Abstract
Somatic embryogenesis (SE) is the most suitable biotechnological tool for the rapid clonal propagation of endangered woody plants, but many bottlenecks limit understanding its molecular and physiological processes in Nothapodytes nimmoniana. Combinations of two-dimensional electrophoresis (2-DE) and mass spectrometry (MaSp) were used to study proteomic expression changes during SE of the forest tree. Callus was induced from mature seed embryos, and embryogenic callus (EC) obtained at very low frequency after about 6 month culture. Globular embryos were induced from the seed embryo-derived EC and the subsequent stages of the SE. Analysis of the extracted proteins from globular, heart/torpedo-shaped, and maturing embryo stages resolved in the 2-DE gels showed increased protein expression across developmental stages of the somatic embryos. The mass spectrometric analysis with database search aided identification of 55 out of 100 and 54 selected protein spots. Identified proteins classified by the cellular role which they perform are involved in aspects of stress responses, energy metabolism, carbon fixation, secondary metabolism, and other metabolic functions, while three proteins are of unknown cellular role. The putative role of the expressed proteins during SE provided insight into the physiology of somatic embryo development in N. nimmoniana.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Somatic embryogenesis (SE) is an in vitro developmental program through which somatic cells generate embryogenic cells that can undergo morphological and biochemical changes to produce somatic embryos capable of regenerating plant (Zimmerman 1993; Quiroz-Figueroa et al. 2006). In dicotyledonary plants, the program involves globular, heart, torpedo-shaped, and cotyledonary stages (Zimmerman 1993). In the early years, studies on the SE of forest trees were on physiological aspects with view to optimize culture conditions for mass propagation and commercial applications. However, efforts have shifted towards understanding the molecular aspects during the last few decades (Isah 2016a). In the recent, emphasis is on describing the program at molecular levels through studying the biological processes that occur during its developmental stages by the use of molecular approaches (Abbasi et al. 2016; Isah 2016a). Proteomic expression changes are among the employed molecular approaches that have advanced the understandings of the structure, genetic organization, and evolution of plant genome (Manjasetty et al. 2012; Agrawal et al. 2013; Gantait et al. 2014). It provides insight into physiological and biochemical changes associated with somatic embryo development as a reflection of molecular changes involved, and have applications in optimizing culture conditions for mass production at commercial scale (Iraqi and Tremblay 2001a; Chen and Harmond 2006; Agrawal et al. 2013).
Nothapodytes nimmoniana is an endangered forest tree and most convenient source for the large-scale isolation of camptothecin (CPT) due to the higher yield over the other plant sources of the alkaloid. In recent years, there has been an upsurge in demand of the chemical by the pharmaceutical industry. This led to an indiscriminate exploitation of N. nimmoniana forest for isolation of the anticancer alkaloid to meet demand of the chemical in production of its analog drugs (Padmanabha et al. 2006; Suhas et al. 2007; Ramesha et al. 2008). Considering the state of the exploitation for isolation of CPT and the need for 7–8 years for its seedlings to reach reproductive stage, biotechnological approaches offer alternative strategy for mass production of the clones (Isah and Mujib 2015a, b; Ali et al. 2016; Prakash et al. 2016; Isah 2015a, 2017; Isah and Umar 2018; Isah et al. 2018). The approach could be employed in the scale-up production of CPT, so as to mitigate indiscriminate harvest of endangered natural population for isolation of the alkaloid (Isah and Mujib 2015a, b). SE is the most suitable biotechnological tool having application in the large-scale production to meet demand of the chemical by the pharmaceutical industry (Lutz et al. 1985; Gupta et al. 1991; Mujib et al. 2014, 2016). However, response to the production of the embryos in the in vitro cultures of N. nimmoniana is poor. Earlier studies have employed the molecular approach of proteomic expression changes in studying physiological metabolic changes associated with the development of in vitro embryos through the various stages in woody plants, with applications in optimizing culture conditions for mass production at commercial scale (Lippert et al. 2005; Pan et al. 2009; Noah et al. 2013). However, the reported studies on the SE in N. nimmoniana are few (Fulzele and Satdive 2003; Khadke and Kuvalekar 2013) despite potential application which it offers in the scale-up production of CPT. In this study, to gain physiological molecular insight about the somatic embryo development in N. nimmoniana, two-dimensional electrophoresis (2-DE) and mass spectrometry (MaSp) were employed to study proteomic expression changes during the SE. It is the first molecular and proteomic study of SE in the species. Among bottlenecks in understanding the molecular, physiological, developmental processes in N. nimmoniana and related species having poorly known genomic data in the bioinformatics databases are the deficiency in genomic information, as encountered in the present study. The results obtained have provided insight into metabolic changes involved in the in vitro embryogenesis in N. nimmoniana across the various developmental stages.
Materials and methods
Cultures establishment and conditions
Authenticated seed samples were obtained from Sanjivini herbals Tamil Nadu India, mature seed embryos excised from the seeds using the strategy reported earlier (Isah and Mujib 2015c). However, only seed embryo-derived callus cultures from single seed source-derived tissue were used for the proteomic expression study. Callus was induced from the isolated seed embryo explants on solid Murashige and Skoog (1962) medium (MS) amended with 2,4-dichlorophenoxy acetic acid (2,4-D) + benzylaminopurine (BAP) (13.56 + 2.22 µM). The induced callus cultures were maintained on medium amended with 2,4-D + BAP (9.04 + 4.44 µM) through regular subculture after every 3–4 weeks for up to 6 months before embryogenic tissue was produced. Globular embryos were induced from the EC when transferred to MS medium amended with BAP + 2,4-D (4.44, 6.66 or 8.88 + 2.26 µM), or plant growth regulators (PGRs)-free. The induced embryos progressed through heart-torpedo-shaped stages when transferred to medium supplemented with BAP (2.22 µM) followed by the early stage maturation on medium amended with gibberellic acid (GA3) (5.20 µM).
In the present study, the culture medium contained 3% sucrose and media pH adjusted to 5.6–5.8 using 1N NaOH or HCl. The media were solidified with 8% Agar (Agar Agar Microbiology Mumbai, India) before autoclaved at 121 °C for 21 min. All of the cultures were maintained under the culture room conditions of 25 ± 2 °C temperatures, 50–70% relative humidity, and 16 h photoperiod provided by cool fluorescent tubes (Phillip India) having illuminations of 40 W, 50 µmol m−2 s−1 PFD. For protein extraction, somatic embryos at the heart/torpedo-shaped stages were taken as single embryo mass sample as the stages proved difficult to distinguish while separating samples for the embryos at globular and maturation stages.
Alkaloid extraction and high-performance thin-layer chromatographic analysis
CPT extraction was carried out by the use of microwave extraction method reported by Fulzele and Satdive (2005), while high-performance thin-layer chromatographic (HPTLC) analysis by the modified method of Kulkarni et al. (2010). Briefly, 20 µl of the dried embryo tissue samples extracts (globular, heart/torpedo-shaped, and maturing embryos) were loaded on an HPTLC plate (Merck silica gels) using an automated sample loading system, and then allowed to dry. The loaded samples were developed in a glass chamber saturated with ethyl acetate: toluene (7:3) mobile phase solvent system. The obtained chromatographs were scanned using Camag 3 densitometric scanner and the peaks, peak area, and Rf of the spots recorded. A pure reference sample (CPT standard) was procured from Sigma-Aldrich (CAS No. 7689-03-4) and standard solution prepared by dissolving CPT powder in chloroform:methanol (4:1) solvent mixture. Fluorescence was recorded at 366 nm and Rf of the CPT standard evaluated. CPT yield of the embryos at the various stages of development was evaluated from a standard curve of the CPT reference standard drawn from five dilutions of the prepared standard stock solution after developed in a chromatographic chamber. The alkaloid was identified by the Rf values of 8.1–82, absorbance in the UV region of 366 nm, and in comparison with the standard curve having linear regression line equation Y = 0.0018x + 7.2184.
Protein extraction
Extraction of proteins from the globular, heart/torpedo-shaped, and maturing embryo tissue samples was carried out in triplicate sets through the use of phenol method reported by Isaacson et al. (2006). Briefly, 2 g of each of the samples were ground into fine powder in liquid nitrogen and the resultant powder suspended in 10 ml extraction buffer (pH 7.5) that contained 50 mM HEPES, 2% β-mercaptoethanol, 700 mM sucrose, 1 mM PMSF, 50 mM ethylenediaminetetraacetic acid (EDTA) and 100 mM KCl, and 15 ml phenol was added. The resultant solution was mixed in cold room rocker for 30 min at 4 °C and then centrifuged at 3000×g for 10 min at 4 °C. Upper phenolic phase was recovered in separate tube and 15 ml ice-cold 0.1 M ammonium acetate added to the contents before incubated at − 20 °C overnight for precipitation. The obtained proteins were pelleted by centrifugation at 6000×g for 15 min at 4 °C and the resultant pellet washed with methanol, followed by two rounds with acetone. The pellet was dissolved in methanol and then centrifuged at 3000×g and − 20 °C for 10 min after each of the wash steps. This was followed by drying in solubilization buffer that contained 2 M thiourea, 7 M urea, 4% CHAPS and 50 mM dithiothreitol (DTT). Resultant proteins in the samples were quantified using bovine serum albumin as standard (Bradford 1976).
