Abstract
Dichrostachys cinerea (L.) Wight & Arn, which belongs to the Mimosoid clade of the legume subfamily Caesalpinioideae, was introduced into India and has since become invasive across wide areas of the country. It is nodulated, and like all other mimosoids it has indeterminate nodules with its microsymbionts housed in membrane-bound symbiosomes rather than within cell wall-enclosed fixation threads. Fifty-eight bacterial strains were isolated from root nodules on plants growing in soils from 13 sampling sites in India with various agroclimatic conditions. Genetic analysis of 36 strains resulted in diverse RAPD genotypes, with equal composition of Ensifer and Bradyrhizobium as its root nodule microsymbionts. Multi locus sequence analysis (MLSA) of 12 strains using the recA, glnII, atpD and 16S rRNA genes revealed significant genetic diversity forming novel clades and lineages and are potential new species. The D. cinerea strains were variants of local symbionts previously described as rhizobia associated with native and exotic mimosoid trees, as well as rhizobia associated with the non-mimosoid Caesalpinioid shrub Chamaecrista pumila and wild Papilionoid legumes from India. The symbiosis essential genes (nodA and nifH) of the D. cinerea strains were diverse and clustered according to geographical origin. Mosaic combinations of core and sym genes were harbored by both Ensifer and Bradyrhizobium suggesting gradual diversification and microevolution of rhizobia under pressure from the host in combination with edaphic and environmental factors. The dominant microsymbionts of native and invasive legumes, including D. cinerea, in alkaline soils of India are essentially of the ‘E. aridi’ and B. yuanmingense types. Dichrostachys cinerea rhizobia were symbiotically efficient on their homologous host, but also have ability to nodulate the crop Vigna radiata, and hence may be good candidates to be used for inoculants on legume crops as well as on Mimosoid trees (P. cineraria, V. nilotica, V. raddiana, S. senegal) used in sustainable agroforestry practices to enhance soil nitrogen content.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Dichrostachys is a genus in the Mimosoid clade which is nested within the legume subfamily Caesalpinioideae (LPWG 2017; Sprent et al. 2017; de Faria et al. 2022). It comprises 14 species of shrub or small tree and is indigenous to the Old World tropics (Sprent 2005). The species Dichrostachys cinerea (L.) Wight & Arn., commonly known as the sickle bush, is a perennial tree native to Africa and Australia (https://www.cabidigitallibrary.org/doi/https://doi.org/10.1079/cabicompendium.18119). From Africa it was introduced in Cuba in the nineteenth century, later invading unused agricultural lands. It is also invasive in southern African savannas through clonal spread by root suckers (Nielsen et al. 2013; Wakeling and Bond 2007). It is a drought-tolerant plant and can grow in soil with varying pH (4.5–8.5), it grows well on deep, sandy loamy soil, and can survive mean annual rainfalls from 200–400 mm (Coates-Palgrave 1988). Although D. cinerea is an aggressive and devastating invasive species in some countries it has utility e.g. it can be used as biomass for bioenergy generation. It also has several environment-related benefits, such as soil conservation, sand dune stabilization, re-vegetation, and intercropping, and hence it can provide a base for environmentally and economically sustainable agroforestry and silvo-pastoral systems (Heuzé et al. 2015; Sáez and Alfayate 2020). Like most Mimosoids, its capacity for root nodulation means it is able to symbiotically fix atmospheric nitrogen (N), allowing it to be used in co-cultivation systems as a source of N fertilization with other (non-legume) fodder or cash crops (Pule-Meulenberg and Dakora 2009; Nielsen et al. 2013).
