Abstract
The protein ATP-binding cassette subfamily G member 2 (ABCG2) is one of the major factors behind multidrug resistance (MDR) in breast cancer. We performed three-dimensional quantitative structure–activity relationship (3D-QSAR) modelling, docking, and molecular dynamics (MD) simulation to design pyrimidine-based ABCG2 antagonists. The developed QSAR model (r2 = 0.92, q2 = 0.82, and good cross-validated r2 = 0.73) dictate requirement of electrostatic, and hydrophobic fields for modulating bioactivity. Based on this rationale, we designed and screened 1010 new compounds, among them 2 (ND-510 and ND-500) exhibit excellent drug-like features. Comparative molecular docking, MM/GBSA and ADMET profiles were determined to understand the interactive poses, affinity, and drug-likeness of the designed compounds. Furthermore, MD simulations were performed with the ABCG2 receptor, and the results were compared with the two earlier synthesized active compounds. The outcomes of the study will help researchers to develop new antagonists for treatment of MDR breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
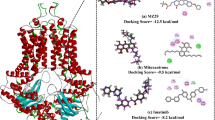
Breast cancer is one of the leading causes of premature mortality in females worldwide (Sharma 2021; Costantino et al. 2021). Multidrug resistance (MDR) is one of the major impediments to the chemotherapeutic agents or drugs treating different type of cancers. Several studies show that the main mechanism behind the MDR is the over expression of efflux transporter proteins from the family of ATP-binding cassette (ABC) on the plasma membrane of cancer cells (Chen et al. 2016; Kusuhara and Sugiyama 2007; Estrella et al. 2013). According to human genome organization report, ATP-binding cassette (ABC) transporter proteins are the main cause of MDR (Dean et al. 2001; Bailey-Dell et al. 2001). ABC transporters encode are divided into seven families from A to G (Chen et al. 2016) and ABCG2 is one of the dominant proteins, which responsible for breast cancer resistance. The structure of this half transporter consists of a membrane spanning domain and a nucleotide binding domain, which has walker A and B motifs connected to ATP-binding cassette (Fig. 1) (Ejendal and Hrycyna 2002). The name ‘hydrophobic vacuum cleaner’ is also given to the human ABCG2 as it is responsible for removing the non-polar molecules from the cells (Homolya et al. 2011). ABCG2 contributes to the efflux mechanism which involves the substrate binding to the transporters, hydrolysis of ATP molecules and the energy released is also utilized in conformational changes of the transporter domains (Khunweeraphong et al. 2017). The entire mechanism leads to the flip-flop motion through which substrates are evicted out from the cytoplasm to extracellular spaces (Bhardwaj et al. 2019). These features make them potential pharmaceutical targets in building new inhibitors.
Today, computed assisted drug design reached its zenith due to its cost effectiveness, rapidness, and less manual effort. For example, QSAR is one of the most important method due to its high and fast throughput and good hit rate (Neves et al. 2018). The structural properties of compounds such as chemical descriptors and biological activities provide clues via QSAR studies which could help (with the help of electrostatic negative and positive fields requirement) to design new molecules (Neves et al. 2018; Ahamad et al. 2019). We and others (Silbermann et al. 2019, 2021; Tadayon and Garkani-Nejad 2019) have designed several carbocyclic and heterocyclic compounds using in-silico techniques in the past. Our own experience dictates that heterocyclic compounds bearing N/O/S units are excellent candidate for the drug discovery. Among a large pool of nitrogen bearing compounds, pyrimidine derivatives are well known for diverse their bioactivity. Owing to its high structural similarity with nucleobases, several pyrimidine derivatives have been reported in literature with anticancer, antitubercular, antimicrobial activities (Silbermann et al. 2020; Krapf et al. 2018, 2017; Sivasankarapillai et al. 2020; Parkali et al. 2021; Singh et al. 2022).
In the present work, we carried out 3D-QSAR study to identify electrostatic, steric, and hydrophobic field factors responsible for bioactivity leading to the identification of 1010 compounds. In this study, the work was initiated and developed various QSAR models (Gad 2014) herein, we are discussing one of the best field-based 3D-QSAR model. The electrostatic, steric and hydrophobic fields introduced to the compound by the insertion of ChEMBL, Zinc databases, VEHICLe (ring systems) and eMolecules building blocks (Irwin and Shoichet 2005; Bento et al. 2014). The study proceeded to design new compounds by substitution of field effects at R1 and R2 position and generated 1010 compounds dataset SPARK-Cresset (Bauer 2018). Furthermore, molecular docking-based virtual screening studies of 26 pyrimidine derivatives and newly designed (ND) 1010 compounds were screened for the assessment of effectiveness of binding energies to the active cleft of ABCG2. The binding cleft of ABCG2 and two hits namely ND-510 and ND-500 were proposed. Both the ND compounds were selected based on the interactive pocket and the docking poses/scores.
To know the drug-likeliness and absorption, distribution, metabolism, excretion/toxicity (ADME/T) properties the compounds were analysed by QikProp—Schrödinger. The compounds were further analysed for the comparative Molecular Mechanics energies combined with Generalized Born and Surface Area continuum Solvation (MM/GBSA) studies by Prime—Schrödinger to acquire the free binding energies.
Consequently, conformational stability of the docked complexes of both Comp-9 & Comp-18 derivatives and ND-510 & ND-500 compounds was examined by MD simulations exploiting Desmond v4.1 allied various parameters such as potential energy (PE), root-mean-square deviation (RMSD), radius of gyration (Rg), root-mean-square fluctuation (RMSF), solvent accessible surface area (SASA), hydrogen bond monitoring, protein–ligand contacts, and torsion angles (Kanipakam et al. 2021; Katari et al. 2016). Thus, in the current study, both designed compounds ND-510 and ND-500, were proposed better than the Comp-9 and Comp-18. Furthermore, the proposed ND-510 and ND-500 compounds, which are putative potential ABCG2 inhibitors, also provide future insight into the experimental validations in the anticancer regiment. The mechanism of ABCG2 and the workflow of the study is illustrated (Fig. 1).
