Abstract
Medium-chain fatty acids (MCFAs) are an ideal feedstock for biodiesel and a range of oleochemical products. In this study, different combinations of CnFATB3, CnLPAAT-B and CnKASI from coconut (Cocos nucifera L.) were coexpressed in transgenic Arabidopsis thaliana by a Cre/LoxP multigene expression system. Transgenic lines expressing different combinations of these genes were designated FL (FatB3 + LPAAT-B), FK (FatB3 + KASI) and FLK (FatB3 + LPAAT-B + KASI). The homozygous seeds of transgenic Arabidopsis thaliana expressing high levels of these genes were screened, and their fatty acid composition and lipid contents were determined. Compared with its content in wild-type A. thaliana, the lauric acid (C12:0) content was significantly increased by at least 395%, 134% and 124% in FLK, FL and FK seeds, respectively. Meanwhile, the myristic acid (C14:0) content was significantly increased by at least 383%, 106% and 102% in FL, FLK and FK seeds, respectively, compared to its level in wild-type seeds. Therefore, the FLK plants exhibited the best effects to increase the level of C12:0, and FL expressed the optimal combination of genes to increase the level of 14:0 MCFA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Medium-chain fatty acids (MCFAs), defined as fatty acids with fewer than 18 carbons, most of which are 12, 14, and 16 carbons in length, are natural compounds present in plant tissues (Schönfeld and Wojtczak 2016). In industries, MCFAs are an ideal source for biodiesel and a wide range of oleochemical feedstocks, including pharmaceuticals, personal care products, lubricants and detergents (Martínez-Vallespín et al. 2016; Jovancevic et al. 2017). The metabolic engineering of high biomass crops for increased lipid content and desired fatty acid compositions has been proposed as a novel platform to produce low-cost, energy-dense lipids such as triacylglycerol (TAG) (Chapman et al. 2013; Troncoso-Ponce et al. 2013). However, the impact of increasing MCFA levels by genetic transformation is theoretically limited.
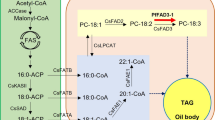
Fatty acid (FA) and TAG synthesis in plants is complex and regulated by different factors in different cellular compartments, and each intermediate biochemical reaction is catalyzed by specialized enzymes (Durrett et al. 2008). The first committed step of de novo FA biosynthesis in plastids is catalyzed by acetyl-coenzyme A (CoA) carboxylase (ACCase), which converts acetyl-CoA to malonyl-CoA, which is then condensed by a set of beta-ketoacyl-acyl carrier protein synthases (KASs) that are involved in FA chain elongation from C4 to C18 (Gornicki and Haselkorn 1993). Among the several types of KASs, CnKASI, which was cloned from the coconut endosperm, is the most essential and catalyzes the production of C16:0-ACP from C4:0-ACP in six iterative cycles of C2 condensation (Yuan et al. 2014; Yang et al. 2016).
After de novo FA synthesis occurs in the plastid, the transfer of nascent FA to the endoplasmic reticulum (ER) for TAG assembly requires the hydrolysis of acyl-ACP to release free FA. This reaction is catalyzed by acyl-ACP thioesterases. Plant acyl-ACP thioesterases are separated into two classes, termed FatA and FatB3. Generally, FatA enzymes preferentially hydrolyze 18:1-ACP; whereas, saturated acyl-ACP is the preferential substrate of FatB3 thioesterases (Dussert et al. 2013a). Many reports revealed high FatB3 transcription levels in lipid-rich tissues containing high saturated FA levels, which implied a connection between FatB3 function and saturated FA biosynthesis (Joët et al. 2009). In fact, CnFatB1 and CnFatB2, which are two paralogs of FatB3 from the coconut endosperm, showed specificity toward 14:0 and 16:1 MCFAs; whereas, CnFatB3 exhibited a substrate preference for C12:0 and C14:0 (Jing et al. 2011). Most importantly, in plants that store MCFAs, CnFatB3 is the only paralog that encodes an MCFA-specific thioesterase, which is able to prematurely truncate the elongating acyl-ACP within the plastid and allow its export into the cytoplasm (Toni and Anthony 2001).
One of the two main routes of TAG assembly in the ER leads to the accumulation of TAG acylated with MCFAs. Through this assembly pathway, lysophosphatidic acid acyltransferase (LPAAT), which is separated into two classes of genes termed LPAAT-A and LPAAT-B, is involved in the transfer of acyl-CoA to the second position of glycerol. LPAAT-Bs display a preference for distinct, unusual saturated or unsaturated acyl groups that are mostly expressed in storage organs and have been reported in several different plants, including meadowfoams (Limnanthes alba and Limnanthes douglasii) (Brough et al. 1997) and garden nasturtium (Tropaeolum majus) (Taylor et al. 2010). Consistent with this, CnLPAAT-B from the coconut endosperm also efficiently transfers medium-chain fatty acyl-CoA substrates to the sn-2 position during the biosynthesis of TAG (Dussert et al. 2013b; Yuan et al. 2015a).