Two-dimensional gel electrophoresis
The 2-DE was carried out by the method of O’Farrell (1975). Briefly, immobiline dry strip gels (17 cm, pH 3–10; Bio-rad) were rehydrated at 20 °C for 14 h in 200 µl of sample that contained 250 µg proteins. Isoelectric focusing was carried out in PROTEAN IEF apparatus (Bio-Rad). The voltages applied were 250 V for 1 h, 500 V for 1 h, 1000 V for 2 h, 2000 V for 2 h followed by a linear increase of 8000 V for 18 h and 500 V for 1 h. The strips were then subjected to reduction in buffer that contained 50 mM Tris (pH 8.8), 8 M urea, 20% glycerol, 2% SDS, and 130 mM DTT. This was followed by alkylation for 15 min in buffer that contained Tris (pH 8.8), 8 M urea, 20% glycerol, 2% SDS, and 135 mM iodoacetamide. The 2-DE was carried out in a Dodeca cell (Bio-rad PROTEAN Plus) to separate the focused proteins using 12% SDS at constant voltage of 200 V. Obtained 2-DE gels were stained with colloidal Coomassie brilliant blue dye followed by destain with milli-Q water. The experiments were performed in triplicate sets using the extracted protein samples. Resultant gels were scanned with Epson Scanner (GE Healthcare Bioscience Pvt Ltd., New Delhi, India) before stored in 10% acetic acid until further analysis.
Gel image analysis and peptide extraction
The 2-DE gel images were analyzed with the image processing software (2D Platinum). Pattern of the spot distribution within and between sample groups was studied by visualizing the 3D structure of each of the spots to identify proteins and non-proteins spots; non-protein spots were eliminated from the gels, while protein spots retained. To figure the significant differences between the sampled groups, spot intensity of the gel images bearing the protein spots was subjected to the one-way analysis of variance and abundance estimated from volume percentage. The spots were normalized within replicate sample groups, and gels matched through the match set. Only spots that showed significant and reproducible biological changes between replicate gels of the various embryonic stages with a fold change ≥ 1.5 and p value of 0.05 or less were considered differentially expressed and selected for the MaSp analysis. Selected spots were excised from the gels manually using a sharp blade and transferred into centrifuge tube individually. Excised gels were then rinsed with Milli-Q water three times and chopped into smaller pieces within the centrifuge tube. The gel pieces were destained with 40 mM ammonium bicarbonate followed by 40 and 100% acetonitrile. This was followed by rehydration with 5 mM DTT in ammonium bicarbonate and alkylation with 20 mM iodoacetamide in 40 mM ammonium bicarbonate. In-gel digestion of the reduced spots was carried out using 12.5 ng/ml trypsin (modified sequencing grade trypsin–Promega, USA) in 50 mM ammonium bicarbonate by incubating the digested gels in an ice bath for 45 min. A volume of ammonium bicarbonate was added to the pieces and incubated overnight at 37 °C. The obtained peptides were extracted from gel pieces by incubation with formic acid (5%) for 10 min at 37 °C followed by the use of extraction buffer before the final extraction in 100% acetonitrile. The supernatant obtained (from the three steps) was pooled together and dried down by spinning the tubes in speed vac and the resultant residue stored at − 20 °C until MaSp analysis.
Mass spectrometry, database search, and functional categorization of the identified proteins
Peptides extracted from each of the spots were individually mixed with volume of saturated matrix solution, i.e 10 mg/ml α-cyano-4-hydroxycinnamic acid (Sigma-Aldrich), in 50% acetonitrile (Merck)/0.1% trifluoroacetic acid (Sigma-Aldrich). It was then spotted on silver MALDI target plate and dried at room temperature. Mass spectral data of the sampled spots were obtained with MALDI MS-ABI Sciex 7000 TOF/TOF system (Framingham Massachusetts, USA) calibrated in positive reflective mode with an accelerating voltage of about 20 kv for MaSp mode. Molecular ions that displayed common signal were submitted to the MaSp/MaSp analysis using specific mode precursor ion fragment at laser frequency of 50 Hz. Resultant MaSp/MaSp spectra were acquired in positive reflective mode following collision-induced dissociation and the data analyzed with Flex Analysis 2.4 data explorer 4.5. Peptides MaSp/MaSp were analyzed using Mascot (Matrix Science, London, UK) with the parameters: product ion mass tolerance of 150 ppm, monoisotopic mass value, one mixed cleavage, and fragment mass tolerance of ± 0.3 Da with methionine oxidation and an iodoacetamide derivative of cysteine specified as variable and fixed modifications. The Mascot was set up to search the national center for biotechnology (NCBInr) database (http://www.ncbi.nlm.nih.gov/protein) with the parameters: taxonomy; Viridiplantae, assuming the use of digestion enzyme trypsin and variable modifications with up to two missed cleavages. Only significant hits defined through the MASCOT probability analysis (p < 0.05) and peptides identified with individual ion scores greater than 40 were considered as true identity. Putative cellular functional categorization of the identified protein spots was performed by the use of Kyoto encyclopedia of genes and genomes (KEGG) which is available (http://www.genome.jp/kegg/kegg2.html) combined with the Uniprot database (http://www.uniprot.org/proteomes/).
Results and discussion
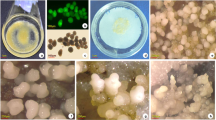
SE offers alternative biotechnological strategy for the rapid clonal production of endangered forest trees that produce anticancer molecule CPT. However, the program occurs at low frequency in the in vitro cultures of woody plants, and limited information is known about the prevailing molecular and physiological processes (Guan et al. 2016). During its developmental stages, molecular changes are involved in the production and proliferation of embryo mass and understanding them is key to improvement of in vitro embryogenesis in N. nimmoniana. In the recent, there is increasing consideration towards improving culture conditions to optimize production of the embryos. In this study, to obtain somatic embryos, callus was induced from isolated mature seed embryos explant on solid MS medium amended with 2,4-D + BAP (12.06 + 4.44 µM) within 2–9 weeks culture (Fig. 1a–c). The regular subculture of the callus on the solid medium supplemented with 2,4-D + BAP (9.04 + 4.44 µM) after every 3–4 weeks for up to 6 months resulted in the formation of embryogenic tissue (Fig. 1c–e) at very low frequency. Initially, the callus cultures cultivated on the PGRs combinations became slightly friable and whitish, followed by compact and yellowish-brown appearance within the culture duration. From the EC, globular embryos were induced when the callus cultures were transferred to medium amended with BAP + 2,4-D (4.44, 6.66 or 8.88 + 2.26 µM), or without PGRs within 3–5 weeks culture (Fig. 1f, g). Nodular features were observed on the EC, which differentiated into globular embryos on medium amended with BAP (2.22 µM) at high frequency after 3 weeks culture over the other concentrations tested. The embryos formed in clusters of nodular aggregates at periphery of the EC showed slow development, appeared earlier on BAP + NAA (8.88 + 2.69 µM)-supplemented medium than the other PGRs tested. The induced globular embryos did not progress to the other stages when maintained on the induction medium, but, upon transferred to medium amended with BAP (2.22 µM), it passed through heart/torpedo-shaped stages within 4–7 weeks culture (Fig. 1h, i). This suggests that a reduction in BAP concentration is essential for the development of globular embryos to the other stages in N. nimmoniana. The produced embryos showed the early stage maturation on medium supplemented with GA3 (5.20 µM) within 3–5 weeks culture (Fig. 1j), but the majority did not show adequate maturation for conversion into plantlets. In the present study, because the somatic embryos at the embryo maturation stage (cotyledonary stage) showed a lack of physiological maturity, they are referred to as “maturing embryos.”