In India D. cinerea is naturalized in dry deciduous forests in western to southern parts of the country including the states of Andhra Pradesh, Delhi, Haryana, Goa, Gujarat, Karnataka, Kerala, Madhya Pradesh, Maharashtra, Orissa, Punjab, Rajasthan, Tamil Nadu and Uttar Pradesh (Bhandari 1990, CABI Compendium https://doi.org/10.1079/cabicompendium.18119). It was originally planted along with other legumes and grasses for the restoration of degraded land with reduced organic matter and poor moisture retention capacity e.g. Asola-Bhatti Wildlife Sanctuary situated in Delhi (Pant and Pant 2017). Little is known about the rhizobia that nodulate D. cinerea, but effective root nodules on D. cinerea were formed by Rhizobium sp. strain NGR234 (now named E. fredii NGR234) isolated from nodules of Lablab purpureus in Papua New Guinea (Trinick 1980), and by R. fredii USDA257 (now named E. fredii USDA257) isolated from wild Glycine soja in China (Pueppke and Broughton 1999). However, it failed to nodulate with the promiscuous Beta-rhizobium strain, Paraburkholderia phymatum STM815T (Moulin et al. 2014). We have not encountered any published study related to the characterization of N-fixing rhizobia naturally associated with it in India or elsewhere. Therefore, the aim of present study was to isolate and characterize the root nodule bacteria (RNB) associated with D. cinerea in different agroclimatic zones of India specifically from soils of Rajasthan (RJ), Haryana (HR), Punjab (PB), Madhya Pradesh (MP), Puducherry (PY) and Meghalaya (ML). It was also considered to be of interest to compare its microsymbionts with those from native (Mimosa hamata, M. himalayana, Prosopis cineraria, Vachellia jacquemontii, V. nilotica, V. leucophloea, and Senegalia senegal) and invasive (M. pudica, Leucaena leucocephala) Mimosoid species in the Thar desert as well as from other parts of India, wherein most of the aforementioned genera were predominantly nodulated by diverse species of Ensifer (Gehlot et al. 2012, 2013, 2016; Sankhla et al. 2017; Choudhary et al. 2017, 2018, 2020; Chouhan et al. 2022). In consideration of its invasiveness, this investigation was initiated with the possibility that like other invasive legumes, such as L. leucocephala (Chouhan et al. 2022), the invasiveness of D. cinerea is assisted by an ability to nodulate with a broad diversity of native microsymbionts (species of Ensifer, Bradyrhizobium, Rhizobium and Mesorhizobium) similar to those previously reported from a wide range of native or exotic legumes growing in the Thar desert and other agroclimatic regions of India. Or, that it may have a specific preference in its selection of microsymbionts, and as with another invasive legume, M. pudica (Gehlot et al. 2013), its preferred symbiont has accompanied it from its native environment.
2 Materials and Methods
2.1 Trapping of rhizobia in root nodules of Dichrostachys cinerea from soils of various sites
Healthy seeds of D. cinerea (collected from plants growing in the Botanical Garden, J.N.V. University, Jodhpur) were surface sterilized by washing in 90% (v/v) alcohol and an antifungal agent [0.1% (w/v) BavistinR] for 1 min followed by five rinses in sterile distilled water. Seeds were further surface sterilized in 4% (v/v) sodium hypochlorite (NaOCl) for 1 min and washed 6 times with autoclaved distilled water to remove the residues of sterilizing chemicals. Surface sterilized seeds of D. cinerea were germinated on moist sterile filter paper and sowed in sterile pots filled with soils collected from different sampling sites in RJ, HR, PB, MP, ML and PY (Table 1, Fig. 1). Pots were placed in a greenhouse under controlled conditions (28–30 °C and natural sunlight giving 14 h light and 10 h of darkness) and moistened regularly with autoclaved tap water. After 8 to 10 weeks plants were harvested, and nodulation was checked as described earlier (Sankhla et al. 2017).
2.2 Nodule sampling for microscopy, and isolation and purification of rhizobia
From each harvested D. cinerea plant 2–3 root nodules were selected for rhizobial isolation, while other nodules were fixed in glutaraldehyde and prepared for light and transmission electron microscopy (TEM) according to de Faria et al. (2022). For isolation of rhizobia, adhered soil particles were removed with water and prior to surface sterilization they were wrapped in muslin cloth; the nodules were then surface-sterilized firstly in 90% (v/v) alcohol for 1 min, transferred to 0.1% (w/v) BavistinR for 30 s, and rinsed 4–5 times in autoclaved distilled water to remove traces of fungicide. Finally, the nodules were transferred to 4% (v/v) sodium hypochlorite (NaOCl) for 3 min and then washed 6 times in sterile water. The surface sterilized nodules were swabbed onto Yeast Extract Mannitol Agar- Congo red (YEMA-CR) medium to check that sterilization of the nodule surface was 100%, and then crushed into 1–2 drops of sterile distilled water followed by streaking of the white exudate onto YEMA-CR medium. The streaked master plates were incubated at 28 °C for up to 7 days and rhizobia-like colonies (i.e. white or transparent or translucent, circular, smooth margin, raised, EPS producing, mucilaginous or gummy and not taking up the red dye) were picked up for purification. Single colonies were streaked in a four-way pattern on fresh media until pure colonies were obtained (Howieson and Dilworth 2016). Pure cultures were maintained for further analysis, but also preserved as glycerol stocks in -80 °C.
