Paul, Minnesota, USA
Kang L, Hao H, Yang Z, Li X, Kang G (2009) The advances in the research of apple ring rot. Chin Agric Sci Bull 25(09):188–191 (in Chinese)
Koganezawa H, Sakuma T (1980) Fungi associated with blister canker and internal bark necrosis of apple trees. Bull Fruit Tree Res Station C (Morioka) 7:83–99
Koganezawa H, Sakuma T (1984) Causal fungi of apple fruit rot. Bull Fruit Tree Res Station C (Morioka) 11:49–62
Kuwatsuka K (1921) J Okitsu Hortic Soc (Engei no Kenkyu) 17:190–195, in Japanese
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Lin Y, Huang L, Suolang L, Gao X, Chen Y, Kang Z (2011) A rapid laboratory evaluation system for apple ring rot. Acta Phytophylacica Sinica 38(1):37–41 (in Chinese)
Liu JK, Phookamsak R, Doilom M, Wikee S, Li YM, Ariyawansa H, Boonmee S, Chomnunti P, Dai DQ, Bhat JD, Romero AI, Zhuang WY, Monkai J, Jones EBG, Chukeatirote E, Ko Ko TW, Zhao YC, Wang Y, Hyde KD (2012) Towards a natural classification of Botryosphaeriales. Fungal Divers 57:149–210
Lv D, Zhang J, Zhang Z, Zhou Z, Chen X, Du X, Qu S (2012) The relationship between rDNA-ITS sequences and biological characteristics of the apple ring rot pathogen Botryosphaeria berengeriana de Not f. sp. piricola (Nose). Fungal Genom Biol 2:104
Maharachchikumbura SSN, Guo LD, Cai L, Chukeatirote E, Wu WP, Sun X, Crous PW, Bhat DJ, McKenzie EHC, Bahkali AH, Hyde KD (2012) A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Divers 56:95–129
Marques MW, Lima NB, de Morais MA, Michereff SJ, Phillips AJL, Câmara MPS (2013) Botryosphaeria, Neofusicoccum, Neoscytalidium and Pseudofusicoccum species associated with mango in Brazil. Fungal Divers 61:195–208
Miura M (1917) Ringo no Byoki. Shokabo, Tokyo, pp 106–109 (in Japanese)
Morgado LN, Noordeloos ME, Lamoureux Y, Geml J (2013) Multi-gene phylogenetic analyses reveal species limits, phylogeographic patterns, and evolutionary histories of key morphological traits in Entoloma (Agaricales, Basidiomycota). Persoonia 31:159–178
Muggia L, Prerez-Ortega S, Fryday A, Spribille T, Grube M (2014) Global assessment of genetic variation and phenotypic plasticity in the lichen-forming species Tephromela atra. Fungal Divers 64:233–251
Nose T (1933) On the ring rot of pears and the causal organism, especially on its perfect generation Physalospora piricola. Ann Agric Exp Sta Chosen 7(2):156–163 (in Japanese)
Nylander JAA (2004) MrModeltest v2. Program Distributed by the Author. Uppsala University, Evolutionary Biology Centre
Ogata T, Sano T, Harada Y (2000) Botryosphaeria spp. isolated from apple and several deciduous fruit trees are divided into three groups based on the production of warts on twigs, size of conidia, and nucleotide sequences of nuclear ribosomal DNA ITS regions. Mycoscience 41:331–337
Park EW (2005) An infection model of apple white rot based on conidial germination and appressorium formation of Botryosphaeria dothidea. Plant Pathol J 21:322–327
Pavlic D, Slippers B, Coutinho TA, Wingfield MJ (2009) Multiple gene genealogies and phenotypic data reveal cryptic species of the Botryosphaeriaceae: a case study on the Neofusicoccum parvum/N. ribis complex. Mol Phylogenet Evol 51:259–268
Peng B, Liu L, Wu H, Tian L, Zhou Z, Gu Q (2011) The intraspecific genetic diversity of pathogenic fungi of apple ring rot. Sci Agric Sin 44(6):1125–1135 (in Chinese)
Phillips AJL, Alves A, Abdollahzadeh J, Slippers B, Wingfield MJ, Groenewald JZ, Crous PW (2013) The Botryosphaeriaceae: genera and species known from culture. Stud Mycol 76:51–167