a Isolated aseptic mature seed embryos explant, b cultured mature seed embryo explant on semi-solid MS medium added with 2,4-D + BAP (13.56 + 2.22 µM) showing callus induction, c proliferation of the callus induced from the seed embryos explant, d proliferated callus transition to the embryogenic callus (EC) after about 5–6 months culture, e obtained embryogenic tissue, f induction of globular embryos from the EC (arrowheads) on MS medium added with BAP (4.44 µM), g globular embryos induced on BAP (8.88 µM) amended medium, h, i heart/torpedo-shaped embryos obtained on medium added with BAP (2.22 µM), j maturing embryos on medium added with BAP (2.22 µM), and k maturing embryos showing the lack of physiological maturation on MS medium amended with GA3 (5.20 µM). Scale bar (a–d, f and k = 1.0 cm, e and g = 0.7 cm, h–j = 0.45 cm)
Proteomic expression study is among emerging biotechnological approaches having application in studies aimed at understanding physiological processes during in vitro embryogenesis in higher plants (Manjasetty et al. 2012). It could provide molecular insight into changes involved in the in vitro embryo development in endangered woody plants, with application in optimizing culture condition(s) for mass propagation and commercial application. To get some insight into the physiological changes involved in somatic embryo development in N. nimmoniana, proteins were extracted from the embryo tissue samples and 2-DE performed showed visible and reproducible various spots’ distributions across developmental stages of the somatic embryos (Fig. 2). Analysis of the obtained 2-DE gel images resulted in the selection of 154 of the resolved protein spots that showed over 1.5 fold change in expression levels during at least one of the embryogenesis stages (GE, HT, and ME), out of which MaSp/MaSp combined with database homology search aided identification of 55 (electronic supplementary data). Because N. nimmoniana genome is not sequenced and limited bioinformatics data are available about related species in the NCBInr database, the homology search performed resulted in low identification success rate. In this study, the poor identification success rate was attributed to the low mascot score obtained with many of the processed proteins spots, particularly with the analyzed peptides matches/mass spectra of weak intensity which, in the overall, imply uncertainty about identity of the spots (Table 1; Fig. 3). This is a common problem in the proteomic study of plants that do not (or its related species) have much of genomics information in the bioinformatics databases. The identified protein spots are indicated on the representative 2D map of Fig. 2 and Table 1 and electronic supplementary data file. In some of the encountered cases, more than one protein spots were found to correspond to the same enzyme, but different isomers as, for instance, in superoxide dismutase (SOD), glyceraldehyde-3-phosphate dehydrogenase, enolase, α-tubulin, ascorbate peroxidase, dehydrins, and late embryogenesis abundant (LEA) proteins. The identified proteins based on the cellular functional categorization are involved in the aspects of stress responses (35%), energy metabolism (25%), carbon fixation (9%), secondary metabolism (6%), and other cellular and metabolic processes (20%), while 5% of the proteins are of unclassified cellular role (Fig. 3). In this study, the largest group of the identified proteins are those involved in stress response, energy metabolism, and other cellular metabolic functions, while low number of proteins involved in photosynthesis and secondary metabolism was identified. This indicates the higher role of cellular metabolic processes during SE in N. nimmoniana. An interesting aspect of the results is the higher expression of proteins (shown by maximum numbers of proteins spots in the 2DE gels) during embryo maturation stage (Fig. 2) and upregulation of most of the identified protein spots during the stage that, in the overall, imply higher metabolic state of the embryos.
a Putative cellular functional categorization of the expressed proteins spots (identified by mass spectrometry) from the extracted protein samples of GE, TH, and ME stages as performed using Kyoto Encyclopedia of Genes and Genomes (KEGG) which is available at (http://www.genome.jp/kegg/kegg2.html) combined with the Uniprot database (http://www.uniprot.org/proteomes/), and b representative mass spectra obtained by MaSp/MaSp showing one among the many protein spot that had spectra of weak intensity and that turned out low mascot score which implied uncertainty about identity of many of the processed proteins spots
Role of stress
Oxidative stress involving oxidative burst produces reactive oxygen species (ROS) that modulates cell division and metabolism towards SE (Zavattieri et al. 2010). Production of the ROS and scavenging enzymes plays a part in the different stages, and change in the isoforms can be used as a marker (Kairong et al. 1999; Gupta and Datta 2003). In this study, three Cu–Zn SOD isoforms (spots 22A, 22B, and 22C) that confer protection to cells against ROS-induced oxidative stress showed the upregulated expression in the maturing embryos about globular and heart/torpedo-shaped stages. An MnSOD (spot 22D) showed upregulated expression during heart/torpedo-shaped embryo stages relative to the globular, but became downregulated in the maturing embryos. The higher expression of Cu–Zn SOD isoforms during the embryo maturation implies a role in scavenging ROS generated by high metabolic state to tolerable cellular level needed for its development. In Lycium barbarum, activity of SODs showed the elevated levels during the early stages of the SE and was maximal at the time multicellular embryos were formed but, declined with the further development (Kairong et al. 1999). However, in Persian walnut, along with other antioxidant enzymes, it showed upregulated expression in torpedo-shaped embryos and then downregulated during the cotyledonary stage. Various cotyledonary embryos showed similar activity of the enzyme and, a Cu–Zn SOD isoform showed upregulated expression during embryo maturation (Jariteh et al. 2015). In this study, differential expression of SOD isoforms observed showed Cu–Zn as the most expressed in all the stages, while MnSOD conferred protection against ROS at higher levels during globular and heart/torpedo-shaped stage but less in the maturing embryos (spot 22D).
To promote the SE, activity of SODs generates an intracellular accumulation of H2O2 which could serve as ‘second cellular messenger’ that induce gene expression and synthesis of proteins promoting progression to the various stages. This may occur through regulating the transcriptional level of antioxidant enzymes and superoxides (Kairong et al. 1999; Apel and Hirt 2004; Isah and Mujib 2012). The higher intracellular accumulation of the H2O2 induces damages to membrane lipids and other macromolecules. Catalase (CAT), ascorbate peroxidase (APX), and glutathione-S-transferase (GST) participate in regulating levels of the H2O2 to tolerable cellular concentration needed for the embryogenesis. Two CAT isoforms (spot 27A and 27B) showed higher expression during globular-to-embryo maturation stage, a trend similar to that observed with the SODs. This suggests an efficient role in scavenging H2O2 generated by the activity of SOD isoforms to sufficient cellular levels needed for the development of the in vitro embryos. A similar expression pattern was shown by APX isoforms (spot 21A, 21B). In Persian walnut, differential expression of CAT isoforms was observed during the primary and secondary SE and increased in the early stages of embryo formation (Jariteh et al. 2013, 2015). However, in Quercus suber, upregulated expression of APX occurred across the stages with slight downregulation during intermediary stages relative to the globular embryos, and GST was proposed to play a role in ROS detoxification in the proliferating somatic embryos (Gomez-Garay et al. 2013). Glutathione-S-transferase (spot 23A) showed higher expression levels during globular and heart/torpedo-shaped embryo stages, but became remarkably downregulated in the maturing embryos. Glutathione-S-transferase was among the expressed proteins during SE in Vitis vinifera (Marsoni et al. 2008) and showed pronounced upregulated expression in the embryogenesis stages in Eleutherococcus senticosus, suggesting an antioxidant role (Shohael et al. 2007). However, downregulated expression occurred in the globular and cotyledonary embryos of Gossypium hirsutum (Ge et al. 2014).
The stress response involved in the development of somatic embryos need a heat-shock system to play a part during the developmental reprogramming, with common elements of auxin playing a role (Zavattieri et al. 2010). Heat-shock proteins that belongs to Hsp 70 (spot 25A, B, and C) and Hsp 23 (spot 25D) categories showed higher expression in the globular and heart/torpedo-shaped stages, except spot 25A that became downregulated. This was followed by downregulation of Hsp 23 in the maturing embryos, suggesting differential expression of HSPs during somatic embryo development in N. nimmoniana. The higher and differential expression of the Hsps could be due to the increased and variable stress levels associated with somatic embryo development or for a role in conferring stability to other proteins during developmental stages as increased protein synthesis was observed across the stages, reflected by the number of protein spots detected in the 2-DE map in Fig. 2. This is in conformity with the role of Hsp as chaperones essential for cell recovery and function whose expression is higher during stressful condition to prevent protein denaturation (Zhu et al. 1993); Hsps were ascribed a role during SE in Vitis vinifera and Picea abies (Businge et al. 2013). The expression of chaperones and proteins involved in the somatic embryo development that includes LEA proteins (spot 31A, 31B, and 31C) having cellular roles as DNA-binding and repair, chaperone function, Ca2+ binding, as structural components of cytoskeleton and abiotic stress tolerance, may show changes at the developmental stages. In this study, the LEA proteins (spot 31A and 31B) were expressed in all the stages with the highest in the embryo maturation stage, while another LEA protein (spot 31C) was not expressed in the globular embryos, but, in the heart/torpedo-shaped and maturing embryos with upregulation in the later stage. The upregulated expression of the proteins could be for a role in facilitating embryo growth, maturation, and stress defense, particularly during the maturation stage. The abiotic stress of in vitro culture can induce the expression of different categories of LEA proteins, including dehydrins (spots 36A and 36B) that play a role in abiotic stress response. Many stress factors can induce their synthesis in somatic tissues, and the pattern of accumulation is under developmental control (Bomal et al. 2002; Burrieza et al. 2012). In this study, higher expression of dehydrins (spots 36A and 36B) was observed across the stages for a possible role in abiotic stress tolerance needed for the embryogenesis. This is also in conformity with the essential role of stress in embryo development through the various stages in N. nimmoniana. Many dehydrins were identified in the embryogenic cultures of sugarcane (Burrieza et al. 2012). A stress-inducible protein (spot 39C) showed higher expression across the stages for a possible role in reorganizing physiological and metabolic processes involved in the embryos development, and higher expression shown by enzymes involved in antioxidant defense systems.