Regarding MCFA engineering, previous studies have made some progress on the modification of seed oils to increase MCFA content and predominantly focused on the engineering of lauric acid (C12:0) (Reynolds et al. 2017). In the MCFA biosynthesis process, specialized thioesterase (FatB3) is an essential enzyme that prematurely truncates the standard fatty acid elongation cycle within the plastid and allows the export of MCFAs into the cytoplasm. In the cytoplasm, MCFAs are incorporated into TAG by three key acyltransferases: glycerol-3-phosphate acyltransferase (GPAT), lysophosphatidic acid acyltransferase (LPAAT) and diacylglycerol acyltransferase (DGAT), which avoid the incorporation of lipid head groups into the cell membrane. Even so, the synergistic combination of multiple genes is needed to increase the incorporation of MCFAs into TAG.
Cre/LoxP multigene expression system is a site-specific DNA excision system, which originated from Escherichia coli phage P1 and mediated by the Cre recombinase, recognizes and interacts with the two 34-bp lox sites to direct the excision, integrate or inverse the intervening DNA sequence depending on the orientation of lox sites towards each other (Hoess and Abremski 1985). Proper functioning of Cre-mediated recombination is independent of host cofactor or accessory proteins. Additionally, Cre-recombinase specifically recognize the loxP sequence of 34 bp in length and excises a DNA segment flanked by two direct repeats of loxP, leaving a single copy of loxP (Kasai and Harayama 2016). This system has been proven to be a powerful marker rescue tool, and overcomes the limitations of sexual crossing in eukaryotic cells (Dohlemann et al. 2016).The function of the Cre/LoxP system was recently well documented and considered as a more efficient multigene recombination system (Mizutani et al. 2012).
The coconut (Cocos nucifera L.) endosperm accumulates a high saturated oil content (approximately 91.4%), and more than 60% of its total fatty acid composition is MCFAs; the coconut endosperm is especially rich in lauric acid (C12:0; 47.8%) and myristic acid (C14:0; 19.4%) (Patil and Benjakul 2018). In this study, CnKASI, CnFATB3 and CnLPAAT-B from coconut palm were introduced into a Cre/LoxP multigene expression system and transformed into Arabidopsis. The lauric acid (C12:0) content increased in all FL, FK and FLK transgenic Arabidopsis seeds when compared with that in wild-type seeds. The myristic acid (C14:0) content also increased. These results suggested that CnKASI, CnFatB3 and CnLPAAT-B synergistically improve the MCFA yield. Additionally, the lipid content was increased in all FL, FK and FLK transgenic Arabidopsis seeds compared with that in the wild-type seeds. The combination of CnFatB3 and CnKASI had the optimum combinatorial expression effect on lipid yield. As far as we know, this is the first report to reveal synergy among MCFA-associated genes from coconut palm. This study will provide a potential approach to achieve high MCFA content by metabolic engineering in plants.
Materials and methods
Plant materials
Coconuts (Cocos nucifera L.) were obtained from the Coconut Research Institute, Chinese Agricultural Academy of Tropical Crops, Hainan, PR China (Yuan et al. 2015b). The samples were randomly harvested, immediately frozen in liquid nitrogen, and stored at − 70 ℃ for RNA extractions. All chemicals, endonucleases and other required enzymes were obtained from Sigma (St. Louis, MO, USA) or TaKaRa (Dalian, China) unless otherwise stated. Arabidopsis thaliana ecotype Columbia was used in this study. Arabidopsis plants were cultivated in a growth chamber at 23 ℃ with a 16-h photoperiod (16 h of 150 μE m2 sec1 light and 8 h of darkness).
Construction of target genes in donor vectors
The acyl-ACP thioesterases FatB3 (AEM72521.1), LPAAT-B (Q42670.1) and KASI (JX275887) from coconut, named CnFatB3, CnLPAAT-B and CnKASI, respectively, were cloned as in our previous studies (Yuan et al. 2017, 2015a, b). The three genes were then subcloned into the pYL322-d1 and pYL322-d2 vectors. The seed-specific promoter napin, Pro-at5g5400 and Pro-at5g1646 (Jeong et al. 2014) were employed to drive the expression of CnFatB3, CnLPAAT-B and CnKASI, respectively. The PCR primers used to amplify these genes and promoters are shown in Supplementary Table 1.