2.3 Phenotypic profiling
Various phenotypic tests were performed on selected strains (identified based on their recA gene sequences). The temperature tolerance of the strains was tested by inoculating them onto YEMA and incubating at high temperatures (35 °C, 40 °C, 45 °C and 48 °C). The salt tolerance of the strains was analyzed by streaking them onto YEMA supplemented with a range of salt (NaCl) concentrations: 0.5%, 1%, 2% and 3% (w/v). The pH tolerance of strains was tested by inoculating them onto YEMA adjusted to a range of pH values: 5, 6, 8, 10 and 11 (Sankhla et al. 2017). The acid or alkaline production by strains was tested in YEM broth with Bromo thymol blue (BTB) indicator (Somasegaran and Hoben 1994). Resistance or sensitivity of strains towards seven antibiotics was determined using HiMedia antibiotic discs (single concentration). The metabolic ability of strains to utilize sole carbon (sugar) sources was determined using 21 HiMedia sugar discs in broth containing Andrade pH indicator (Tak et al. 2020).
2.4 Genotypic profiling
Purified bacterial strains were activated in Tryptone yeast (TY) broth in an incubator shaker (120 rpm) at 28 °C for DNA isolation using the method [constituent’s phenol, chloroform, STE (Sodium chloride-Tris–EDTA) buffer and TE buffer] of Cheng and Jiang (2006). The purity and concentration of DNA were checked by NanoDrop and DNA of 100 to 1000 ng µl−1 was stored at 4 °C for further molecular studies (Tak et al. 2016, 2020), such as DNA fingerprinting, and amplification of housekeeping (recA, glnII, atpD and 16S rRNA) and symbiotic (nodA and nifH) genes. The RPO1 primer-based Rapid Amplification of Polymorphic DNA (RAPD) profiling was obtained (Richardson et al. 1995) using thermal cycling conditions described in Table S1 for categorizing strains into RAPD genotypes (groups and individual).
2.5 Amplification of housekeeping (recA, atpD, glnII and 16S rRNA) and symbiotic (nodA and nifH) genes
For identification of bacterial strains at the molecular level the conserved recA (encoding recombination protein A) core gene was successfully amplified using two set of primers, recA6F and recA555R (amplicon size 550 bp), and TSrecAF and TSrecAR (600 bp), for Ensifer and Bradyrhizobium, respectively (Table S1). The Multi locus Sequence Analysis (MLSA) of selected strains was performed by amplifying additional protein-coding housekeeping genes (atpD and glnII) and the universal molecular chronometer 16S rRNA gene using different pairs of primers and thermal cycling conditions (Table S1). The symbiosis essential gene nodA (encodes N-acyltransferase nodulation protein) was amplified using two sets of primers nodA1 and nodA2 (650 bp), and nodAf.brad and nodAr.brad (550 bp). The nifH gene, which encodes nitrogenase reductase protein and is essential for N fixation, was amplified (product size 750 bp) using forward (nifHF) and reverse (nifHI) primers. Details about primers used and thermal cycling conditions used for gene amplification are given in Table S1.
2.6 Sequencing and phylogenetic analysis
Sanger sequencing of purified PCR products of different genes was achieved through an external company (AgriGenome Labs. Pvt. Ltd., Kochi, Kerala, India) that provided results in ABI and FASTA format. Chromatograms and sequences were viewed, edited, and trimmed using Gene Tool lite version 1.0 (Double Twist Inc., Oakland, CA, USA) software. Nucleotide sequences of strains were analyzed for percentage sequence similarity using BLASTn (Nucleotide Basic Local Alignment Search Tool) of NCBI (National Center for Biotechnology Information). Sequences from the present study and those of closely related, reference and type strains (as per the LPSN list of valid and not validly published type strains of a species in a genus) were downloaded from the NCBI nucleotide in FASTA format, and then aligned using CLUSTALW (Thompson et al. 1994) of MEGA7 (Kumar et al. 2016). Phylogenetic trees (individual or concatenated) were reconstructed in MEGA7 using the Maximum Likelihood method and General Time Reversible (GTR + G + I) model with 1000 bootstrap values.
2.7 Nodulation assay to compare symbiotic efficacy of the rhizobial strains
A selection of genetically characterized Ensifer and Bradyrhizobium strains were authenticated on their host D. cinerea and cross inoculated onto Vigna radiata. Surface sterilized seeds were germinated on moist filter paper and transferred (around 5–6 seeds/pot) under aseptic conditions into sterile potting mixture (3:1 soilrite: river sand) in autoclaved plastic pots (Tak et al. 2020). Each seedling was inoculated with 1 ml of bacterial suspension prepared in 1% sucrose. Pots set up in triplicates for each treatment and control were placed in a glasshouse under controlled conditions (28–30 °C) and maintained for 4–6 weeks. Plants were watered with sterile tap water and nourished with N-free nutrient solution (Yates et al. 2004). After 4–6 weeks harvested plants were examined for number of root nodules, shoot fresh weight (g plant−1), shoot dry weight (g plant−1), and compared with un-inoculated control plants either fed with a nutrient solution supplemented with nitrate in the form of KNO3 (0.1%) or without any added N (Yates et al. 2004; Sankhla et al. 2017; Tak et al. 2020) to determine relative symbiotic efficacy.