Materials and methods
Platform and software utilized
The ABCG2 structure modelling, 3D-QSAR modelling, designing of new molecules, molecular docking, and MD simulations were carried out on High Performance Computing (HPC) Dell server with 128 GB RAM, and CUDA enabled NVIDIA (Model: Nvidia Tesla v100) graphics processing units (GPUs) 1 GPU Memory: Total 16 GB X 4, pre-installed with Schrodinger LLC, New York, 2019 and Cresset v4.0.2. The AutoDock v4.2 installed on Centos LINUX platform consisting of 64 GB RAM.
Homology modelling and validation
The primary FASTA sequence consisting of 655 amino acids were procured from UniProt database (Q9UNQ0) which includes the relevant information. Suitable template was selected based on BLASTP search and a computational reliable modelling was performed using Prime-advanced homology modelling using Schrodinger and the same reliable 3D structure was considered as native protein (6FFC) in the study (Jackson et al. 2018). The model was further subjected for evaluation using B-factor and Ramachandran plot (PROCHECK) generated through Prime, to render the stereo-chemical reliability (Supplementary Fig. S1) (Laskowski et al. 1993).
QSAR modelling
The 2,3D structures of the pyrimidine compounds selected from the literature and drawn structures on Forge v10.4.2 module. Using customised thresholds, the alignment protocol was chosen based on the maximum common substructure implemented to match the training and test set compounds. For atomic positions of duplicate conformers, the maximum number of conformations for each molecule was set to 500 and the RMSD (root-mean-square deviation) cut off was set to 0.5 Å. The XED force field set the gradient cut off for conformer minimization to 0.1 kcal/mol and the energy window to 3 kcal/mol. Low energy conformations with the best matching were used as a reference for the 3D-QSAR model. Using the random selection process, the initial training set of 26 compounds were divided into training and test sets. The maximum distance for sample points was set to 1.0 Å during QSAR modelling generation. With 50 percent field similarity and 50 percent Dice volume similarity. The conformations were also annotated with their measured field points, which were derived in a 3D field point pattern using the XED (eXtended Electron Distribution) force field. Positive and negative electrostatic fields, as well as ‘shape' (van der Waals) and ‘hydrophobic' fields, are among these four molecular fields. The pattern of the field point represents the shape, hydrophobicity, and electrostatics of the compound in a condensed form.
Validation and visualization of model
The predictive ability of the derived 3D-QSAR model was confirmed by different statistical tests, which include correlation coefficient (r2), cross-validation regression coefficient (q2), and similarity score (Sim). The (q2) were calculated by PRESS (prediction error sum of squares) and the SSY (sum of squares of deviation of the experimental values from their mean), defined as:
where Yexp denotes the compound’s experimental biological activity in the training set, Ypred denotes the compound’s expected activity in the training set, and Ymean denotes the training set compounds’ activity mean values. The model’s robustness was also verified by calculating the coefficient of prediction, or r2 test, using the following equation:
where Ypred test denotes the expected behaviour of the test set compound, Ytest denotes the experimental activity of the test set compound, and Ymean denotes the mean values of the training set compound’s activity.
To optimise the operation, the model was calculated using the LOO (leave one out) process and leave one out cross-validation (LOOCV) is the most successful approach. The training set is done with a data size of (N–1) and the remaining data are tested (N represents the entire dataset). The Bayesian method was used to visualise the training dataset qualitatively, which assisted in understanding the hydrophobic, electrostatic, and shape features that underpin the structure–activity relationship of a selected group of compounds. The activity cliff description, on the other hand, specifies the active compounds favourable and unfavourable hydrophobicity, positive and negative electrostatic sites, and favourable form.
Identification of field points and visualization
To better understand the structure–activity relationship (SAR) process of pyrimidine analogues 3D-QSAR model was utilized. The training set compound’s coefficient and variance were also investigated to achieve the previously described activity-related field points. The model showed the regions, indicating that local fields have a significant impact on biological activity of the compounds. The greater the connection between the electrostatic/steric fields, the larger the points, and thus, proportional to higher affinity values. The QSAR model points were superimposed on the reference compounds to capture the space field point's localisation. The positive steric coefficient, which showed a steric bulk leading to higher activity, was a good predictor of the established QSAR model in the structural analysis. The activity effects of substituents were influenced by other positive and negative electrostatic coefficient influences.
Scaffold-hopping designing of new molecules
The 26 compounds were chosen from the previously published structures to construct the 3D-QSAR model (Silbermann et al. 2020). All the molecules evaluated using Hoechst 33,342 and Pheophorbide A Assays, toward ABCG2 and the range of activities were wide enough to allow for the creation of an efficient model. Marvin Sketch was used to create the 2D chemical structures, for all the molecules and they were turns subjected to molecular mechanics energy minimization using the MMFF94 force field. At the semi-empirical stage, the 3D geometries structures of all the compounds were also optimised. The Forge v10.4.2 of Cresset was used to measure the protonation state assuming a pH of 7.0, and Spark v10.4.0 was used for scaffold hopping of the molecule.
The loaded molecules displayed in the wizard together with an option to control the score weighting that is used with multiple reference molecules in an editable form. Herein, we used multiple molecules to make one chemical series and scored against an active molecule from a pyrimidine series.
Field constraints are particularly useful when you are particular to field point which is critical to define the activity which was implemented by Spark to filter out fragments with undesirable properties, transfer of electrons as in H-bonding or metal binding and the electrostatic character of the compound interaction. Lastly, databases ChEMBL common, Zinc databases, VEHICLe (ring systems) and eMolecules building blocks that are usable reagents into the corresponding R-group were used to mark an experimental setup that is required for the ND molecules and the calculations were performed. For this crucial step the calculation method pull-down menu was used to select the desired method.