Energy metabolism
Many genes that are expressed during somatic embryo development are involved in regulating developmental processes, and involve the biosynthesis of compounds, cell division, elongation, and differentiation. The most pressing need of cells during the processes and proper function is energy and metabolic intermediates that are provided by glycolysis and tricarboxylic acid cycle (TCA). In this study, enzymes involved in glycolysis and TCA cycle were the second most expressed for their possible role in providing cellular energy needed for metabolic processes associated with somatic embryo development in N. nimmoniana. The most expressed glycolytic enzyme is enolase with three identified proteins spots (4A1, 4A2, and 4A3). Enolase converts 2-phosphoglycerate to phosphoenolpyruvate in the glycolytic pathway, while, during gluconeogenesis, it is important in starch accumulating seeds. Accumulation of its transcripts was found maximal in the torpedo stage embryos of Arabidopsis (Andriotis et al. 2010) and was regarded mature embryo marker in Picea glauca (Lippert et al. 2005). Glyceraldehyde 3-phosphate dehydrogenase (spot 30A, 7B) that converts glyceraldehyde 3-phosphate into glycerate 1, 3-bisphosphate plays a role in ROS signaling in plants, while phosphoglycerate kinase (spot 28A) catalyzes reversible transfer of phosphate group from 1,3-bisphosphoglycerate to adenosine diphosphate to produce 3-phosphoglycerate and adenosine triphosphate, besides an ascribed role as an α-cofactor. In this study, the two enzymes showed upregulated expression in the maturing embryos about the other stages. Phosphoglycerate kinase was among the expressed proteins during secondary SE in cassava (Baba et al. 2008). The 3-phosphoglycerate generated by the activity of phosphoglycerate kinase is transformed to 2-phosphoglycerate through 2,3-bisphosphoglycerate intermediates involving internal transfer of phosphate group in reaction catalyzed by phosphoglycerate mutase (spot 9A). Although the cellular role of phosphoglycerate mutase in plants is unelucidated, it was proposed to be involved in defense response (Shin et al. 2009). Phosphoglycerate mutase was among embryo-specific proteins expressed during SE in Cyclamen persicum (Lyngved et al. 2008), suggesting a possible role in embryo development in N. nimmoniana. Pyruvate decarboxylase involved in the decarboxylation of pyruvate generated by glycolytic pathway into acetaldehyde and CO2 in cytoplasm and mitochondria of plant cells (spot 19A), which showed higher expression across embryogenesis stages with the most in the heart/torpedo-shaped to maturing embryo stages, suggesting more of role in the development of mature embryos than others. In the cotyledonary somatic embryos of cotton, its downregulation was involved in reduced respiration, and occurred due to the low energy production, and for nutrient storage needed for future germination (Ge et al. 2014). However, during SE of Quercus suber, it showed the upregulated expression between proliferating, cotyledonary to mature embryo stages (Gomez-Garay et al. 2013). Malate synthase (spot 15A) that catalyzes condensation and later hydrolysis of acetyl-Co enzyme A and glyoxylate to form malate and CoA showed a little change in globular and heart/torpedo-shaped stage, but upregulated in the maturing embryos. During the early stage of castor bean seed maturation, it was among the expressed at higher level, but downregulated in mature seeds, suggesting a role in embryo maturation (Gonzalez 1990). Its role in N. nimmoniana somatic embryo development is likely for facilitating embryo maturation as most of the embryonic tissues showed only early stage maturation. It could also be for a role in somatic embryo development through energy production as enzymes involved in energy production showed upregulated expression during embryo maturation. The NAD-dependent malate dehydrogenase (spot 29A) that catalyzes the oxidation of malate to oxaloacetate by reducing nicotinamide adenine dinucleotide (NAD+) to NADH in many metabolic pathways that includes TCA cycle and gluconeogenesis showed upregulated expression in maturing embryos about other SE stages in N.nimmoniana (spot 29A). It was ascribed a critical role in embryonic development and heterotrophic metabolism in Arabidopsis (Beeler et al. 2014), and was among energy production enzymes expressed at higher level during SE in date palm (Sghaier-Hammamia et al. 2009). The NADH–ubiquinone oxidoreductase (spot 16D1) that plays a role in oxidative phosphorylation which provides high energy needed for cellular processes showed upregulation in its expression during heart/torpedo-shaped embryo stages about the globular but, downregulated in maturing embryos. This suggests its higher role in embryogenesis during heart/torpedo-shaped stages, besides the primary role in energy production. It also suggests that oxidative phosphorylation provides less energy needed for metabolic activities involved in embryo maturation in N. nimmoniana, but higher in the heart/torpedo-shaped stages. Its expression was observed in the embryogenic cultures of Medicago truncatula (Imin et al. 2004) and during embryo maturation in Larch (Teyssier et al. 2014). Chloroplast ATP synthase 1 β subunit (spot 18A), another enzyme that plays a role in cell energy production through synthesis of ATP at the expense of electrochemical proton gradient formed by light-dependent electron flow in the mitochondrial thylakoid membrane, was among the higher expressed proteins in all the stages. This is in conformity with the increased cellular demand for energy needed for high metabolic processes during somatic embryo development and upregulated expression shown by enzymes involved in energy production. Its higher expression suggests high chloroplastic ATP generation needed for metabolic process- associated somatic embryo development in N. nimmoniana.
Cellular function of nucleotide sugar precursor synthesis needed for cell wall biogenesis is performed by UDP-glucose pyrophosphorylase (spot 24A), besides its role in plant growth and development through participation in carbohydrates biosynthesis regulation (Winter and Huber 2000). The proteins showed higher expression with the advance in embryogenesis stages, suggesting possible role in somatic embryo development through carbohydrate metabolism needed to build cell wall polymers essential for cell division. The high expression had been observed in the somatic embryos of C. persicum (Rode et al. 2012) and in polyethylene glycol-treated mature embryos of C. papaya (de Moura Vale et al. 2014). Phosphoenolpyruvate carboxykinase (spot 20A) showed higher expression across the embryogenesis stages for the possible embryo-specific role which it plays in carbohydrate metabolism. Its expression in the embryos also suggests a likely role in converting glycerol 3-phosphate product of photosynthesis to starch for storage in the chloroplast or glucose and sucrose as upregulated expression of photosynthetic enzymes and UDP-glucose pyrophosphorylase was observed. In soybean, during the induction of globular embryos, transcripts of a gene encoding phosphoenolpyruvate carboxykinase were among the expressed (Thibaud-Nissen et al. 2003).
During embryo maturation, metabolic activities including respiration are slowed to prepare embryos for quiescence, and lack of the maturation is a feature of recalcitrant seeds. The similarity is also found in somatic embryos, although, not at comparable physiological level to the in vivo embryo maturation (Iraqi and Tremblay 2001b). In this study, upregulated expression of energy metabolism and antioxidant enzymes were observed during later stages of somatic embryo development, indicating their possible higher metabolic state. In Araucaria augustifolia, Balbuena et al. (2009) attributed higher expression of enzymes involved in energy metabolism during late stages of zygotic embryogenesis to shift from stress-associated metabolism to make molecular oxygen available as the final hydrogen acceptor needed to increase the levels of respiration in mature seeds. However, during secondary SE of cassava, higher expression of enzymes involved in energy production was attributed to the higher metabolic state essential for maintaining intense cell division in the developing embryos (Baba et al. 2008). In this study, high expression of antioxidant and energy metabolism enzymes observed could be attributed to the higher metabolic state of the embryos which coupled with the ROS generation from metabolic processes needed for proper development possibly played a role in lack of physiological maturation and conversion of most maturing embryos to plantlets (Fig. 1k).