Construction of multigene expression vectors
To generate the pFatB3–KASI–380DTH, pFATB3–LPAAT-B–80DTH and pFatB3–KASI–LPAAT-B–380DTH vectors to produce transgenic plants, the CnFatB3, CnLPAAT-B and CnKASI gene expression cassettes were introduced into the multiple gene expression vector pYLTAC380DTH by the homologous recombination method (Lin et al. 2003). In brief, a mixture of the donor plasmid, pYL322-d1/NAPIN-FATB3, and the receptor plasmid, pYLTAC380DTH, was introduced into E. coli strain NS3529, which expresses Cre recombinase. The obtained recombinant plasmids were digested using the restriction enzyme NotI (Thermo Fisher, USA) to verify target gene assembly into the receptor plasmids. The receptor plasmid FatB3–380DTH and the donor plasmid pYL322-d2/16460-KASI were mixed and transformed into competent NS3529 cells on the second round. Colonies were screened, and recombination plasmids were extracted for digestion with PI-SceI. The digested plasmids were transformed into competent TransT1 cells. Recombinant colonies were selected for PCR determination. The donor plasmid PYL322-d1/5400-LPAAT-B and the second recombination plasmid FatB3–KASI–380DTH were assembled by the same method on the third round.
Arabidopsis transformation and selection
The recombined vectors were transported into Agrobacterium strain GV3101 cells by electroporation. The transformants were verified by PCR and used for plant transformation. Arabidopsis was transformed by the floral dip method (Bent and Clough 1998). The transformants were first selected by growth with hygromycin (30 mg/L) (Zhang et al. 2006) and then confirmed by PCR amplification of the genomic DNA using the following primers: Hpt-F: TGTCCTGCGGGTAAATAGC and Hpt-R: ATGTTGGCGACCTCGTATT. The conditions for PCR amplification were 98 ℃ for 30 s, 32 cycles of 98 ℃ for 10 s, 69 ℃ for 30 s, 72 ℃ for 40 s and a final extension step of 72 ℃ for 7 min. Homozygous lines in the T3 generation were identified by segregation analyses.
Quantitative real-time PCR (qRT-PCR) analysis
The seeds of wild-type and transgenic Arabidopsis were collected after 40 flowering when the seeds were fully matured. Total RNA was extracted from seeds using the instructions with the RNA extraction kit (Takara, Dalian, China). The transcript levels of the three genes in Arabidopsis seeds were determined using qRT-PCR. qRT-PCR amplification was performed by the use of a SYBR@ PremixEx Taq™II Kit according to the manufacturer’s instructions (TaKaRa, Dalian, China) with β-actin as an internal control. The reactions were performed in triplicate, including a template-free reaction and a reverse transcriptase-free reaction. The qRT-PCR primers used to amplify CnFatB3, CnKASI and CnLPAAT-B are shown in Supplementary Table 2.
Fatty acid composition detection
Total lipids were extracted in triplicate from the mature seeds of single transgenic Arabidopsis and wild-type Arabidopsis plantlets using dichloromethane:methanol (2:1). Fatty acid methyl esters were recovered using N-hexane. Analysis of the fatty acid methyl esters was performed using gas chromatography (GC), and methyl heptadecanoate (C17:0) obtained from Sigma (St. Louis, MO, USA) was used as an internal standard. FAMEs were analyzed using an Agilent Technologies GC 19091 N-1331 column with HP-INNOWax (30.0 m × 0.25 mm × 0.25 um) and a flame ionization detector. FAMEs in the mixture were identified by comparing their retention times with those of 15 FAME standards (Nu-Check Prep, USA).
Statistical analysis
Three biological triplicates were used for every sample to ensure reproducibility, and the data provided as mean ± standard deviation. Student’s t test was used to analyze significant differences between the transgenic and nontransgenic (with empty vector) groups. Differences with p < 0.05 and p ≤ 0.01 were regarded as statistically significant and extremely significant, respectively.
Results
Construction of a Cre/LoxP recombination system carrying CnFatB3, CnLPAAT-B and CnKASI multiple genes inserts
The CnFatB3, CnLPAAT-B and CnKASI genes were amplified from the previously described plasmids (Yuan et al. 2017, 2015a, b). The seed-specific promoters napin, At5400 (defined as Pro-at5g5400) and At16460 (defined as Pro-at5g16460) were cloned from Arabidopsis thaliana genomic DNA. In our study, napin was used to promote CnFatB3 expression, and Pro-at5g5400 and Pro-at5g1646 were used to promote CnLPAAT-B and CnKASI expression, respectively (Fig. 1a–c). The NOS terminators were derived from the plant expression vector pCAMBIA1300S. The CnFatB3 and CnLPAAT-B expression modules were recombined to form a combinatory dual-gene coexpression system (CnFatB3–LPAAT-B, designated FL). The CnFatB3 and CnKASI expression modules were constructed as a dual-gene coexpression system (CnFatB3–KASI, FK) as well. Similarly, the CnFatB3–LPAAT-B expression system and the CnKASI expression module were used to produce a triple gene coexpression system (CnFatB3–LPAAT-B–KASI, FLK) (Fig. 1 d–f).