Molecular docking experiment
The docking studies were performed using molecular modelling software AutoDock and cross-validated with Flare. As per previously studied protocols, pyrimidine derivatives and ND compounds were docked into the human ABCG2 modelled structure (Ahamad et al. 2018; Pasqualetto et al. 2020). The studies showed that the structure is having various binding cavities but due to the importance of the cavity 1 and cavity 2 was placed for grid binding site. By eliminating all the water molecules and adding polar hydrogens/Kollman charges, the structure was refined. The Gasteiger Huckel charges of energy-minimized ligand conformations were calculated and saved as the default format. For each docking review, the number of runs were set to 150. With a starting population of 150 randomly placed individuals and a maximum number of 25,000 generations, the Lamarckian Genetic Algorithm (LGA) was used. Inside the X, Y, and Z axes, a grid box of 60 × 60 × 60 was developed, with the grid point spacing set to 0.375 and all other docking parameters left at normal. The alignment of docked complexes analysed using PyMOL viewer (DeLano 2002) and 2D diagrams showing protein–ligand interactions were obtained by ligand interaction module of Schrödinger.
ADME/T screening
The QikProp module of Schrödinger was used to evaluate the pharmacological properties, toxicity, Lipinski estimation, and absorption parameters of the best compounds obtained.
MD simulations
The MD calculations for predicting the consistency of the protein–ligand docking complex, as well as the native structure of ABCG2, were performed using Desmond v4.1 (Desmond 2021). All the docked complexes were placed in a Simple Point Charge (SPC) water model, and the structures were neutralised using counter ions and with a physiological salt concentration of 0.15 M. The Periodic boundary conditions, the Particle Mesh Ewald (PME) method for electrostatics, a 10 threshold for Lennard–Jones interactions, and the SHAKE algorithm for restricting movement of all covalent bonds involving hydrogen atoms were all used to design the device. The native system and the ABCG2-ligand docked complexes were simulated and prepared for a multistep protocol. For a maximum of 2000 steps with solute restraints, the energy was minimised using a hybrid method of steepest descent (10 steps) and the LBFGS (Limited-Memory Broyden–Fletcher–Goldfarb–Shanno) algorithm, which was followed by a similar energy minimization for 2000 steps without solute restraints. Then, at 10 K temperature, a 12 fs simulation in the NVT ensemble for restraining non-hydrogen solute atoms was repeated, followed by a 12 fs simulation in the NPT ensemble for restraining non-hydrogen solute atoms. At temperature 300 K, 24 fs simulations in the NPT ensemble were restrained with solute non-hydrogen atoms, and 24 fs simulations in the NPT ensemble were restrained with no constraints. During the initial simulations, Berendsen thermostats and barostats were used to monitor the temperatures and pressures. In the NPT ensemble, a Nose–Hoover thermostat at 300 K and Martyna–Tobias–Klein barostats at 1.01 bar pressure were used to simulate the relaxed method for 100 ns with a time stage of 2 fs. With a time, interval of 4.8 ps, each trajectory (20, 832 trajectories) was registered. The PE, RMSD, RMSF, SASA, Rg, protein–ligand contacts, hydrogen bond monitoring, and the torsion angles were monitored through the period of simulation runtime for checking the stability of the docked complexes.
Results and discussion
Modelled ABCG2 structure and validation
The 3D structure of ABCG2 was designed using Prime-advanced homology modelling by Schrödinger and the missing residues at the positions 47–57 and 302–327 were disclosed. The PDB: 6FFC was utilized as a template which consists of 655 amino acids with two chains (Supplementary Fig. S2) (Jackson et al. 2018). The target and the template were aligned using multiple sequence alignment module embedded in the Prime. Further, loop refinement was performed with all the parameters set to default, and top model was short-listed amongst the five best structures. The RMS angle deviation, RMS bond deviation, torsion deviation 3.31, 0.021, 1.82, and 0.010, and the side chain deviation was set, respectively. The Ramachandran plot was used to produce the statistical protein report, revealing that the residues were falling in the permitted and favourable regions. The stereo-chemical properties of the selected model displayed a satisfactory distribution of Psi (φ), Phi (ψ) angles and the same was used for further analysis.
3D-QSAR model development and statistical analysis
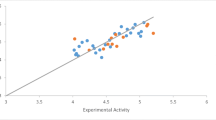
The 3D-QSAR model of the pyrimidine compounds were predicted against the ABCG2 receptor. The 3D structure of pyrimidine (26) were divided into training and test sets for the QSAR model construction. The created dataset’s highly active and low active training and test set compounds and their corresponding biological activity (IC50) was used and represented as a dependent variable. Using the random procedure, the dataset was divided into two subsets: one with 18 compounds designated as training set, and the other with 8 compounds designated as test set (Table 1). The good regression coefficient (r2 = 0.92) and cross-validation regression coefficient (q2 = 0.82) values and the cross-validated with the training set, the model shows good predictive and descriptive capabilities. On the other hand, the test set displayed accurate estimation and excellent cross-validated values (r2 = 0.73). The fitness plot was used to look at the characteristics of the developed QSAR model and cross-validation data points (Fig. 2A).
3D-QSAR outcome: A The fitness plot plotted between predicted and activity illustrated training (green color) and test set (blue color) molecules, B The alignment of 26 molecules datasets which utilized in QSAR models, C Molecular representation of aligned training and test set molecule with their respective molecular field points, D The activity cliff summary of electrostatics and hydrophobic favourable and unfavourable region of the best biological active molecule molecular regions
After the alignment of 26 compounds, the 3D-QSAR model was generated using field points and chemical descriptors. The aligned compounds (Fig. 2B) and their respective molecular field points were labelled as negative field points in cyan colour, indicating molecular regions interacting with positive or H-bond donors of the ABCG2 (Fig. 2C). The molecular regions that interact with the target's negative or H-bond acceptors are indicated by the colour red in positive field points. The gold colour represents a hydrophobic field point, which shows areas with high polarizability/hydrophobicity, whereas the yellow colour represents van der Waals field points (Fig. 2C).
Activity cliff visualization analysis
The findings of the “Activity Cliff Summary of Electrostatics” study aided in understanding the molecular regions where all pairs of compounds were compared. It showed more positive fields and less negative ones resulting in increased anticancer activity. The “Activity Cliff Summary of Hydrophobics” showed the areas of biological activity where hydrophobic interaction is either beneficial (green) or harmful (magenta). Further the “Activity Cliff Summary of Shape” is determined to identify regions where steric bulk was either excellent (green) or good (yellow). The bad steric (magenta) reflects an unfavourable form that results in decreased bioactivity (Fig. 2D).