3 Results
3.1 Nodulation of Dichrostachys cinerea and nodule anatomy
Dichrostachys cinerea (DC) were found growing profusely in the Botanical garden of the Department of Botany, JNVU, Jodhpur, Rajasthan, India (Fig. S1a). After the monsoon rainfalls these plants grow profusely through root suckers. It has an axillary or extra-axillary spike inflorescence of 2–5 cm long with pink- and yellow-coloured flowers. The apical part of the inflorescence contains yellow hermaphrodite spike flowers, and the lower half part bears pink sterile spike flowers (Fig. S1b). Seeds are glossy, small (4–6 × 3–4.5 mm) and dark brown (Fig. S1c). Two to three months old plants were harvested from pots, and the root system bearing small, round, branched, and indeterminate type of nodules was observed (Fig. S1d). The anatomy and ultrastructure of D. cinerea nodules were examined using light microscopy and TEM of sections taken from fixed and resin-embedded nodules originally sampled through trap-experiments (Fig. 2a-d). The nodules were similar to those previously studied on other mimosoid species, such as those in the genera Mimosa and Vachellia (Gehlot et al. 2013; Sankhla et al. 2017; Choudhary et al. 2020) in that they are indeterminate with single or multilobed meristems (Fig. 2a), and the N2-fixing zone has both infected and uninfected cells (Fig. 2b). In addition, the invasion zone is relatively small with occasional infection threads (Fig. 2c, d).
Structure and ultrastructure of Dichrostachys cinerea nodules examined using light microscopy (a, b) and transmission electron microscopy (TEM) (c, d). (a) Longitudinal section (LS) of a multi-lobed nodule with two meristems indicated (m). The N2-fixing infected zone (iz) consists of dark-stained cells interspersed with non-stained uninfected cells. (b) High magnification view of mature N-fixing cells (*) and uninfected cells (uc) within the infected zone. (c) TEM of an infection thread (IT) entering a host cell in the invasion zone; the IT has originated from the host cell wall between two host cells (*). The wall of the IT (arrow) and the host cell wall (*) are both immunogold labelled with a monoclonal antibody (JIM5) that recognizes a pectin epitope. Note that the host cell receiving the IT is metabolically very active with numerous plastids (p) and mitochondria (m). b = bacteroid. (d) Bacteroids (b) within an infected cell in the N-fixing zone; note that there can be up to four bacteroids per symbiosome (*). A remnant of an IT (arrow) originating from a pocket of bacteria in an intercellular space (is) can also be observed; the walls of this are immunogold labelled strongly with JIM5. Bars = 500 µm (a), 50 µm (b), 1 µm (c, d)
3.2 Isolation and purification of potentially symbiotic bacteria from root nodules of Dichrostachys cinerea
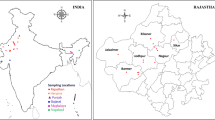
A total of 58 bacterial strains were isolated from root nodules of D. cinerea grown in rhizospheric as well as non-rhizospheric soils collected from thirteen sites covering five states namely RJ, HR, PB, MP, ML and one union territory PY. The maximum numbers of soil sampling sites were from RJ covering five districts (Jaisalmer, Jodhpur, Nagaur, Barmer and Dausa). The soils from the various sampling sites ranged from alkaline (pH 9) to acidic (pH 4.9). The geographical coordinates of the soil collection sites are listed in Table 1 and marked on a map of India (Fig. 1). The 58 bacterial strains purified from root nodules of D. cinerea are listed in Table 1 with their site of isolation. Fast-growing bacterial strains with colony characters such as white, opaque, raised, entire margins, smooth surface, non-mucilaginous, EPS-producing were identified as species of Ensifer (10 strains) based on molecular characterization (recA gene sequencing and BLASTn). Slow-growing strains with white, opaque, raised, gummy, entire margins, and less mucilaginous colony morphology were identified as Bradyrhizobium (10 strains).