Heterologous expression of multigene combinations in A. thaliana
Three multigene combinations, FL, FK and FLK, were transformed into Arabidopsis thaliana using the floral dip method. T1 generation transgenic plants were screened based on hygromycin resistance. PCR was employed to identify the genes in T2 generation transgenic Arabidopsis. As shown in Fig. 2a–c, the three multigene combinations were successfully integrated into the Arabidopsis genome. Hygromycin screening was then applied to further confirm homozygous T2 generation transgenic plants.
Identification of transgenic plants by PCR and qRT-PCR. a–c Agarose gel electrophoresis showing the PCR identification of T1 generation transgenic Arabidopsis. M: DNA marker. Lane 1: Wild type. Lane 2: FKL. Lanes 3 and 4: FKL. a Lane 5: FK. Lane 6: FL. b Lanes 5 and 6: FK. c Lanes 5 and 6: FL. d–f Relative expression levels of CnFatB3, CnLPAAT-B and CnKASI mRNA in the transgenic FL, FK and FLK plants, respectively
To reveal the expression levels of three genes in different transgenic plants, qRT-PCR was employed to detect the expression of CnFatB3, CnKASI and CnLPAAT-B in the three transgenic lines. The results indicated that CnFatB3, CnKASI and CnLPAAT-B, were significantly overexpressed in the three transgenic lines compared with their expression in wild-type Arabidopsis (Fig. 2d–f). Interestingly, CnLPAAT-B showed the highest expression level in the three transgenic lines. Finally, the three lines with the highest expression levels of each combination of genes were chosen for fatty acid profile analysis. These were lines 3, 5 and 10 from the FL plants; lines 9, 12 and 75 from the FK plants; and lines 3, 17, and 2 from the FLK plants.
Multigene combinations improve total fatty acids content
The percentage of the total fatty acid content in 50-mg seeds was calculated and regarded as the content relative to that of C17:0, which was used as an internal standard. As expected, the relative total fatty acid content was significantly increased when seeds from transgenic Arabidopsis plants were compared to those from wild-type, and the total fatty acid content was increased by at least 47%, 70% and 56% in the FL, FK and FLK seeds, respectively, compared to that in the wild-type seeds. Moreover, several lines exhibited an extremely significant increase in total fatty acid content. Line 12 from the FK plants had the highest fatty acids content among all of the measured lines and contained 9.687 ± 1.102 μg total fatty acid/50 mg seeds (while, wild-type content is 4.821 ± 0.241) (Fig. 3).
Analysis of the differences in total fatty acids content in different transgenic plants compared with that in wild-type. FKL: FatB3–KASI–LPAAT-B, FL: FatB3–LPAAT-B, FK: FatB3–KASI. All experiments were performed in triplicate. *Significant difference according to Student’s t test at p < 0.05. **Extremely significantly different, p < 0.01
Multigene combinations alter fatty acid composition
The total fatty acid composition of mature seeds from the T3 homozygous transgenic lines expressing three combinations of genes and wild-type Arabidopsis was analyzed by gas chromatography (GC). The results (Fig. 3) showed that the levels of lauric acid (C12:0) and myristic acid (C14:0) were significantly increased in seeds from all three transgenic lines compared to those in seeds from wild-type plants. The relative amount of lauric acid (C12:0) increased by at least 395%, 134%, and 124% in seeds from the FLK, FL and FK transgenic Arabidopsis lines, respectively, compared with that in seeds from the wild type. Similarly, the myristic acid (C14:0) levels increased by at least 383%, 106% and 102%, respectively, in the FL, FLK and FK transgenic lines (Fig. 4a–c). Notably, there was also a significant difference in the medium-chain fatty acid content between different transgenic lines expressing the same combination of genes. Different sites of integration during genetic transformation and the unequal expression levels of each gene could be the main causes for this phenomenon (Table 1).