Visualization of field contributions for predicted activity
The pyrimidine analogues fitted to the established field-based 3D-QSAR model in structural field point regions controlling predicted activity and field contributions to predict activity were investigated. The electrostatics positive (green) represent favourable contributions with increased electrostatic field activity, whereas the electrostatics negative (orange) represent unfavourable electrostatic contributions, which showed decreasing predicted activity.
Designing of new pyrimidine derivatives by Scaffold-hopping
The scaffold-hopping study was carried out to know the electrostatic fields/steric hindrance due to the substitution of a particular group in a molecule which gives us a visual idea in binding the molecule with protein. The electrostatic fields or steric hindrance are either enhanced or diminished by substituting polar and non-polar groups in a molecule. The electrostatic field is usually improved by replacing polar groups such as –OH, –NO2, –NH2 and halogen etc. The electrostatic field points have been presented by red colour (positive field) and cyan colour (negative field). However, golden colour represents lipophilicity that is increased in the molecule by substituting alkyl groups and aryl groups such as methyl, ethyl, propyl, and benzene ring. The greater is the field points; greater would be the electrostatic fields/steric hindrance. Consequentially binding capacity of the studied molecules will be increased or reduced.
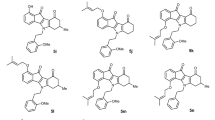
In the present study, the earlier reported biologically active Comp-9 and Comp-18 (Fig. 3A, B), and the ND compounds (ND-510 & ND-500) (Fig. 3D, F) were selected to perform the scaffold-hopping analysis for the comparative study to assign the binding efficacy with a particular protein (Fig. 3A, B). For this purpose, we utilized Cresset software. The pictorial elucidation of scaffold hopping of reported compounds (Comp-9 and Comp-18) and ND compounds (ND-510 & ND-500) have been demonstrated in (Fig. 3A–F). Due to positive field points, the −OH group formed hydrogen bonds with the ABCG2 protein. However, the –NO2 group at position R1 created a large negative field points that helped the molecule form a polar bond with protein, as shown in (Fig. 3C and D). The scaffold–hopping study revealed that the electrostatic field points was enhanced by the substitution of triazole derivatives at position R1 and hydroxyl (–OH) group at position at R2 in compound ND-510 as shown in (Supplementary Table 1). In case of compound ND-500, it was observed that a large positive field points (red colour) are produced around –CH2OCH2OH group at position R2 (Fig. 3E, F and Supplementary Table 2). Due to the increased positive fields (red) and negative fields (cyan), the tendency of the molecule to bind the donor group of proteins enhanced which was confirmed the molecular docking. Nitrogen of the triazole group of the compound ND-510 formed polar bond and substitution by −OH at position 2 generates a positive field as represented by red colour and binds with the acceptor group of protein. As evidenced by the molecular docking the −OH group also formed a polar bond. It was also observed that the positive field point is produced around –NH group by substitution of hydroxyl group and triazole derivatives at positions R1 and R2, respectively. Under the influence of this positive field –NH group formed another polar bond with ABCG2 protein, as is confirmed from the molecular docking.
Designed new molecules. A Comp-9 field effects negative field points (cyan), positive field points (red), B Representation Comp-18, C Alignment of ND-510 compounds in one frame at position R1 substituted, D Representation of Molecular field of ND compound 510 at substitution position of R1, E Representation of field effects and field points of ND-500 molecule in one frame at position R2 substituted, F Representation of Molecular field of ND compound 500 at substitution position of R2. Negative field points (cyan), positive field points (red), hydrophobic field points (gold) and Van der Waal field points (yellow)
In contrast to the ND compounds, in earlier reported Comp-9 and 18 positive and negative field points are observed appreciably around –NH and –NO2 groups (Comp-9), and –NH and –CN groups (Comp-18). In case of Comp-9 due to negative field points –NO2 groups formed Hydrogen bond with ABCG2 protein. However, the NH group formed another hydrogen bond due to positive field points. In Comp-18, the nitrogen of pyrimidine ring under the influence of negative field points formed a polar bond. However, the negative field points due to –CN group is not enough to form a polar bond. Thus, it can be stated from the scaffold-hopping study that the ND compounds (ND-510 & ND-500) are more efficient than the reported compounds (Comp-9 & Comp-18).
Virtual screening and molecular docking
Virtual screening was performed for the 26 pyrimidine compounds against the binding cleft of ABCG2. A series of pyrimidine compounds were utilized to perform virtual screening to propose a potent hit compound by AutoDock4 and the results were cross-validated with the Flare module of Cresset software. The best biological active compounds docked binding energy of Comp-9 and Comp-18 were − 9.4 kcal/mol and − 10.2 kcal/mol, respectively. Further same compound docked by the Flare module of Cresset and found quite high energies of Comp-9 and Comp-18 were attending LF-rank scores of − 9.45 kcal/mol and − 10.97 kcal/mol, respectively (Table 1).
Comp-9 and Comp-18 docking pose: The docking results of ABCG2–Comp-9 complex formed three H-bonds with Val124, Asp127, and Arg378, are forming stable confirmations (cyan dotted line showed weak H-Bond and green dotted line showed strong H-bond interaction) (Fig. 4A). Additionally, the residues Gly104, Val106, Leu107, Ile108, Asn109, Gly122, Tyr123, Val125, Gln126, Asp128, Thr371, Ser372, Phe373, Tyr469, Phe470, Leu471, and Gly472 provided the stability towards ABCG2 with several Van der Waal’s interactions. The complex Comp-18 ABCG2 at the interactive site revealed formation of one H-bond with residue Thr214, and the residues Cys43, Tyr44, Thr82, Met131, Lys172, Val173, Ser176, Leu209, Asp210, Glu211, Pro212, Thr213, Gly215, Leu216, Ile239, Phe240, Ser241, Ile242, His243, Gln244, Pro245, Arg246, and Tyr247 were involved in Van der Waal interactions (Fig. 4B).