3.3 Salt, pH and temperature tolerance of rhizobia
Selected bacterial strains were further characterized phenotypically. Six Ensifer strains isolated from RJ and PB showed up to 1% salt (NaCl) tolerance while four Ensifer strains isolated from HR and MP survived up to 2% NaCl (Table S2). In contrast, Bradyrhizobium strains were comparatively sensitive to salt stress and could not grow in 1% NaCl. Ten Ensifer strains showed a pH tolerance range from 6 to 11 while ten Bradyrhizobium strains grew in a pH range of 5 to 11 (Table S2). Bradyrhizobium strains isolated from PY (DC-PY35, DC-PY36 and DC-PY37) and ML (DC-ML38 and DC-ML39) are tolerant to temperatures from 28 to 35 °C while strains from RJ (DC-RJ11 and DC-RJ13) and MP (DC-MP30, DC-MP31 and DC-MP32) were able to grow at temperatures from 28 to 48 °C. All the Ensifer strains showed tolerance up to 48 °C. In the BTB test all the Ensifer strains were acid producers while the Bradyrhizobium strains were slightly acidic to slightly alkaline producers plus a few strains showed a neutral response.
3.4 Metabolic profile: carbon utilization and intrinsic antibiotic resistance of rhizobia
Of the six Ensifer strains tested (Fig. S2) all utilized Arabinose and Xylose, while Ensifer strain DC-MP27 utilized a maximum of 13 sugars. In comparison, the eight tested Bradyrhizobium strains utilized fewer sugars except for DC-RJ11. The intrinsic antibiotic resistance (IAR) profiles of the Ensifer (10) and Bradyrhizobium (10) strains are presented in Table S3. All the tested Ensifer strains were resistant to 15 µg Erythromycin except DC-RJ12 that showed a minimum zone of inhibition. The tested Bradyrhizobium strains showed resistance against 5 µg Ciprofloxacin and 30 µg Tetracycline except two strains (DC-MP31 and DC-PY36) that showed a minimum zone of inhibition against Tetracycline (Table S3).
3.5 DNA fingerprinting and identification of strains based on recA gene sequences
Of the 58 purified rhizobia 36 strains were selected for genetic fingerprinting using the RPO1 primer based on colony morphology and site of collection. A total of five genetic groups (Groups I to V) were formed including 16 strains, while the remaining 20 strains had unique banding patterns representing individual genotypes (Table S4). Location dependent banding patterns were observed; Groups I to III consist of strains from RJ, Group IV from HR, while Group V included strains from PB. This grouping indicates that although some strains that are affiliated to a particular geographical area are genetically similar most of them are highly diverse. Based on the RPO1 genetic groups, individual genotypes, and/or their geographical origin (i.e. from sampling sites representing all the different States and Union territories of India in the present study) 20 strains of D. cinerea were selected for recA gene sequence-based identification and phylogenetic analysis. From the BLASTn results ten were identified as Ensifer and ten as Bradyrhizobium. NCBI GenBank accession numbers are listed in Table S5.
3.6 Phylogenetic analysis of Ensifer and Bradyrhizobium recA genes
A maximum likelihood phylogenetic tree was constructed using the recA gene sequences of ten Ensifer (DC-HR18, DC-HR20, DC-MP27, DC-MP29, DC-PB24, DC-PB26, DC-RJ2, DC-RJ8, DC-RJ12 and DC-RJ16) and ten Bradyrhizobium (DC-ML38, DC-ML39, DC-MP30, DC-MP31, DC-MP32, DC-PY35, DC-PY36, DC-PY37, DC-RJ11 and DC-RJ13) strains isolated from D. cinerea (Fig. 3). The recA phylogeny revealed five Ensifer recA-types comprising two lineages (EL-I and EL-II) and three clades (EC-I, EC-II and EC-III), the latter two clades (EC-I an EC-II) were placed in the ‘Ensifer aridi’ (preferred name and not validly published; Rocha et al. 2020) cluster. This phylogenetic clustering indicated a geographical pattern with strains from soils of a particular state generally grouping together, although a few RJ strains clustered with strains from HR. The strains from RJ, MP, and HR shared similarities with ‘E. aridi’ while two PB strains (DC-PB24 and DC-PB26) shared close similarities to E. kostiensis.