Pringle A, Baker DM, Platt JL, Wares JP, Latge JP, Taylor JW (2005) Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59:1886–1899
Qu J, Li X, Zhang Y, Fan K (2007) Evaluation of fungitoxicity of tebuconazole against Alternaria mali and Physalospora piricola on apple in laboratory and in field. Chin J Pestic Sci 9(2):149–152 (in Chinese)
Slippers B, Crous PW, Denman S, Coutinho TA, Wingfield BD, Wingfield MJ (2004a) Combined multiple gene genealogies and phenotypic characters differentiate several species previously identified as Botryosphaeria dothidea. Mycologia 96:83–101
Slippers B, Fourie G, Crous PW, Coutinho TA, Wingfield BD, Carnegie AJ, Wingfield MJ (2004b) Speciation and distribution of Botryosphaeria spp. on native and introduced Eucalyptus trees in Australia and South Africa. Stud Mycol 50:343–358
Slippers B, Fourie G, Crous PW, Coutinho TA, Wingfield BD, Wingfield MJ (2004c) Multiple gene sequences delimit Botryosphaeria australis sp. nov. from B. lutea. Mycologia 96:1030–1041
Smith H, Crous PW, Wingfield MJ, Coutinho TA, Wingfield BD (2001) Botryosphaeria eucalyptorum sp. nov., a new species in the B. dothidea-complex on Eucalyptus in South Africa. Mycologia 93:277–285
Swofford DL (2003) PAUP*. Phylogenetic analysis using parsimony (* and other methods). Version 4. Sinauer Associates, Sunderland, Massachusetts
Tang W, Ding Z, Zhou Z, Wang Y, Guo L (2012) Phylogenetic and pathogenic analyses show that the causal agent of apple ring rot in China is Botryosphaeria dothidea. Plant Dis 96:486–496
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol 31:21–32
Udayanga D, Liu X, Crous PW, McKenzie EHC, Chukeatirote E, Hyde KD (2012) A multi-locus phylogenetic evaluation of Diaporthe (Phomopsis). Fungal Divers 56:157–171
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc: Guide Methods Appl 18:315–322
Xu C, Wang C, Sun X, Zhang R, Gleason ML, Eiji T, Sun G (2013) Multiple group I introns in the small-subunit rDNA of Botryosphaeria dothidea: implication for intraspecific genetic diversity. PLoS One 8:e67808
Yamamoto W (1961) Species of the genera of Glomerella and Guignardia with special reference to their imperfect stages. Sci Rep Hyogo Univ Agric 5(1):1–12 (in Japanese)
Zhang G, Li B, Dong X, Wang C, Li G, Guo L (2011) Microanatomy conformation of apple branch tumors caused by Botryosphaeria dothidea. Acta Phytopathol Sinica 41(1):98–101 (in Chinese)
Zhou Z, Hou H, Wang L, Zhu F (2010) Trunk apple ring rot artificial inoculation method and the identification of cultivar resistance. J Fruit Sci 27(6):952–955 (in Chinese)
Acknowledgments
We are grateful for help in sample collecting by Prof. Zengqiang Zhou (Zhengzhou Institute of Pomology, Henan, China) and Prof. Meng Zhang (Henan Agricultural University, Henan, China). We thank Prof Pedro W. Crous (CBS-KNAW Fungal Biodiversity Centre, The Netherlands.) and Dr Eric H.C. McKenzie (Landcare Research, Auckland, New Zealand) for exchanging the authentic cultures and giving suggestion in nomenclature. This work was supported by National Natural Science Foundation of China (31371887, 31171797), the 111 Project from Education Ministry of China (B07049), Specialized Research Fund for the Doctoral Program of Higher Education (20130204110002) and China Agriculture Research System (CARS-28).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Xu, C., Wang, C., Ju, L. et al. Multiple locus genealogies and phenotypic characters reappraise the causal agents of apple ring rot in China. Fungal Diversity 71, 215–231 (2015). https://doi.org/10.1007/s13225-014-0306-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-014-0306-5