Docking confirmations and representation of H-bond forming residues of earlier synthesised compounds and the ND compounds towards the binding pocket of ABCG2 receptor amino acids in ball and stick model and ligands in stick model, A Comp-9-ABCG2 complex, B Comp-18-ABCG2 complex, C ND-510-ABCG2 complex and D ND-500-ABCG2 complex
The screening and the docking analysis were performed by virtual screening of 1010 compounds, from the derived 3D-QSAR/scaffold hopping compounds. The results also revealed the binding potency, molecules active poses, and their mechanism of action.
ND-510 and ND-500 docking pose: The docking experiment of ND-510 towards the ABCG2 binding cleft was stabilized with five H-bonds by residues Tyr44, Leu102, Gln126, Asp217, and Val130 (Fig. 4C). The H-bond interaction between ligand and protein. Apart from H-bond formation, the residues Val46, Gly80, Asp98, Pro99, Ser100, Gly101, Ser103, Gly104, Asp105, Val106, Ala115, Phe117, Gly122, Asp127, Asp128, Val129, Met131, Gly132, Thr133, Leu134, Thr135, Thr213, Gly215, Leu216, Ser218, Ser219, Arg246, Tyr247, Ser248, Ile249, Arg465, Val466, Phe470, and Tyr469 are seen in the centroid region forming hydrophobic bonds with ABCG2 (Table 2). The affinity of ND-500 compound revealed that the complex binding orientation was strengthened with the formation of two H-bonds with the amino acid residues Ser241 and Arg263 headed for ABCG2 receptor (Fig. 4D). The residues Gly83, Gly84, Gly85, Lys86, Thr133, Ile201, Ser205, Leu216, Ser218, Ala221, Lys233, Gln234, Thr237, Ile238, Ile242, Tyr247, Ser248, Ile249, Lys251, Ser261, Gly262, and Leu264 were involved in Van der Waal’s interactions (Table 2).
The comparative results of the screening and docking resulted in two best compounds namely, ND-510 and ND-500 with good affinity towards the ABCG2 binding pocket. The configurations were characterized as a combined score of Van der Waals forces, hydrogen bonding, π interaction, and other relevant parameters, and signified in the form of a docking score. The higher score indicates a higher chance of ligand–protein binding and the compounds, ND-510 (R1) and ND-500 (R2) rendered the LF-rank scores of − 14.416 kcal/mol and − 16.472 kcal/mol, respectively (Fig. 4C, D). The results showed that the compounds have good docking score in comparison to standard compounds i.e., high binding affinity of these prioritized compounds. Additionally, a 2D diagram was generated to reveal different molecular interactions. The findings rendered a molecular level understanding to infer that the proposed compounds ND-510 and ND-500 are promiscuous and could probably act as potential antagonists towards the active groove of ABCG2.
Prime-MM/GBSA analysis
The binding free energy (∆G) was examined in Schrödinger Prime-MM/GBSA module on the compound and the designed hits complex with ABCG2. The ∆G values of two compounds namely, Comp-9 and Comp-18 were observed in the acceptable range of − 36.41 kcal/mol and − 29.18 kcal/mol (Fig. 5A–B), whereas ∆G values of ND compounds ND-510 and ND-500 were observed with values of -60.83 kcal/mol and -61.01 kcal/mol, respectively (Table 2 and Fig. 5C–D). The outcome of docking and binding energies portrayed the residues Tyr44, Leu102, Val124, Gln126, Asp127, Val130, Thr214, Asp217, Ser241, Arg263, and Arg378 played an essential role in stabilizing the protein–ligand complex and these amino acids are involved in hydrogen bond (pink arrow interaction between ligand and protein amino acids) and hydrophobic interactions by Schrödinger–ligand interaction diagram.
In contrast, few residues in the study showed a good binding affinity towards the active pocket of ABCG2. The overall results revealed that the proposed hits have the edge over the ABCG2 binding cleft, which is attributable to a stronger binding ability. The additional Van der Waals interacting residues and the binding strengthens the bond, thus a tighter fit to the binding groove of ABCG2, which may hinder the substrate accessibility and the biological function and therefore, could be exploited for new anticancer drugs discovery (Fig. 5A–D).
Predicted pharmacokinetic properties
The pharmacological properties of compounds, such as flexibility, molecular size, and hydrophobicity, may significantly impact their molecular behaviour in living systems. With a good combination of permeability and solubility properties, good bioavailability can be achieved. As a result, it is critical to predict Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADME/T) profiling during hit detection and optimisation.
For selecting and prioritising drug candidates, ADME/T and pharmacological properties are critical. The 26 ES compounds of ABCG2 was studied for drug-like properties. The QikProp module’s performance indicated that the compounds’ pharmacological properties were not violated. The drug compounds come into contact with a variety of membrane barriers as well as the target cell. These permeability predictions will assist in the understanding of cell-based bioassays; for example, the permeability over human skin was predicted to be in the range of − 3.0–1.2. The results show that all compounds have an ideal LogP range of − 2.0–6.5, indicating a good balance of permeability and solubility and good oral bioavailability. The capacity of the compound to dissolve in a lipophilic (non-aqueous) medium was also calculated, with a threshold QplogKhsa of − 1.5–1.5, SASA of 500 to 600, and log Kp of 0.0 to 175.0, respectively (Supplementary Table 3). In comparison to all the 26 compounds, the results revealed that the ND compounds, namely ND-510 and ND-500, were well within the acceptable range, indicating high bioavailability (Table 2A and B).
Lipinski rule violation: The compounds were further examined for the Lipinski rule of five parameters. The results revealed that the compounds showed threshold molecular weight > 250 and < 370, Hydrogen bond acceptor (2.5–5.5) and Hydrogen bond donor (1–2). The results also showed that the compounds followed the Rule of Five (0–1). On the other hand, the ADME/T screening of ND compounds ND-510 and ND-500 imply that these compounds may be eliminated more easily from the body and fitted with the Lipinski’s properties thereby having good human oral absorption drug-likeliness in the biological systems (Table 1 and Supplementary Table 4).