Maximum Likelihood phylogenetic tree of ten Ensifer and ten Bradyrhizobium strains isolated from host Dichrostachys cinerea with type strains reconstructed using recA gene sequences. Bootstrap values calculated for 1,000 replications and above 50% value indicated at internodes. The scale bar represents 5% nucleotide substitution per site. Abbreviations: B., Bradyrhizobium; E., Ensifer; BC, Bradyrhizobium clade; EC, Ensifer clade; BL, Bradyrhizobium lineage; EL, Ensifer lineage and superscripted T, Type strain. (GenBank accession numbers are in parenthesis)
Similarly, Bradyrhizobium strains isolated from different agroclimatic regions presented a clear geographical pattern in their grouping. The seven Bradyrhizobium recA-types comprised one clade (BC-I) and six lineages (BL-I to BL-VI) (Fig. 3). Strains belonging to the novel clade BC-I (DC-MP30, DC-MP31, DC-RJ11 and DC-RJ13) and lineage BL1 (DC-MP32) were isolated from arid and semi-arid regions of RJ and MP states, and all were closely related to B. yuanmingense in Bradyrhizobium Mega Clade-I. Two Bradyrhizobium strains (DC-PY35 and DC-PY36) that were isolated from the coastal region of PY were distinct from other D. cinerea strains. Strain DC-PY35 diversified from B. zhanjiangense whereas DC-PY36 clustered close to it. Another PY strain DC-PY37 occupied a position near B. ivorense within Bradyrhizobium Mega Clade-II. Two Bradyrhizobium strains, DC-ML38 and DC-ML39, both isolated from the wet subtropical climatic conditions and acidic soils of Shillong, ML, were divergent from B. elkanii and B. embrapense, respectively in Mega Clade-II.
3.7 Multi locus sequence analysis (MLSA) of Ensifer and Bradyrhizobium
In the four (rrs-glnII-atpD-recA) gene concatenated phylogeny (Fig. 4) of six selected Ensifer strains (DC-HR18, DC-MP27, DC-PB24, DC-RJ2, DC-RJ12 and DC-RJ16) five MLSA types (MLSA T-I to V) were formed, and congruence was observed in the phylogenetic positioning of the strains vis a vis the individual housekeeping gene based phylogenies. From the five MLSA types three appear to be novel and are potentially new species of Ensifer. Strains DC-RJ2 and DC-HR18 from (T-I) were identical to ‘E. aridi’ LMR001 (isolated from Vachellia gummifera, Morocco; Le Quéré et al. 2017; Rocha et al. 2020) whereas the strain DC-MP27 (T-II) formed a lineage close to a group of Indian ‘E. aridi’-type strains. Both in the individual and concatenated core gene phylogenies strain DC-PB24 (T-III) clustered close to E. kostiensis (isolated from Senegalia senegal, Sudan) and strains LL-HR123 and LL-PB121 isolated from L. leucocephala grown in HR and PB soils, respectively, by Chouhan et al. (2022). Strain DC-RJ12 (T-IV) shared close similarity with Ensifer sp. CPTN45 isolated from root nodules of Chamaecrista pumila grown in soils from Tamil Nadu, India (Rathi et al. 2018), while strain DC-RJ16 (T-V) formed a novel lineage and was divergent to Ensifer sp. LL-RJ68 (isolated from L. leucocephala, RJ, India).
Maximum Likelihood phylogenetic tree of six Ensifer strains isolated from host Dichrostachys cinerea with type strains and other Ensifer strains isolated from Indian soil reconstructed using concatenated rrs-glnII-atpD-recA gene sequences. Bootstrap values calculated for 1,000 replications and above 50% value indicated at internodes. The scale bar represents to 2% nucleotide substitution per site. Following abbreviations represents to: E., Ensifer; MLSA, multi locus sequence analysis; T, Type and superscripted T, Type strain
The maximum likelihood concatenated four (recA-glnII-atpD-rrs) gene phylogeny of six (DC-ML38, DC-ML39, DC-MP30, DC-PY35, DC-PY37 and DC-RJ13) Bradyrhizobium strains resembled the individual housekeeping gene phylogenies. However, the Bradyrhizobium MLSA types (T-Ia, T-Ib, T-II, T-III, T-IV and T-V) formed in the concatenated phylogenetic tree (Fig. 5) gave better taxonomic resolution revealing the apparent novelty of the strains and hinting at their potential to belong to new species. MLSA types T-Ia (DC-RJ13) and T-Ib (DC-MP30) formed a distinct novel clade with C. pumila symbionts from soils of RJ and with strain LL-MP86 from L. leucocephala (MP, India). One PY strain, DC-PY35, clustered in Mega Clade-I close to B. zhanjiangense while strain DC-PY37 clustered in Mega Clade-II close to B. ivorense. The Shillong Bradyrhizobium strains DC-ML38 and DC-ML39 formed novel lineages in Mega Clade-II and were closely related to B. embrapense and strain EHNEHU6 isolated from root nodules of Eriosema chinense (ML, India).