Other cellular and metabolic functions
The development of somatic embryos involves dedifferentiating somatic cells into embryogenic through the induction and expression stages involving molecular and physiological changes regulated by the expressed genes. The changes are based on the influence of expressed gene products that determine the growth and development of cultures, including polyamines which play a role in growth and development through their role in cell division and morphogenesis. Spermidine synthase (spot 6A), a key enzyme involved in the biosynthesis of polyamines whose activity is regulated at transcriptional and translational level and the activity correlates with the cellular levels of polyamines (Martin-Tanguy 2001), showed upregulated expression in the maturing embryos about the other stages, and was among the higher expressed proteins in the embryos. Spermidine was reported to play a significant role in promoting embryogenic differentiation in the in vitro cultures (Silveira et al. 2006) and correlated increase in the polyamine levels promoted somatic embryo development (Minocha et al. 2004). Their continual biosynthesis is an essential part of increased metabolic state during differentiation and development of somatic embryos. Annexins (spot 37A and 37B), which are multigene super-family of cytosolic calcium-dependent membrane binding proteins that play a role in cellular functions such as membrane cytoskeleton interaction, calcium channel activity and signal transduction, exocytosis, and endocytosis fundamental to cellular processes as secretion and repair, growth, and differentiation (Gerke and Moss 2002), showed upregulated expression in the embryo maturation stage with one of the spots (37B) expressed at higher levels over the other (37A), suggesting that their differential expression is involved in SE in N. nimmoniana. Alpha-tubulin (spot 5F1 and 5F2) that plays a role in cell division and elongation through organelles and daughter chromosome segregation during mitotic division essential for morphogenesis showed transient upregulated expression across embryogenesis stages for possible role in cell division and elongation. Alpha-tubulin was among the expressed proteins during SE in Picea glauca (Lippert et al. 2005) and secondary SE in Manihot esculentum (Baba et al. 2008). In addition, the cellular processes involved in somatic embryo development such as glycolysis and ROS production generate toxic electrophilic methylglyoxal whose reaction with the intracellular macromolecules that include protein and nucleic acid can lead to their inactivation. Lactoylglutathione lyase (spot 10A) involved in their detoxification showed higher expression across embryogenesis stages in similar way to enzymes involved in glycolytic pathway and antioxidant defense. Translation initiation factor (spot 12A) that binds to small ribosomal subunit during initiation of translation in protein synthesis by helping stabilizing the formation of functional ribosome around a start codon to give translation initiation a regulatory mechanism showed higher expression across embryogenesis stages, particularly during the embryo maturation. This could be due to the increased cell protein synthesis needed for embryo development and maturation that involves the covalent bonding of synthesized amino acids facilitated by peptidyl-prolyl isomerase proteins such as cyclophilins. Recent findings have implicated many cellular functions to the cyclophilins, including cell division, regulation of transcription, playing part in photosynthesis, as molecular chaperones, signaling molecules, and cellular stress tolerance (Kumari et al. 2013). In this study, cyclophilins showed upregulated expression (spot 2A) in the maturing embryos, suggesting their role in embryo development and stress tolerance needed for SE. Glutamine synthetase (spot 7A) that plays an essential role in nitrogen metabolism by catalyzing condensation of ammonia and glutamate to form glutamine showed downregulated expression between globular and maturing embryo stages, suggesting decreased nitrogen metabolism during embryogenesis in N. nimmoniana. In carrot, accumulation of its isoforms, transcripts, and activity was the highest during globular embryo stage, but declined over later stages of somatic and zygotic embryogenesis, suggesting similar nitrogen assimilation pattern in the embryos (Higashi et al. 1998). However, accumulated expression occurred during the early stages of cotyledonary embryo development in Brazilian pine (Balbuena et al. 2009), while an intense expression was observed in somatic than zygotic embryos of date palm (Sghaier-Hammamia et al. 2009).
The endogenous substances that regulate somatic embryo development include auxin whose concentration plays a role during embryogenic competence acquisition and expression (Jimenez 2001). Plants use strategies to regulate its levels by hydrolysis of amide-linked conjugates that act as storage or inactive forms of the hormone. Hydrolysis of the indole-3-acetic acid (IAA)—amide conjugates—results in free IAA in reaction mediated by auxin amidohydrolase (Woodward and Bartel 2005). An auxin hydrolase (spot 17A) showed higher expression during the stages and peaked during embryo maturation, an unusual feature of late developmental stages of embryogenesis. In Abies alba, embryo maturation was associated with an increase in the endogenous concentration of IAA, and suppression of the hormone biosynthesis inhibited somatic embryo development (Vondrakova et al. 2011). In this study, higher expression of enzymes involved in auxin regulation observed could be explained by the lack of physiological maturation shown by the embryos as auxin is needed for the proper development of somatic embryos. Physiological quality of the somatic embryos is determined by maturation, germination, and subsequent development into seedling which accumulation of storage proteins plays a role, and among them include 11S globulins precursor isoform 4 (spot 32B). This protein showed higher expression from globular to maturing embryo stages, suggesting role in mature embryo development. Storage proteins identified in the somatic embryos of C. persicum were proposed as markers of embryogenesis (Winkelmann et al. 2006).
In this study, a higher metabolism that was shown by the somatic embryos through upregulated expression of various proteins in the maturation stage further supported the results on antioxidant and energy metabolism that ʻpointedʼ to lack of physiological maturation. During zygotic embryogenesis in Araucaria augustifolia, a woody species that produces recalcitrant seeds (as in N. nimmoniana) which are shed with high moisture, and shows little desiccation tolerance, proteins involved in metabolic processes were of high abundance in the late stages of embryos development due to the active metabolic state (Balbuena et al. 2009), which further supports the results of the study.
Carbon fixation
In plants, several proteins and other co-factors assemble to execute energy conversion reaction of photosynthesis; reaction center captures the energy of photons using pigment molecules to turn them into usable form (Isah 2015b). Once the light energy is absorbed by pigment molecule or passed to them by resonance transfer from surrounding light harvesting complex, two electrons are released into the electron transport chain. The transported electrons within the chain get raised to an excited state and their return to ground state couples to energy production. In this study, photo system (PS) II reaction center M proteins (PsbM) (spot 32A) are light-driven water: plastoquinone oxidoreductase that uses light energy for abstracting electrons from water, thus, generating oxygen and proton gradient which are later used to form ATP. The protein comprises core antenna complex that captures photons and an electron transfer chain for converting photonic excitation into charge separation. A subunit is involved in dimerization of PSII, and at the monomer–monomer meet is the only protein–protein contacts observed between PsbM subunits and lipid, the chlorophyll and carotenoids that contribute to dimerization of PSII. Photo system II protein D1 (PsbA) (spot 16D) together with the D2 reaction center heterodimer binds P680, the primary electron donor of PSII. The later electron acceptors share non-heme iron with each subunit binding pheophytin, quinone, other chlorophylls, carotenoids, and lipids, and provides most of the ligands for the Mn4–Ca–O5 complex cluster of oxygen-evolving complex. A Cl (− 1) ion association with the D1 and D2 is the need for oxygen evolution. Chlorophyll a/b-binding proteins of the PS II (spot 33A) associates with the chlorophylls and xanthophylls to make up part of antenna complex. The excitation energy of the PS II due to stress condition is balanced by the chlorophyll a/b-binding protein (Liu and Shen 2004; Li et al. 2004). In this study, a higher expression was shown by the components of photosynthetic apparatus making up the photo systems and chlorophyll a/b-binding protein during the developmental stages of the embryos from globular to maturation stage. Given that the embryonic tissues showed increased visible pigmentation over the stages (Fig. 1h–k), it suggests higher carbon fixation to provide carbon compounds and monomers for the synthesis of cell wall polymers, and that photosynthesis plays role in the development of the embryos in N. nimmoniana. It also suggests that a role is played by enzymes involved in carbon fixation to provide products used in the energy generation for embryo development given the expression of enzymes involved in carbon fixation identified in the present study (spot 34A and 35A). The large and small sub units of Ribulose—1,5-bisphosphate carboxylase/oxygenase (RubisCO) (spot 34A, 35A) showed higher expression from globular to maturing embryo stage in similar way to the other identified carbon-fixing enzymes. Higher carbon fixation in the maturing embryos facilitates synthesis of 3-carbon compounds that can be converted to the other forms of carbohydrates for reserve accumulation and respiration to provide cellular energy needed for metabolic processes. It also suggests that the embryos developed photosynthetic capacity/autotrophy during the advanced stages of development. Although enzymes involved in photosynthetic energy production showed higher expression across embryogenesis stages, particularly in the embryo maturation and as supported by the increased visible pigmentation of the embryos, the majority did not convert to plantlets, suggesting a higher metabolic state that possibly led to the lack of physiological maturation (Fig. 1k).