MD simulations interpretation
The MD simulation protocol provided exact binding orientations and the interacting residues of the docked complexes of the system embedded with water molecules, atoms, volume, temperature, and pressure. The membrane model of DPPC (325 K) was placed automatically around the ABCG2 and saved the model in full system with OPLS-2005 force field. An orthorhombic box with distance of grid set to 10 × 10 × 10 Å and volume of ~ 1300 Å was set. The system was solvated by addition of salts with 0.15 M and the system was prepared before initiating the simulations (Supplementary Fig. S2). The simulations were initiated with the full system analysis of the docked complexes with 68,000 and 67,980 atoms for the Comp-9 and Comp-18 docked complexes, whereas 79,748 atoms and 73,546 atoms were seen for ND compounds ND-510 and ND-500, respectively, with ABCG2 receptor. The MD simulation period run of 100 ns was executed for four docked complexes and 20, 832 trajectories were analysed for PE, RMSD, RMSF, SASA, Rg, H-bond, protein–ligand contacts, torsion angle, SSE%, and interaction fraction.
Potential energy, RMSD, and RMSF analysis
The potential energy was analysed for the Comp-9 and Comp-18 docked complexes, which revealed a stable configuration throughout the 10,000 ps run time. The complex Comp-9 and Comp-18 demonstrated energy consistency with − 159,000 kcal/mol and − 157,000 kcal/mol (Fig. 6B and Supplementary Fig. S4B). In comparison, the ND compounds ND-510 and ND-500 portrayed PE range of − 161,000 kcal/mol and − 160,000 kcal/mol (Fig. 6A and Supplementary Fig. S4A). The outcome of energy analysis suggested that the ND-510 and ND-500 complexes were consistent, and the system was inacceptable physiological, environmental condition compared to Comp-9 and Comp-18. The RMSD was measured to get the average change in the displacement of a selection of atoms for a particular frame with respect to a reference frame. To define the structural conformations throughout the simulations the range should be from 1 to 3 Å is acceptable for small and globular proteins. The average RMSD of the Comp-9 and Comp-18 complexes were noted to be 4.5 Å and 4.8 Å, respectively. The ND-510 and ND-500 presented the average RMSD of 2.8 Å and 3.0 Å, respectively. The RMSD analysis also rendered that the ABCG2 was deviating with a high drift of 9 Å in case of earlier synthesized compounds whereas; the protein was fallen steady in ND compounds around 5 Å. Therefore, the Comp-9 and Comp-18 complex with ABCG2 was originated in all proper and better binding poses. Interestingly, ND-510 and ND-500 complexes displayed that were relatively more stable throughout the simulation time than the (Fig. 6C, D and Supplementary Fig. S4C, 4D).
MD simulation analysis. A Potential energy plot of ABCG2–ND-510 complex, B Potential energy plot of ABCG2-Comp-9 complex, C RMSD graph plotted against ABCG2 and ND-510 compound, D RMSD plot of ABCG2-Comp-9 complex, E RMSF plot of showing b-factor and ligand contacts ABCG2-ND-510 complex, F RMSF plot of ABCG2-Comp-9 complex
The RMSF determines fluctuations and characterizes local changes along with the protein chain of each residue with ABCG2 receptor. The average RMSF of Comp-9 and Comp-18 complexes were observed in the range of 4.1 Å and 4.3 Å, whereas ND-510 and ND-500 presented an average RMSF of 3.3 Å and 3.0 Å, respectively. The peaks in the RMSF indicate areas of the protein that fluctuate. Hence, it was evident from the RMSF analysis that fluctuations were inclined out of threshold compared to ND molecules. The analysis also presented that the ligand contact was comparatively less in the compounds than ND molecules, which suggesting the stronger affinity towards the binding cleft of the ABCG2 receptor (Fig. 6E, F and Supplementary Fig. S3E, 3F).
SSE%, SASA, and Rg analysis
The protein secondary structure elements percentages (SSE%) like alpha-helices and beta-sheets, loops and strands are monitored throughout the simulation run time for all the four docked complexes. The SSE% reports rendered that the helix and sheets were distributed correctly in Comp-9, Comp-18, ND-510, and ND-500 complexes, by each residue over the simulation time. In comparison, the ND compounds revealed more stable structural plasticity than the compounds towards the ABCG2.
The SASA and the measure of the compactness of structures were monitored and revealed that Comp-9 and Comp-18 had a range of 200 Å2 and 220 Å2. The ND-510 and ND-500 showed a stable SASA range of 75 Å2 and 80 Å2, respectively. The comparative SASA analysis comprehended that the ND compounds showed more accessibility towards water molecules with ABCG2 (Fig. 7 and Supplementary Fig. S5). The PSA, MolSA, Van der Waals surface area, and solvent accessible surface area of the ND-510 and ND-500 compounds were more stable than the Comp-9 and Comp-18 (Fig. 7 and Supplementary Fig. S5).
Rg analysis was carried for all the complexes for further investigation. The ligand–protein complex showed its principal atom and moment of inertia that dictates the protein compactness (Rawat and Biswas 2012). The analysis displayed that the Rg of Comp-9 and Comp-18 was presenting kinking trend with decrease ranges of 4.5 Å and 4.3 Å. Contrastingly, the Rg of ND-510 and ND-500 showed a smooth trend and subsequently increased ranges of 4.8 Å and 5.0 Å, respectively. From the analysis of SASA and Rg, the results suggested that the compactness, area of stability was more consistent for ND-510 and ND-500 on ABCG2 than the Comp-9 and Comp-18 (Fig. 7A, B and Supplementary Fig. S5A, 5B).