Compositions analysis of individual fatty acid content in transgenic plants. FLK (a), FL (b) and FK (c) compared with that of wild-type plants. FKL: FatB3–KASI–LPAAT-B, FL: FatB3–LPAAT-B, FK: FatB3–KASI. All experiments were performed in triplicate. *Significant difference according to Student’s t test at p < 0.05. **Extremely significantly different, p < 0.01
In addition, the levels of other fatty acids in the transgenic lines expressing three gene combinations were also determined. Unlike the consistent increase in C12:0 and C14:0 across the transgenic lines, the content of the other fatty acids was different in the three transgenic lines. For example, in the FL lines, the very-long-chain fatty acid (VLFA) content, including C22:1, C24:0 and C24:1, was significantly increased compared to that in the wild type. In the FK transgenic line, the C18:1 and C18:2 fatty acid content was significantly increased in two transgenic lines compared to that in the third transgenic line and the wild type (Fig. 4a–c). However, in the FLK lines, the change of VLFA and UFA was not significant.
Combinatorial expression effect of CnFatB3, CnLPAAT-B and CnKASI
To further unravel the optimal combinatorial expression effect among CnFatB3, CnLPAAT-B and CnKASI, the transgenic lines expressing each combination of genes with the most pronounced effect were selected for further comparison. These were lines 17, 8, and 3; lines 3, 1, and 2 and lines 9, 2, and 3 from the FLK, FL and FK transgenic lines, respectively. Interestingly, the highest lauric acid (C12:0) content was detected in the FLK transgenic lines. The lauric acid content was 19% and 116% higher than that in the FL and FK transgenic lines, respectively. In contrast, the C14:0 content was highest in the FL transgenic line and at least 58% and 92% higher than that in the FLK and FK transgenic lines, respectively (Fig. 5). Above all, with regard to the biosynthesis of C12:0, the FLK transgenic line expressed the optimal combination of the genes, and to increase the C14:0 content, the combination of genes in the FL lines should be used. Additionally, the FLK lines showed increased C18:1 fatty acid levels, and the FL lines exhibited C16:0, C16:1, C24:0 and C24:1 accumulation.
Discussion
The endosperm of coconut (Cocos nucifera L.), an important oil-yielding tropical crop, is rich in MCFAs. This peculiar property makes the coconut endosperm an attractive model to unveil MCFA biosynthesis mechanisms. CnFatB3, CnLPAAT-B and CnKASI play vital roles in the metabolism of MCFAs (Yuan et al. 2015a, b, 2017); however, increasing the expression of a single gene in a complex metabolic pathway such as fatty acid synthesis is relatively ineffective at changing the final product unless concomitant changes are made in other enzymes (Dehesh et al. 2001). In this study, three genes, CnFatB3, CnLPAAT-B and CnKASI, were introduced to construct different Cre/LoxP multigene combination transgenic lines, designated CnFatB3–LPAAT–KASI (FLK), CnFatB3–LPAAT (FL), and CnFatB3–KASI (FK), to determine their combinatorial expression effects on MCFA and lipid biosynthesis. To our knowledge, this is the first time the harmony between MCFA-associated enzymes in coconut has been clarified.
The expression levels of CnFatB3, CnLPAAT-B and/or CnKASI were examined in FLK, FL and FK transgenic Arabidopsis. The gene expression levels were extensively fluctuant in the seeds of plants expressing different combinations of the genes. Different lines of expressing same combination of genes also showed different expression patterns. These results suggested that combinatorial vectors may be inserted into disparate genome sites, resulting in diverse gene expression profiles. The expression of CnLPAAT-B was the highest in all FLK and FL lines and the expression of CnFatB3 was similar in all FK lines. Moreover, the transcription levels of any particular gene were different with different combinations of genes as well. For instance, the CnFatB3 mRNA level was highest in FK seeds and, in contrast, relatively low in FL seeds. Differences in the transcription of a particular gene might be due to the interaction of multiple coexpressed genes in the same vector.
In addition, the correlation between gene expression level and the effects on fatty acid and lipid biosynthesis has been unraveled. In general, high expression levels were accompanied by a remarkable increase in MCFA yield. For example, line #17-8-3; line# 3-1-2; and line #9-2-3, which had the highest gene transcription levels in the FLK, FL and FK transgenic plants, respectively, expressed high levels of 12:0 and 14:0 fatty acids. Coincidently, the other two lines (line #3-4-5, line #10-6-4) with decreased mRNA levels exhibited relatively low 12:0 and 14:0 fatty acid content. Further analysis revealed that the transcription level and lipid content were also positively correlated. To some extent, when the transcription levels of a particular gene were positively correlated with the corresponding enzyme level, this influenced catalysis.