Secondary metabolism
Plant secondary metabolites play an essential role in physiological and biochemical processes in cells, tissues, and organs and their biosynthesis are regulated by the developmental stage adaptation and environmental conditions involved in protective response during stresses (Kliebenstein 2004; Ahuja et al. 2010; Batra and Sharma 2013). CPT is a monoterpene indole anticancer alkaloid biosynthesized through the terpenoid indole alkaloid biosynthetic pathway that involves strictosidine synthase, tryptophan decarboxylase, and secologanin synthase, and is isolated from N. nimmoniana in the highest yield quantity than other natural sources (Govindachari and Viswanathan 1972; Yamamoto et al. 2000; Yamazaki et al. 2003a). It is the most promising anticancer drug of plant origin, but the biosynthetic pathway and regulatory mechanism of production are not clear (Isah 2016b). In this study, strictosidine synthase (spot 38A) catalyzed the condensation of secologanin and tryptamine to produce strictosidine in the CPT biosynthetic pathway. Tryptamine then provided indole moiety of terpenoid indole alkaloids through decarboxylation of tryptophan by tryptophan decarboxylase (spot 38B) in reaction that serves as branching point from primary metabolism to secondary metabolic pathways. Secologanin moiety was then derived from the geraniol, while geraniol 10-hydrolase hydroxylates geraniol at the C-10 position to form loganin and secologanin in reaction catalyzed by secologanin synthase (spot 38C) (Yamamoto et al. 2000; Yamazaki et al. 2003a). In the present study, the enzymes showed higher expression across the stages, in particular, secologanin synthase and tryptophan decarboxylase whose expression was relatively higher than the strictosidine synthase. Accumulation of strictosidine synthase proteins and transcripts in the stem and roots of Ophiorrhiza pumila correlated with the expression of CPT biosynthesis and organ development (Yamazaki et al. 2003b). Similarly, accumulation of CPT was found to be higher in the somatic embryos (Fulzele and Satdive 2003) and as supported by the results of HPTLC analysis carried out in the present study. The accumulated CPT increased between the various stages of somatic embryo development with 495 ± 0.14a, 687 ± 0.18b, and 804 ± 0.16c µg CPT/g embryo DW for the globular, heart/torpedo-shaped, and maturing embryo stages respectively. This reflects the expression of CPT biosynthesis enzymes identified in the study.
Conclusion
SE was reported in N. nimmoniana over a decade ago, but yet, there has been no progress on the production and application in the clonal propagation of the tree so far, despite the potentials, it offers in scale-up production of CPT. In this study, application of 2-DE and MaSp has aided understanding some physiological, metabolic processes involved in the somatic embryos development in N. nimmoniana at proteome expression levels through the roles of stress, energy production, and other metabolic processes by expression of the associated proteins identified. Results of the study have provided some insight into the lack of physiological maturation shown by the embryos at the maturation stage, which could be attributed to their higher metabolic state. As SE offers an alternative technology for studying the development of embryos in the in vivo, due to their genetic and physiological, developmental similarity, further research on the physiology could provide proper information on recalcitrance exhibited by N. nimmoniana seeds, with application in conservation of the tree to mitigate indiscriminate harvest of its endangered natural population for the anticancer alkaloid. Information from the studies could also be of use in understanding metabolism, molecular markers of the various stages, with application in optimizing culture conditions for mass propagation and commercial production of CPT, especially in a scale-up production. Given the low number of the expressed proteins involved in embryo maturation stage identified in the present study, the use of maturation-inducing agents such as polyethylene glycol could provide insight on optimizing culture condition of the embryos through enhancing embryo maturation, in particular. The low number could also be attributed to the limited bioinformatics data available in the NCBInr database (http://www.ncbi.nlm.nih.gov/protein) for the successful identification of the proteins as the number of expressed protein spots during embryo maturation stage was relatively higher compared to the other stages (Fig. 2). Studies on the role(s) of stress in the induction and expression of the somatic embryos will aid the optimization of condition culture factors for enhanced production of the in vitro embryos. This is the first molecular study of SE in N. nimmoniana, and could open further studies aimed at identifying culture conditions for the induction and expression of the embryogenesis, with possible application in scale-up production of CPT to mitigate indiscriminate harvest of endangered natural population for the anticancer alkaloid.
Abbreviations
- SE:
-
Somatic embryogenesis
- EC:
-
Embryogenic callus
- 2-DE:
-
Two-dimensional electrophoresis
- MS:
-
Murashige and Skoog medium
- 2,4-D:
-
2,4-Dichlorophenoxy acetic acid
- NAA:
-
Naphthalene acetic acid
- IAA:
-
Indole 3-acetic acid
- BAP:
-
Benzylaminopurine
- GA3 :
-
Gibberellic acid
- DTT:
-
Dithiothreitol
- IEF:
-
Isoelectric focusing
- NCBInr:
-
National Center for Biotechnology
- MaSp:
-
Mass spectrometry
- NAD+ :
-
Nicotinamide adenine dinucleotide
- ROS:
-
Reactive oxygen species
- CPT:
-
Camptothecin
- GE:
-
Globular embryos
- TH:
-
Heart/torpedo-shaped embryos
- ME:
-
Mature/maturing embryos
- HPTLC:
-
High-performance thin-layer chromatography
References
Abbasi BH, Ali H, Yücesan B, Saeed S, Rehman K, Khan MA (2016) Evaluation of biochemical markers during somatic embryogenesis in Silybum marianum L. 3 Biotech 6(1):71. https://doi.org/10.1007/s13205-016-0366-1
Agrawal GK, Sarkar A, Righetti PG et al (2013) A decade of plant proteomics and mass spectrometry: translation of technical advancements to food security and safety issues. Mass Spectrom Rev 32:335–365. https://doi.org/10.1002/mas.21365
Ahuja I, de Vos RCH, Bones AM, Hall RD (2010) Plant molecular stress responses face climate change. Trends Plant Sci 15:664–674. https://doi.org/10.1016/j.tplants.2010.08.002
Ali M, Isah T, Mujib A, Dipti T (2016) Climber plants: medicinal importance and conservation strategies. In: Shahzad A, Sharma S, Siddiqui SA (eds) Biotechnological strategies for the conservation of medicinal and ornamental climbers. Springer International Publishing, Switzerland, pp 101–138. https://doi.org/10.1007/978-3-319-19288-8
Andriotis VM, Kruger NJ, Pike MJ, Smith AM (2010) Plastidial glycolysis in developing Arabidopsis embryos. New Phytol 185:649–662. https://doi.org/10.1111/j.1469-8137.2009.03113.x
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Ann Rev Plant Biol 55:373–399. https://doi.org/10.1146/annurev.arplant.55.031903.141701
Baba AI, Nogueira FC, Pinheiro CB et al (2008) Proteome analysis of secondary somatic embryogenesis in cassava (Manihot esculenta). Plant Sci 175(5):717–723. https://doi.org/10.1016/j.plantsci.2008.07.014
Balbuena TS, Silveira V, Junqueira M (2009) Changes in the 2-DE protein profile during zygotic embryogenesis in the Brazilian Pine (Araucaria angustifolia). J Proteomics 72(3):337–352. https://doi.org/10.1016/j.jprot.2009.01.011
Batra P, Sharma AK (2013) Anti-cancer potential of flavonoids: recent trends and future perspectives. 3 Biotech 3(6):439–459. https://doi.org/10.1007/s13205-013205-013-0117-5
Beeler S, Liu HC, Stadler M et al (2014) Plastidial NAD-dependent malate dehydrogenase is critical for embryo development and heterotrophic metabolism in Arabidopsis. Plant Physiol 164(3):1175–1190. https://doi.org/10.1104/pp.113.233866
Bomal C, Le VQ, Tremblay FM (2002) Induction of tolerance to fast desiccation in black spruce (Picea mariana) somatic embryos: relationship between partial water loss, sugars, and dehydrins. Physiol Plant 115(4):523–530. https://doi.org/10.1034/j.1399-3054.2002.1150406.x
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1–2):248–254. https://doi.org/10.1016/0003-2697(76)90527-3