Protein–ligand contacts and torsion angles
After simulations, the stability of the Comp-9 and Comp-18 compared with the ND compounds using protein–ligand contacts, total contact energies (Supplementary Fig. S6), and the torsion angles generated all the trajectories. The complex ND-510 and ABCG2 formed hydrogen bonds with the residues Cys43, Tyr44, Leu102, Gln126, Val130, Lys233, Ile239, Gln244, and Pro245 (Fig. 8A). The complex was also strengthened by forming eight water bridges with the residues Cys43, Val130, Ser176, Thr213, Lys233, Ile239, Gln244, and Arg246. The complex ND-500 and ABCG2 formed hydrogen bonds with Gly84, Gly85, Thr202, Ser218, Lys233, Gln234, Thr237, Ser241, Lys251, Ala260, Ser261, and Arg263 (Supplementary Fig. S7A). The amino acid residue Gly84, Gln234, Lys251, Ser261, and Arg263 formed ionic bonds with ABCG2. The complex was also strengthened by the formation of twenty water bridges with the residues Gly84, Gly85, Thr202, Asp203, Pro204, Leu216, Ser218, Ala221, Asn222, Lys233, Gln234, Thr237, Ile238, Ser241, Gln244, Ser248, Lys251, Ala260, Ser261, Gly262, and Arg263.
The complex Comp-9 and ABCG2 formed eight hydrogen bonds with the residues Val 124, Asp 127, Ser176, Leu209, Asp217, Arg236, His350, and Arg378 (Fig. 8B). The amino acid residue Tyr44, Val124, Val125, Gln126, and Lys172 formed an ionic bond, and the residues Tyr44, Val124, Val125, Gln126, Asp127, Asp171, Lys172, Asp175, Ser176, Asp210, Pro212, Asp217, Arg236, and Thr237 strengthened by the formation of fourteen water bridges. The complex Comp-18 and ABCG2 formed six hydrogen bonds with the residues Thr214, His457, Glu458, Val536, Leu539, and Leu540 (Supplementary Fig. S7B). The residues Arg56, Glu60, Lys168, Thr214, Leu454, His457, Ser461, Leu478, Val536, Leu539, and Leu540 were strengthened by the formation of eleven water bridges. The ND compounds increased the binding affinity and entropy gain in the complex formation by forming good number of hydrogen bonds with inhibiting the binding cleft of ABCG2.
The torsion angles were more stable within the allowed range for the ND-510 and ND-500 compared to Comp-9 and Comp-18 during the simulation period. The results also rendered that every rotatable bond in the ligand was consistently stable in ND compounds (Supplementary Fig. S8). A dial and bar plots accompany each rotatable bond torsion of the four docked complexes, with the dial/radial plot describing the torsion’s conformation during the race. The start of the simulation is depicted in the plot’s middle, and the time progression is plotted radially outward. The probability density of the ligand torsion towards the receptor dictating the protein-bound conformation is shown in the bar plots, which summarises the data from the dial plots.
Overall, from MD simulations analysis, PE, RMSD, RMSF, hydrogen bonding patterns, water bridges, protein–ligand contacts and torsion angles of ABCG2 docked complexes revealed stability of ND compounds in natural physiological, environmental conditions and better when compare with compounds. Additionally, the ND compounds were portraying good pharmacokinetic properties that showed better interactions towards the binding cleft of ABCG2. From the study the designed antagonist molecules ND-510 and ND-500, could be useful to develop novel inhibitors, and, in turn, helps to inhibit the overexpression levels of ABCG2 in cancer conditions.
Conclusion
We developed a robust 3D-QSAR model to identify potential pyrimidine-based ABCG2 inhibitors. The results of comparative molecular docking and MD simulations study indicated that compounds ND-510 and ND-500 as the most promising candidates. Finally, drug-likeness, ADMET, and related properties studies indicated that all the designed compounds are good candidates and have a pharmacokinetic vision due to balance of permeability, solubility, and good oral bioavailability. Therefore, these designed new pyrimidine derivatives could be evaluated as a new drug against MDR breast cancer.
References
Ahamad S, Hassan MI, Dwivedi N (2018) Designing of phenol-based β− carbonic anhydrase1 inhibitors through QSAR, molecular docking, and MD simulation approach. 3 Biotech 8(5):1–18
Ahamad S, Islam A, Ahmad F, Dwivedi N, Hassan MI (2019) 2/3D-QSAR, molecular docking and MD simulation studies of FtsZ protein targeting benzimidazoles derivatives. Comput Biol Chem 78:398–413
Bailey-Dell KJ, Hassel B, Doyle LA, Ross DD (2001) Promoter characterization and genomic organization of the human breast cancer resistance protein (ATP-binding cassette transporter G2) gene. Biochim Biophys Acta BBA Gene Struct Expr 1520(3):234–241
Bauer MR (2018) Using Spark to design macrocycle BRD4 inhibitors
Bento AP, Gaulton A, Hersey A, Bellis LJ, Chambers J, Davies M, Krüger FA, Light Y, Mak L, McGlinchey S (2014) The ChEMBL bioactivity database: an update. Nucleic Acids Res 42(D1):D1083–D1090
Bhardwaj B, Baidya A, Amin S, Adhikari N, Jha T, Gayen S (2019) Insight into structural features of phenyltetrazole derivatives as ABCG2 inhibitors for the treatment of multidrug resistance in cancer. SAR QSAR Environ Res 30(7):457–475
Chen Z, Shi T, Zhang L, Zhu P, Deng M, Huang C, Hu T, Jiang L, Li J (2016) Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: a review of the past decade. Cancer Lett 370(1):153–164
Costantino C, Alba D, Cimino L, Conforto A, Mazzucco W (2021) The role of vaccination and screening in limiting the worldwide disease burden of preventable female cancers: a review. Women 1(1):16–28
Dean M, Hamon Y, Chimini G (2001) The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res 42(7):1007–1017
DeLano WL (2002) Pymol: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr 40(1):82–92
Desmond S (2021) Schrödinger. Retrieved from https://www.schrodinger.com/desmond. Accessed 6 Oct 2021