The use of a seed-specific promoter with high efficiency is a valid way to improve crop seed production. In this study, napin, Pro-at16460 and Pro-at5400 were employed to promote CnFatB3, CnKASI and CnLPAAT-B gene expression, respectively. High expression levels of CnFatB3, CnKASI and CnLPAAT-B suggested that these promoters were highly efficient and seed specific, which is concordant with the previous reports of expression in embryos and endosperms (Jeong et al. 2014). Specifically, the expression of CnLPAAT-B was still relatively high in FLK and FL, two different transgenic Arabidopsis plants expressing different combinations of genes. This implied that Pro-at5400 was a more powerful promoter than the napin and Pro-at16460 promoters.
To verify the combinatorial expression effects of CnFatB3, CnLPAAT-B and CnKASI on fatty acid biosynthesis, 15 kinds of fatty acids species were examined in all FLK, FL and FK transgenic plants. Instructively, the levels of 12:0 and 14:0 fatty acids, two important MCFAs, were significantly increased in transgenic lines expressing all three combinations of genes compared to their levels in wild-type plants. Although the overexpression of the single CnFatB3, CnLPAAT-B and CnKASI genes improves the 12:0 and 14:0 fatty acid content, e.g., the amount of 12:0 fatty acid increased by at least 104.1%, 133% and 30% and that of 14:0 fatty acid increased by 29.6%, 13% and 80% in the CnLPAAT-B, CnKASI and CnFatB3 transgenic plants, respectively, when compared to their expression in wild-type plants, their increased levels in plants overexpressing single genes were lower than those in the FLK, FL and FK combination plants (Yuan et al. 2015a, b, 2017). These results suggest the combinatorial expression effect of the CnLPAAT-B, CnKASI and CnFatB3 enzymes on MCFA biosynthesis. Among the FLK, FL and FK transgenic plants, the FLK plants, which synchronously expressed the CnFatB3, CnLPAAT-B and CnKASI genes, exhibited the most effective increase in 12:0 fatty acid levels. In addition, the FL plants, which expressed the CnFatB3 and CnLPAAT-B genes, exhibited the most vigorous enhancement of the 14:0 fatty acid yields. These results suggest that the CnFatB3, CnLPAAT-B and CnKASI enzymes have a strong combinatorial expression effect on 12:0 fatty acid biosynthesis and that the functions of CnFatB3 and CnLPAAT-B are concordant with the increase in the 14:0 fatty acid content. Based on previous reports, the accumulation of C12:0 and C14:0 in FKL of this study should be due to the combination of these three determinants, which play crucial roles on MCFAs biosynthesis.
Except for the C12:0 and C14:0 fatty acids, the levels of other fatty acids, such as C20:2, C22:1, C24:0 and 24:1 fatty acids, were significantly different in the FLK, FL and FK plants compared with the wild-type plants. These findings are consistent with previous reports. For instance, CnFatB3 exhibited a preference for C16:0 and C18:0 fatty acids (Yuan et al. 2017). The expression of transgenic CnKASI and CnLPAAT-B increased the levels of C6:0, C8:0 and C16:0 fatty acids (Yuan et al. 2015a, b). Unexpectedly, there is a decrease in C18:2 with an increase in C18:3 appearing in several transgenic lines, since unsaturated fatty acids are more concentrated on sn-2 position in typical vegetable oil, the exact reason need to be clarified further. Moreover, total fatty acid content in the FLK, FL and FK plants were significantly increased compared with those in the wild-type plants. When analyzing the harmony of CnFatB3, CnLPAAT-B and CnKASI in lipid assembly, the lipid contents of the FLK, FL and FK plants were compared, and the highest lipid content was found in the FL transgenic plants. Therefore, CnFatB3 and CnKASI are indispensable genes that drive lipid assembly. Hence, CnFatB3, CnLPAAT-B and CnKASI are crucial enzymes involved for not only MCFA biosynthesis but also lipid assembly.
In conclusion, in this work, the combinatorial expression effects of gene expression in plants stably transformed with CnFatB3, CnLPAAT-B and CnKASI in a constitutive expression system carrying tissue-specific and inducible promoters were determined. A significant combinatorial expression effect on MCFA levels, especially 12:0 and 14:0 fatty acid biosynthesis, and TAG accumulation were shown when CnFatB3, CnLPAAT-B and/or CnKASI were coexpressed in Arabidopsis. This work revealed that the incorporated CnFatB3, CnLPAAT-B and CnKASI genes worked in harmony in the 12:0 fatty acid biosynthesis pathways; while, the combination of CnFatB3 and CnLPAAT-B expression was optimal for 14:0 fatty acid biosynthesis. Thus, CnFatB3, CnLPAAT-B and CnKASI could be underlying powerful candidate genes used in the MCFA production industry. When multiple genes were stably coexpressed, the accumulated MCFA levels significantly exceed their currently reported levels, and these results could provide new insight into genes with distinct roles in fatty acid and TAG metabolism and assist in technology to increase in the levels of particular fatty acids by the addition or knockdown of other genes.