Burrieza HP, Lopez-Fernandez MP, Chiquieri TB et al (2012) Accumulation pattern of dehydrins during sugarcane (var. SP80. 3280) somatic embryogenesis. Plant Cell Rep 31(12):2139–2149. https://doi.org/10.1007/s00299-012-1323-z
Businge E, Bygdell J, Wingsle G et al (2013) The effect of carbohydrates and osmoticum on storage reserve accumulation and germination of Norway spruce somatic embryos. Plant Physiol 149:273–285. https://doi.org/10.1111/ppl.12039
Chen SX, Harmon AC (2006) Advances in plant proteomics. Proteomics 6:5504–5516. https://doi.org/10.1002/pmic.200600143
de Moura Vale EM, Heringer AS, Barroso T et al (2014) Comparative proteomic analysis of somatic embryo maturation in Carica papaya L. Proteome Sci 12(1):37. https://doi.org/10.1186/1477-5956-12-37
Fulzele DP, Satdive RK (2003) Somatic embryogenesis, plant regeneration, and evaluation of camptothecin content in Nothapodytes foetida. In Vitro Cell Dev Biol Plant 39(2):212–216. https://doi.org/10.1079/IVP2002368
Fulzele DP, Satdive RK (2005) Comparison of techniques for the extraction of the anticancer drug camptothecin from Nothapodytes foetida. J Chromatogr A 1063(1–2):9–13. https://doi.org/10.1016/j.chroma.2004.11.020
Gantait S, Debnath S, Ali MN (2014) Genomic profile of the plants with pharmaceutical value. 3 Biotech 4(6):563–578. https://doi.org/10.1007/s13205-014-0218-9
Ge X, Zhang C, Wang Q et al (2014) iTRAQ Protein profile differential analysis between somatic globular and cotyledonary embryos reveals stress, hormone, and respiration involved in increasing plantlet regeneration of Gossypium hirsutum L. J Proteome Res 14(1):268–278. https://doi.org/10.1021/pr500688g
Gerke V, Moss SE (2002) Annexins: from structure to function. Physiol Rev 82(2):331–371. https://doi.org/10.1152/physrev.00030.2001
Gomez-Garay A, Lopez JA, Camafeita E et al (2013) Proteomic perspective of Quercus suber somatic embryogenesis. J Proteomics 93:314–325. https://doi.org/10.1016/j.jprot.2013
Gonzalez E (1990) Expression and localization of malate synthase during maturation and desiccation of castor bean seeds. Protoplasma 154(1):53–58. https://doi.org/10.1007/BF01349535
Govindachari TR, Viswanathan N (1972) The alkaloids of Mappia foetida. Phytochem 11(12):3529. https://doi.org/10.1016/S0031-9422(00)89852-0
Guan Y, Li SG, Fan XF, Su ZH (2016) Application of somatic embryogenesis in woody plants. Front Plant Sci 7:938. https://doi.org/10.3389/fpls.2016.00938
Gupta SD, Datta S (2003) Antioxidant enzyme activities during in vitro morphogenesis of Gladiolus and effect of the application of antioxidants on plant regeneration. Biol Plant 47(2):179–183. https://doi.org/10.1023/B:BIOP.0000022248.62869.c7
Gupta PK, Timmis R, Pullman G et al (1991) Development of an embryogenic system for automated propagation of forest trees. In: Vasil I (ed) Scale-up and automation in plant propagation. Academic press Inc., New York, pp 75–80. https://doi.org/10.1016/B978-0-12-715008-6.50011-7
Higashi K, Shiota H, Kamada H (1998) Patterns of expression of the genes for glutamine synthetase isoforms during somatic and zygotic embryogenesis in carrot. Plant Cell Physiol 39(4):418–424. https://doi.org/10.1093/oxfordjournals.pcp.a029385
Imin N, De Jong F, Mathesius U et al (2004) Proteome reference maps of Medicago truncatula embryogenic cell cultures generated from single protoplasts. Proteomics 4(7):1883–1896. https://doi.org/10.1002/pmic.200300803
Iraqi D, Tremblay FM (2001a) Analysis of carbohydrate metabolism enzymes and cellular contents of sugars and proteins during spruce somatic embryogenesis suggests a regulatory role of exogenous sucrose in embryo development. J Exp Bot 52(365):2301–2311. https://doi.org/10.1093/jexbot/52.365.2301
Iraqi D, Tremblay FM (2001b) The role of sucrose during maturation of black spruce (Picea mariana) and white spruce (Picea glauca) somatic embryos. Physiol Plant 111(3):381–388. https://doi.org/10.1034/j.1399-3054.2001.1110316.x
Isaacson T, Damasceno CM, Saravanan RS et al (2006) Sample extraction techniques for enhanced proteomic analysis of plant tissues. Nat Protocols 1(2):769–774. https://doi.org/10.1038/nprot.2006.102
Isah T (2015a) Natural sources of taxol. Br J Pharma Res 6(4):214–227. https://doi.org/10.9734/BJPR/2015/16293
Isah T (2015b) Adjustments to in vitro culture conditions and associated anomalies in plants. Acta Biol Cracov s Botanica 57(2):9–28. https://doi.org/10.1515/abcsb-2015-0026
Isah T (2016a) Induction of somatic embryogenesis in woody plants. Acta Physiol Plant 38:118. https://doi.org/10.1007/s11738-016-2134-6
Isah T (2016b) Anticancer alkaloids from trees: development into drugs. Pharmacogn Rev 10(20):90. https://doi.org/10.4103/0973-7847.194047
Isah T (2017) Production of camptothecin in the elicited callus cultures of Nothapodytes nimmoniana (J. Graham) Mabberly. Chem Papers 71:1091–1106. https://doi.org/10.1007/s11696-016-0056-9
Isah T, Mujib A (2012) Studies on antioxidant enzymes activity during in vitro morphogenesis of Caladium bicolor Linn. Intl J Modern Cell Mol Biol 1(1):1–9
Isah T, Mujib A (2015a) In vitro propagation and camptothecin production in Nothapodytes nimmoniana. Plant Cell Tiss Organ Cult 121:1–10. https://doi.org/10.1007/s11240-014-0683-1
Isah T, Mujib A (2015b) Camptothecin from Nothapodytes nimmoniana: review on biotechnology applications. Acta Physiol Plant 37:106. https://doi.org/10.1007/s11738-015-1854-3
Isah T, Mujib A (2015c) Enhanced in vitro seedling recovery in Nothapodytes nimmoniana. British Biotechnol J 6(1):2231–2927. https://doi.org/10.9734/BBJ/2015/15368
Isah T, Umar S (2018) Influencing in vitro clonal propagation of Chonemorpha fragrans (moon) Alston by culture media strength, plant growth regulators, carbon source and photoperiodic incubation. J For Res. https://doi.org/10.1007/s11676-018-0794-3
Isah T, Umar S, Mujib A et al (2018) Secondary metabolism of pharmaceuticals in the plant in vitro cultures: strategies, approaches, and limitations to achieving higher yield. Plant Cell Tiss Organ Cult 132:239–265. https://doi.org/10.1007/s11240-017-1332-2
Jariteh M, Ebrahimzadeh H, Niknam V et al (2013) Antioxidant enzymes activities during secondary somatic embryogenesis in Persian walnut (Juglans regia L.). Afr J Biotechnol 10(20):4093–4099. https://doi.org/10.5897/AJB10.1248
Jariteh M, Ebrahimzadeh H, Niknam V et al (2015) Developmental changes in protein, proline and some antioxidant enzymes activities in somatic and zygotic embryos of Persian walnut (Juglans regia L.). Plant Cell Tiss Organ Cult 122:101–115. https://doi.org/10.1007/s11240-015-0753-z
Jimenez VM (2001) Regulation of in vitro Somatic Embryogenesis with emphasis on the role of endogenous hormones. Rev Brasi de Fisio Vegl 13:196–223. https://doi.org/10.1590/S0103-31312001000200008
Kairong C, Gengsheng X, Lin Q et al (1999) The analysis of differential gene expression in early somatic embryogenesis on Lycium barbarum. Plant Cell Tiss Organ Cult 59(3):169–174. https://doi.org/10.1023/A:1006311628358
Khadke S, Kuvalekar A (2013) Direct somatic embryogenesis and plant regeneration from leaf and stem explants of N. foetida: a critically endangered plant species. Int J Plant Anim Environ Sci 3(1):257–264
Kliebenstein DJ (2004) Secondary metabolites and plant/environment interactions: a view through Arabidopsis thaliana tinged glasses. Plant Cell Environ 27(6):675–684. https://doi.org/10.1111/j.1365-3040.2004.01180.x
Kulkarni AV, Patwardhan AA, Lele U, Malpathak NP (2010) Production of camptothecin in cultures of Chonemorpha grandiflora. Pharmacogn Res 2(5):296–299. https://doi.org/10.4103/0974-8490.72327
Kumari S, Roy S, Singh P et al (2013) Cyclophilins: proteins in search of a function. Plant Signal Behav 8(1):e22734. https://doi.org/10.4161/psb.22734
Li XP, Gilmore AM, Caffarri S et al (2004) Regulation of photosynthetic light harvesting involves intra-thylakoid lumen pH sensing by the PsbS protein. J Biol Chem 279(22):22866–22874. https://doi.org/10.1074/jbc.M402461200
Lippert D, Zhuang J, Ralph S et al (2005) Proteome analysis of early somatic embryogenesis in Picea glauca. Proteomics 5(2):461–473. https://doi.org/10.1002/pmic.200400986
Liu XD, Shen YG (2004) NaCl-induced phosphorylation of light-harvesting chlorophyll a/b proteins in thylakoid membranes from the halo-tolerant green alga, Dunaliella salina. FEBS Lett 569(1–3):337–340. https://doi.org/10.1016/j.febslet.2004.05.065