Ejendal K, Hrycyna CA (2002) Multidrug resistance and cancer: the role of the human ABC transporter ABCG2. Curr Protein Pept Sci 3(5):503–511
Estrella V, Chen T, Lloyd M, Wojtkowiak J, Cornnell HH, Ibrahim-Hashim A, Bailey K, Balagurunathan Y, Rothberg JM, Sloane BF (2013) Acidity generated by the tumor microenvironment drives local invasion. Can Res 73(5):1524–1535
Gad SC (2014) QSAR. In: Third E (ed) Wexler PBT-encyclopedia of toxicology. Academic Press, Oxford, pp 1–9
Homolya L, Orbán TI, Csanády L, Sarkadi B (2011) Mitoxantrone is expelled by the ABCG2 multidrug transporter directly from the plasma membrane. Biochim Biophys Acta (BBA) Biomembr 1808(1):154–163
Irwin JJ, Shoichet BK (2005) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model 45(1):177–182
Jackson SM, Manolaridis I, Kowal J, Zechner M, Taylor NM, Bause M, Bauer S, Bartholomaeus R, Bernhardt G, Koenig B (2018) Structural basis of small-molecule inhibition of human multidrug transporter ABCG2. Nat Struct Mol Biol 25(4):333–340
Kanipakam H, Sharma K, Thinlas T, Mohammad G, Pasha MQ (2021) Structural and functional alterations of nitric oxide synthase 3 due to missense variants associate with high-altitude pulmonary edema through dynamic study. J Biomol Struct Dyn 39(1):294–309
Katari SK, Natarajan P, Swargam S, Kanipakam H, Pasala C, Umamaheswari A (2016) Inhibitor design against JNK1 through e-pharmacophore modeling docking and molecular dynamics simulations. J Recept Signal Transduction 36(6):558–571
Khunweeraphong N, Stockner T, Kuchler K (2017) The structure of the human ABC transporter ABCG2 reveals a novel mechanism for drug extrusion. Sci Rep 7(1):1–15
Krapf MK, Gallus J, Wiese M (2017) 4-Anilino-2-pyridylquinazolines and-pyrimidines as highly potent and nontoxic inhibitors of breast cancer resistance protein (ABCG2). J Med Chem 60(10):4474–4495
Krapf MK, Gallus J, Vahdati S, Wiese M (2018) New inhibitors of breast cancer resistance protein (ABCG2) containing a 2, 4-disubstituted pyridopyrimidine scaffold. J Med Chem 61(8):3389–3408
Kusuhara H, Sugiyama Y (2007) ATP-binding cassette, subfamily G (ABCG family). Pflügers Arch Eur J Physiol 453(5):735–744
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26(2):283–291
Neves BJ, Braga RC, Melo-Filho CC, Moreira-Filho JT, Muratov EN, Andrade CH (2018) QSAR-based virtual screening: advances and applications in drug discovery. Front Pharmacol 9:1275
Parkali PM, Shyam Kumar A, Johanna KP, Prodensia TS, Turaga S, Shaiva V, Pujar GV, Joshi SD, Aminabhavi TM, Dixit SR (2021) Molecular docking and three-dimensional quantitative structure-activity relationships for antitubercular pyrimidine derivatives. Polycycl Aromat Compd. https://doi.org/10.1080/10406638.2021.1885455
Pasqualetto G, Schepelmann M, Varricchio C, Pileggi E, Khogali C, Morgan SR, Boostrom I, Rozanowska M, Brancale A, Ferla S (2020) Computational studies towards the identification of novel rhodopsin-binding compounds as chemical chaperones for misfolded opsins. Molecules 25(21):4904
Rawat N, Biswas P (2012) Hydrophobic moments, shape, and packing in disordered proteins. J Phys Chem B 116(22):6326–6335
Sharma R (2021) Global, regional, national burden of breast cancer in 185 countries: evidence from GLOBOCAN 2018. Breast Cancer Res Treat 187(2):557–567
Silbermann K, Stefan SM, Elshawadfy R, Namasivayam V, Wiese M (2019) Identification of thienopyrimidine scaffold as an inhibitor of the ABC transport protein ABCC1 (MRP1) and related transporters using a combined virtual screening approach. J Med Chem 62(9):4383–4400
Silbermann K, Li J, Namasivayam V, Baltes F, Bendas G, Stefan SM, Wiese M (2020) Superior pyrimidine derivatives as selective ABCG2 inhibitors and broad-spectrum ABCB1, ABCC1, and ABCG2 antagonists. J Med Chem 63(18):10412–10432
Silbermann K, Li J, Namasivayam V, Stefan SM, Wiese M (2021) Rational drug design of 6-substituted 4-anilino-2-phenylpyrimidines for exploration of novel ABCG2 binding site. Eur J Med Chem 212:113045
Singh VK, Chaurasia H, Mishra R, Srivastava R, Naaz F, Kumar P, Singh RK (2022) Docking, ADMET prediction, DFT analysis, synthesis, cytotoxicity, antibacterial screening and QSAR analysis of diarylpyrimidine derivatives. J Mol Struct 1247:131400
Sivasankarapillai VS, Somakumar AK, Joseph J, Nikazar S, Rahdar A, Kyzas GZ (2020) Cancer theranostic applications of MXene nanomaterials: recent updates. Nano-Struct Nano-Objects 22:100457
Tadayon M, Garkani-Nejad Z (2019) In silico study combining QSAR, docking and molecular dynamics simulation on 2, 4-disubstituted pyridopyrimidine derivatives. J Recept Signal Transduction 39(2):167–174
Acknowledgements
SA is thankful for the IFTM University, Jamia Millia Islamia and Translational Bioinformatics Group, International Centre for Genetic Engineering and Biotechnology for providing the working environment and excellent infrastructure to peruse the doctorial research work.
Author information
Authors and Affiliations
Contributions
SA contributed to extensive in-silico exercises, MD simulations investigations, and original draft write-up in the manuscript. MIH contributed to corrections in the manuscript. DG contributed to review and corrections in the manuscript. ND contributed major inputs, reviews, and corrections in the manuscript. AI contributed to the final review, correspondence, and corrections in the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
None of the authors have conflicts and declare no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ahmad, S., Hassan, M.I., Gupta, D. et al. Design and evaluation of pyrimidine derivatives as potent inhibitors of ABCG2, a breast cancer resistance protein. 3 Biotech 12, 182 (2022). https://doi.org/10.1007/s13205-022-03231-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03231-1