References
Bent AF, Clough SJ (1998) Agrobacterium germ-line transformation: transformation of arabidopsis without tissue culture. In: Gelvin SB, Schilperoort RA (eds) Plant molecular biology manual. Springer Netherlands, Dordrecht, pp 17–30. https://doi.org/10.1007/978-94-011-5242-6_2
Brough CL, Coventry J, Christie W, Kroon J, Barsby T, Slabas A (1997) Engineering trierucin into oilseed rape by the introduction of a 1-Acyl-sn-Glycerol-3-Phosphate Acyltransferase from Limnanthes Douglasii. In: Williams JP, Khan MU, Lem NW (eds) Physiology biochemistry and molecular biology of plant lipids. Springer Netherlands, Dordrecht, pp 392–394. https://doi.org/10.1007/978-94-017-2662-7_124
Chapman KD, Dyer JM, Mullen RT (2013) Commentary: why don’t plant leaves get fat? Plant Sci 207:128–134. https://doi.org/10.1016/j.plantsci.2013.03.003
Dehesh K, Tai H, Edwards P, Byrne J, Jaworski JG (2001) Overexpression of 3-Ketoacyl-Acyl-Carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol 125(2):1103–1114. https://doi.org/10.1104/pp.125.2.1103
Dohlemann J, Brennecke M, Becker A (2016) Cloning-free genome engineering in Sinorhizobium meliloti advances applications of Cre/loxP site-specific recombination. J Biotechnol 233:160–170. https://doi.org/10.1016/j.jbiotec.2016.06.033
Durrett TP, Benning C, Ohlrogge J (2008) Plant triacylglycerols as feedstocks for the production of biofuels. PlJ 54(4):593–607. https://doi.org/10.1111/j.1365-313X.2008.03442.x
Dussert S, Guerin C, Andersson M, Joet T, Tranbarger TJ, Pizot M, Sarah G, Omore A, Durand-Gasselin T, Morcillo F (2013a) Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. Plant Physiol 162(3):1337–1358. https://doi.org/10.1104/pp.113.220525
Dussert S, Guerin C, Andersson M, Joët T, Tranbarger TJ, Pizot M, Sarah G, Omore A, Durand-Gasselin T, Morcillo F (2013b) Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. Plant Physiol 162(3):1337–1358. https://doi.org/10.1104/pp.113.220525
Gornicki P, Haselkorn R (1993) Wheat acetyl-CoA carboxylase. Plant Mol Biol 22(3):547–552. https://doi.org/10.1007/bf00015984
Hoess RH, Abremski K (1985) Mechanism of strand cleavage and exchange in the Cre-lox site-specific recombination system. J Mol Biol 181(3):351–362. https://doi.org/10.1016/0022-2836(85)90224-4
Jeong H-J, Choi JY, Shin HY, Bae J-M, Shin JS (2014) Seed-specific expression of seven Arabidopsis promoters. Gene 553(1):17–23. https://doi.org/10.1016/j.gene.2014.09.051
Jing F, Cantu DC, Tvaruzkova J, Chipman JP, Nikolau BJ, Yandeau-Nelson MD, Reilly PJ (2011) Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem. https://doi.org/10.1186/1471-2091-12-44
Joët T, Laffargue A, Salmona J, Doulbeau S, Descroix F, Bertrand B, De Kochko A, Dussert S (2009) Metabolic pathways in tropical dicotyledonous albuminous seeds: Coffea arabica as a case study. New Phytol. https://doi.org/10.1111/j.1469-8137.2008.02742.x
Jovancevic N, Dendorfer A, Matzkies M, Kovarova M, Heckmann JC, Osterloh M, Boehm M, Weber L, Nguemo F, Semmler J, Hescheler J, Milting H, Schleicher E, Gelis L, Hatt H (2017) Medium-chain fatty acids modulate myocardial function via a cardiac odorant receptor. Basic Res Cardiol 112(2):13–13. https://doi.org/10.1007/s00395-017-0600-y
Kasai Y, Harayama S (2016) Construction of marker-free transgenic strains of chlamydomonas reinhardtii using a cre/loxp-mediated recombinase system. PLoS ONE 11(8):e0161733. https://doi.org/10.1371/journal.pone.0161733
Lin L, Liu Y-G, Xu X, Li B (2003) Efficient linking and transfer of multiple genes by a multigene assembly and transformation vector system. Proc Natl Acad Sci 100(10):5962–5967. https://doi.org/10.1073/pnas.0931425100
Martínez-Vallespín B, Vahjen W, Zentek J (2016) Effects of medium-chain fatty acids on the structure and immune response of IPEC-J2 cells. Cytotechnology 68(5):1925–1936. https://doi.org/10.1007/s10616-016-0003-1