Maximum Likelihood phylogenetic tree of six Bradyrhizobium strains isolated from host Dichrostachys cinerea with type strains and other Bradyrhizobium strains isolated from India reconstructed using concatenated recA-glnII-atpD-rrs gene sequences. Bootstrap values calculated for 1,000 replications and above 50% value indicated at internodes. The scale bar represents to 2% nucleotide substitution per site. Following abbreviations represents to: B., Bradyrhizobium; MLSA, multi locus sequence analysis; T, Type and superscripted T, Type strain
3.8 Symbiosis essential gene (nodA and nifH) phylogenies of Ensifer and Bradyrhizobium
A phylogenetic analysis was conducted on the symbiosis essential genes nodA and nifH of ten Ensifer and ten Bradyrhizobium strains isolated from D. cinerea in India. The ten Ensifer strains were resolved into four nodA types (T-i to T-iv) (Fig. 6). Two RJ strains (DC-RJ2 and DC-RJ8) (nodA type T-ia and b) grouped with ‘E. aridi’-type strains isolated in India from root nodules of Tephrosia spp., C. pumila and L. leucocephala. One RJ (DC-RJ16) and two HR (DC-HR18 and DC-HR20) strains grouped to form the variant nodA type T-ii, while Ensifer strains from PB (DC-PB24 and DC-PB26) and MP (DC-MP27 and DC-MP29) clustered together with 100% similarity to Ensifer sp. CPG48 (C. pumila, Gujarat, India) in the nodA type T-iii. Three nodA types (T-i to T-iii) can be considered as mega clades of local sym genes which are harbored by a large group of local legumes suggesting these genes are more promiscuous and not specific to any particular host. Only a single strain, DC-RJ12 (nodA type T-iv), from RJ was positioned within the major clade of Indian mimosoid-nodA types closely related to E. arboris, while the remaining Ensifer strains harbored novel nodA types clustering in a mega clade positioned close to E. fredii. Strains such as DC-HR18, DC-HR20, DC-MP27, DC-MP29, DC-RJ2 and DC-RJ8 shared close similarity to ‘E. aridi’ in their nodA and recA gene phylogenies. In the nifH phylogeny (Fig. S3) ten Ensifer strains showed a similar pattern of phylogenetic divergence from type strains as observed in the nodA phylogeny. Strains from RJ (DC-RJ2, DC-RJ8 and DC-RJ16) and HR (DC-HR18 and DC-HR20) clustered into a single clade while strains of PB (DC-PB24 and DC-PB26) and MP (DC-MP27 and DC-MP29) separated into two locational sub-types in the nifH phylogeny (Fig. S3). Strain DC-RJ12 grouped close to Ensifer sp. LL-RJ7 (from L. leucocephala) and Ensifer strains isolated from V. nilotica in HR and Gujarat (Choudhary et al. 2020).
Maximum Likelihood phylogenetic tree of ten Ensifer strains isolated from host Dichrostachys cinerea with type strains and other Ensifer strains isolated from Indian soil reconstructed using nodA gene sequences. Bootstrap values calculated for 1,000 replications and above 50% value indicated at internodes. The scale bar represents to 10% nucleotide substitution per site. Following abbreviations represents to: A., Azorhizobium; E., Ensifer; T, Type and superscripted T, Type strain. (GenBank accession numbers are in parenthesis)
In the nodA phylogeny the ten selected Bradyrhizobium strains dispersed to form six nodA types (T-i to T-vi) (Fig. 7). Locational clustering was observed in the nodA phylogenetic diversity i.e. two RJ strains were closely related to B. yuanmingense and resolved into nodA type T-ia (DC-RJ11) and T-ib (DC-RJ13), while strains from PY (DC-PY35 and DC-PY36) and MP (DC-MP30, DC-MP31 and DC-MP32) resolved into nodA type T-ii and T-iii, respectively, forming novel clades divergent to B. agreste isolated from Glycine clandestina in Australia (Klepa et al. 2021). These two novel clades showed incongruence in their phylogenetic positions in housekeeping and sym gene phylogenies while the phylogenetic position of strain DC-PY37 (T-iv) showed congruence. Strain DC-ML38 (T-v) was divergent from B. elkanii and strain DC-ML39 (T-vi) clustered with previously reported strains from ML, India. The nifH phylogeny of the ten Bradyrhizobium strains was similar to that of nodA (Fig. S4). Ensifer and Bradyrhizobium strains from the present study harbored sym genes overlapping with other local symbionts reported from India.