Lutz JD, Wong JR, Rowe J et al (1985) Somatic embryogenesis for mass cloning of crop plants. In: Henke RR et al (eds) Tissue culture in forestry and agriculture: basic life sciences, vol 32, Springer, New York, pp 105–116. https://doi.org/10.1007/978-1-4899-0378-5_8
Lyngved R, Renaut J, Hausman JF et al (2008) Embryo-specific proteins in Cyclamen persicum analyzed with 2-D DIGE. J Plant Growth Regul 27(4):353–369. https://doi.org/10.1007/s00344-008-9061-8
Manjasetty BA, Büssow K, Panjikar S, Turnbull AP (2012) Current methods in structural proteomics and its applications in biological sciences. 3 Biotech 2(2):89–113. https://doi.org/10.1007/s13205-011-0037-1
Marsoni M, Bracale M, Espen L et al (2008) Proteomic analysis of somatic embryogenesis in Vitis vinifera. Plant Cell Rep 27(2):347–356. https://doi.org/10.1007/s00299-007-0438-0
Martin-Tanguy J (2001) Metabolism and function of polyamines in plants: recent development (new approaches). Plant Growth Regul 100:675–688. https://doi.org/10.1023/A:1013343106574
Minocha R, Minocha SC, Long S (2004) Polyamines and their biosynthetic enzymes during somatic embryo development in red spruce (Picea rubens Sarg.). In Vitro Cell Dev Biol Plant 40(6):572–580. https://doi.org/10.1079/IVP2004569
Mujib A, Ali M, Isah T, Dipti T (2014) Somatic embryo mediated mass production of Catharanthus roseus in culture vessel (bioreactor)—a comparative study. Saudi J Biol Sci 21(5):442–449. https://doi.org/10.1016/j.sjbs.2014.05.007
Mujib A, Ali M, Tonk D, Isah T, Zafar N (2016) Embryogenesis in ornamental monocots: plant growth regulators as signaling element. In: Mujib A (ed) Somatic embryogenesis in ornamentals and its applications. Springer, New York, pp 187–201. https://doi.org/10.1007/978-81-322-2683-3_12
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15(3):473–497. https://doi.org/10.1111/j.1399-3054.1962.tb08052.x
Noah AM, Niemenak N, Sunderhaus S et al (2013) Comparative proteomic analysis of early somatic and zygotic embryogenesis in Theobroma cacao L. J Proteomics 78:123–133. https://doi.org/10.1016/j.jprot.2012.11.007
O’Farrell PH (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250(10):4007–4021
Padmanabha BV, Chandrashekar M, Ramesha BT et al (2006) Patterns of accumulation of camptothecin, an anti-cancer alkaloid in Nothapodytes nimmoniana Graham., in the Western Ghats, India: implications for identifying high-yielding sources of the alkaloid. Curr Sci Bangalore 90(1):95
Pan Z, Guan R, Zhu S, Deng X (2009) Proteomic analysis of somatic embryogenesis in Valencia sweet orange (Citrus sinensis Osbeck). Plant Cell Rep 28(2):281–289. https://doi.org/10.1007/s00299-008-0633-7
Prakash L, Middha SK, Mohanty SK, Swamy MK (2016) Micropropagation and validation of genetic and biochemical fidelity among regenerants of Nothapodytes nimmoniana (Graham) Mabb. employing ISSR markers and HPLC. 3 Biotech 6(2):171. https://doi.org/10.1007/s13205-016-0490-y
Quiroz-Figueroa F, Rojas-Herrera R, Galaz-Avalos RM, Loyola-Vargas VM (2006) Embryo production through somatic embryogenesis can be used to study cell differentiation in plants. Plant Cell Tiss Organ Cult 86:285–301. https://doi.org/10.1007/s11240-006-9139-6
Ramesha BT, Amna T, Ravikanth G et al (2008) Prospecting for camptothecines from Nothapodytes nimmoniana in the Western Ghats, South India: identification of high-yielding sources of camptothecin and new families of camptothecines. J Chromatogr Sci 46(4):362–368. https://doi.org/10.1093/chromsci/46.4.362
Rode C, Lindhorst K, Braun HP, Winkelmann T (2012) From callus to embryo: a proteomic view on the development and maturation of somatic embryos in Cyclamen persicum. Planta 235(5):995–1011. https://doi.org/10.1007/s00425-011-1554-1
Sghaier-Hammamia B, Driraa N, Jorrin-Novo JV (2009) Comparative 2-DE proteomic analysis of date palm (Phoenix dactylifera L.) somatic and zygotic embryos. J Proteomics 73:161–177. https://doi.org/10.1016/j.jprot.2009.07.003
Shin JH, Kim SR, An G (2009) Rice aldehyde dehydrogenase 7 is needed for seed maturation and viability. Plant Physiol 149(2):905–915. https://doi.org/10.1104/pp.108.130716
Shohael AM, Ali MB, Hahn EJ, Paek KY (2007) Glutathione metabolism and antioxidant responses during Eleutherococcus senticosus somatic embryo development in a bioreactor. Plant Cell Tiss Organ Cult 89(2–3):121–129. https://doi.org/10.1007/s11240-007-9220-9
Silveira V, Santa-Catarina C, Tun NN et al (2006) Polyamine effects on the endogenous polyamine contents, nitric oxide release, growth and differentiation of embryogenic suspension cultures of Araucaria angustifolia (Bert.) O. Ktze. Plant Sci 171(1):91–98. https://doi.org/10.1016/j.plantsci.2006.02.015
Suhas S, Ramesha BT, Ravikanth G et al (2007) Chemical profiling of Nothapodytes nimmoniana populations in the Western Ghats, India for anti-cancer compound, camptothecin. Curr Sci 92(8):1142–1147
Teyssier C, Maury S, Beaufour M et al (2014) In search of markers for somatic embryo maturation in hybrid larch (Larix × eurolepis): global DNA methylation and proteomic analyses. Physiol Plant 150(2):271–291. https://doi.org/10.1111/ppl.12081
Thibaud-Nissen F, Shealy RT, Khanna A, Vodkin LO (2003) Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol 132(1):118–136. https://doi.org/10.1104/pp.103.019968
Vondrakova Z, Eliasova K, Fischerova L, Vagner M (2011) The role of auxins in somatic embryogenesis of Abies alba. Central Eur J Biol 6(4):587–596. https://doi.org/10.2478/s11535-011-0035-7
Winkelmann T, Heintz D, Van Dorsselaer A et al (2006) Proteomic analyses of somatic and zygotic embryos of Cyclamen persicum Mill. reveal new insights into seed and germination physiology. Planta 224(3):508–519. https://doi.org/10.1007/s00425-006-0238-8
Winter H, Huber SC (2000) Regulation of sucrose metabolism in higher plants: localization and regulation of the activity of key enzymes. Crit Rev Plant Sci 19(1):31–67. https://doi.org/10.1080/10409230008984165
Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot 95:707–735. https://doi.org/10.1093/aob/mci083
Yamamoto H, Katano N, Ooi A, Inoue K (2000) Secologanin synthase which catalyzes the oxidative cleavage of loganin into secologanin is a cytochrome P450. Phytochem 53:7–12. https://doi.org/10.1016/S0031-9422(99)00471-9
Yamazaki Y, Sudo H, Yamazaki M et al (2003a) Camptothecin biosynthetic genes in hairy roots of Ophiorrhiza pumila: cloning, characterization and differential expression in tissues and by stress compounds. Plant Cell Physiol 44(4):395–403. https://doi.org/10.1093/pcp/pcg051
Yamazaki Y, Urano A, Sudo H (2003b) Metabolite profiling of alkaloids and strictosidine synthase activity in camptothecin producing plants. Phytochem 62(3):461–470. https://doi.org/10.1016/S0031-9422(02)00543-5
Zavattieri M, Frederico A, Lima M et al (2010) Induction of somatic embryogenesis as an example of stress-related plant reactions. Electron J Biotechnol N Am 13(1):15. https://doi.org/10.2225/vol13-issue1-fulltext-4
Zhu JK, Shi J, Bressan RA, Hasegawa PM (1993) Expression of an Atriplex nummularia gene encoding a protein homologous to bacterial molecular chaperone DNA. J Plant Cell 5:341–349. https://doi.org/10.1105/tpc.5.3.341
Zimmerman JL (1993) Somatic embryogenesis: a model for early development in higher plants. Plant Cell 5(10):1411. https://doi.org/10.1105/tpc.5.10.1411
Acknowledgements
Tasiu Isah is grateful to The World Academy of Science (TWAS) for the advancement of science in the developing World Trieste, Italy, and Department of Biotechnology (DBT), Government of India New Delhi for the award of DBT-TWAS Postgraduate Research Fellowship. Technical assistance provided by colleagues and laboratory technologist is acknowledge. Research facilities provided by the Department of Botany, Hamdard University New Delhi, India, are also acknowledged.
Author information
Authors and Affiliations
Contributions
TI conceived the research idea, performed the experiments, and wrote the manuscript. TI, SU, and MPS assisted with intellectual contribution and editing of the manuscript. All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that no conflict of interest exists in the manuscript contents.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Isah, T. Proteome study of somatic embryogenesis in Nothapodytes nimmoniana (J. Graham) Mabberly. 3 Biotech 9, 119 (2019). https://doi.org/10.1007/s13205-019-1637-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1637-4