Mizutani O, Masaki K, Gomi K, Iefuji H (2012) Modified Cre-loxP recombination in Aspergillus oryzae by direct introduction of Cre recombinase for marker gene rescue. Appl Environ Microbiol 78(12):4126–4133. https://doi.org/10.1128/AEM.00080-12
Patil U, Benjakul S (2018) Coconut milk and coconut oil: their manufacture associated with protein functionality. J Food Sci 83(8):2019–2027. https://doi.org/10.1111/1750-3841.14223
Reynolds KB, Taylor MC, Cullerne DP, Blanchard CL, Wood CC, Singh SP, Petrie JR (2017) A reconfigured Kennedy pathway which promotes efficient accumulation of medium-chain fatty acids in leaf oils. Plant Biotechnol J 15(11):1397–1408. https://doi.org/10.1111/pbi.12724
Schönfeld P, Wojtczak L (2016) Short- and medium-chain fatty acids in energy metabolism: the cellular perspective. J Lipid Res 57(6):943–954. https://doi.org/10.1194/jlr.R067629
Taylor DC, Francis T, Lozinsky S, Hoffman T, Giblin M, Marillia E-F (2010) Cloning and characterization of a constitutive lysophosphatidic acid acyltransferase 2 (LPAT2) gene from tropaeolum majus L. Open Plant Sci J. https://doi.org/10.2174/1874294701004010007
Toni V, Anthony JK (2001) Variations in the biosynthesis of seed-storage lipids. Annu Rev Plant Physiol Plant Mol Biol. https://doi.org/10.1146/annurev.arplant.52.1.335\r52/1/335[pii]
Troncoso-Ponce MA, Cao X, Yang Z, Ohlrogge JB (2013) Lipid turnover during senescence. Plant Sci 205–206:13–19. https://doi.org/10.1016/j.plantsci.2013.01.004
Yang T, Xu R, Chen J, Liu A (2016) Beta -Ketoacyl-acyl carrier protein synthase I (KASI) plays crucial roles in the plant growth and fatty acids synthesis in tobacco. Int J Mol Sci 17(8):1287. https://doi.org/10.3390/ijms17081287
Yuan Y, Chen Y, Yan S, Liang Y, Zheng Y, Dongdong L (2014) Molecular cloning and characterisation of an acyl carrier protein thioesterase gene (CocoFatB1) expressed in the endosperm of coconut (Cocos nucifera) and its heterologous expression in Nicotiana tabacum to engineer the accumulation of different fatty acids. Funct Plant Biol 41(1):80. https://doi.org/10.1071/fp13050
Yuan Y, Gao L, Sun R, Yu T, Liang Y, Li D, Zheng Y (2017) Seed-specific expression of an acyl-acyl carrier protein thioesterase CnFatB3 from coconut (Cocos nucifera L.) increases the accumulation of medium-chain fatty acids in transgenic Arabidopsis seeds. Sci Hortic 223:5–9. https://doi.org/10.1016/j.scienta.2017.05.029
Yuan Y, Liang Y, Gao L, Sun R, Zheng Y, Li D (2015a) Functional heterologous expression of a lysophosphatidic acid acyltransferase from coconut (Cocos nucifera L.) endosperm in Saccharomyces cerevisiae and Nicotiana tabacum. Sci Hortic. https://doi.org/10.1016/j.scienta.2015.06.003
Yuan Y, Liang Y, Li B, Zheng Y, Luo X, Dongdong L (2015b) Cloning and function characterization of a β-Ketoacyl-acyl-ACP synthase I from coconut (Cocos nucifera L.) endosperm. Plant Mol Biol Rep. https://doi.org/10.1007/s11105-014-0816-z
Zhang X, Henriques R, Lin S-S, Niu Q-W, Chua N-H (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1:641. https://doi.org/10.1038/nprot.2006.97
Acknowledgements
This research was supported by the Hainan Provincial Natural Science Foundation of China (No. 2019CXTD397), the National Natural Science Foundation of China (NSFC) (No. 31460213 and 31660222) and the Fundamental Research Funds for Chinese Academy of Tropical Agricultural Sciences (No.1630052019001)
Author information
Authors and Affiliations
Contributions
DL and PZ designed the research, YZ, LC and ZZ performed the research, and YZ and LZ wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zheng, Y., Chen, L., Zhu, Z. et al. Multigene engineering of medium-chain fatty acid biosynthesis in transgenic Arabidopsis thaliana by a Cre/LoxP multigene expression system. 3 Biotech 10, 340 (2020). https://doi.org/10.1007/s13205-020-02340-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02340-z