Maximum Likelihood phylogenetic tree of ten Bradyrhizobium strains isolated from host Dichrostachys cinerea with type strains and other Bradyrhizobium strains isolated from Indian soil reconstructed using nodA gene sequences. Bootstrap values calculated for 1,000 replications and above 50% value indicated at internodes. The scale bar represents to 5% nucleotide substitution per site. Following abbreviations represents to: A., Azorhizobium; B., Bradyrhizobium; T, Type and superscripted T, Type strain. (GenBank accession numbers are in parenthesis)
3.9 Symbiotic efficacy of Ensifer and Bradyrhizobium
Strains belonging to different MLSA and nodA types of Ensifer (DC-HR18, DC-MP27, DC-PB24, DC-RJ2 and DC-RJ16) and Bradyrhizobium (DC-ML38, DC-ML39, DC-MP30, DC-PY35, DC-PY37 and DC-RJ13) were cross inoculated onto an important crop legume, Vigna radiata, to determine their symbiotic efficacy (Fig. 8a-c). All tested Ensifer and Bradyrhizobium strains except DC-PY37 nodulated V. radiata. Overall, the Ensifer strains formed more root nodules per plant than the Bradyrhizobium strains, with Ensifer strain DC-HR18 forming the highest number of nodules (Fig. 8a). Shoot fresh weight (Fig. 8b) and shoot dry weight (Fig. 8c) of plants inoculated with Ensifer and Bradyrhizobium strains was higher in comparison to un-inoculated N -minus control plants. A few plants inoculated with Ensifer strains (DC-HR18, DC-MP27, DC-RJ2 and DC-RJ16) and Bradyrhizobium (DC-PY35 and DC-RJ13) even performed better than the N + control. Symbiotically, the best-performing strain on V. radiata was Ensifer sp. DC-RJ16.
Average number of nodules (a), fresh weight of shoot (b) and dry weight of shoot (c) per plant of Vigna radiata inoculated with strains of Ensifer and Bradyrhizobium in comparison to un-inoculated N + and N- control plants. (nodA types are indicated at top of standard deviation bar for parameter fresh weight of shoot)
4 Discussion
4.1 Nodulation of Dichrostachys cinerea in different soils and selection of rhizobia
Gehlot et al. (2012) reported nodulation in the perennial shrub D. cinerea and Chouhan et al. (2020) later observed its nodulation with slow-growing Bradyrhizobium and fast-growing Ensifer strains in arid and semi-arid regions of Rajasthan, India. In the present study we have greatly expanded these aforementioned studies, and have also obtained information about the anatomy of D. cinerea nodules. Nodules formed on D. cinerea were anatomically and ultra-structurally similar to those reported previously on Mimosoid legumes in general (de Faria et al. 2022). This study thus reinforces the notion that Mimosoid legumes have evolved a particular nodule type that is highly conserved within this clade, but also different from all other legume nodule types, including those within their parent subfamily Caesalpinioideae (Sprent et al. 2017). Indeed, it is now considered likely that the high incidence of nodulation in the Mimosoid clade compared to the largely non-nodulated non-mimosoid Caesalpinioideae is due to their rejection of the cell wall-bound “fixation thread” type of bacteroid that is present in the latter, and their adoption of the more intimate and efficient membrane-bound symbiosome that is more redolent of “advanced” SYM-type nodules in the subfamily Papilionoideae (de Faria et al. 2022).
To identify the root nodule microsymbionts associated with D. cinerea, a wide sampling from the state of RJ was performed together with more limited sampling from other states and union territories of India with varied agroclimatic conditions. In addition to analysing the phylogenetic diversity of its microsymbionts, the symbiotic efficacy and the compatibility of wild rhizobia on the crop legume V. radiata was also investigated. The pH of soils collected from different sampling sites ranged from alkaline to neutral to acidic, but D. cinerea nodulated in all the soils used for the trap experiment. In contrary to this Mimosoids such as V. jacquemontii (Sankhla et al. 2017), V. nilotica (Choudhary et al. 2020) and L. leucocephala (Chouhan et al. 2022) failed to nodulate in the acidic soils of ML suggesting that in addition to edaphic and geographical factors (Pires et al. 2018; Rathi et al. 2018), host preferences also play key roles in establishing successful symbiosis. A total of 58 bacterial (both fast- and slow-growing) strains were isolated and purified from root nodules of D. cinerea. DNA fingerprinting of 36 selected bacteria showed considerable genetic diversity in the banding patterns with some location-specific genetic groups suggesting biogeographical factors are least partly responsible (Sprent et al. 2017). Dichrostachys cinerea nodulated with strains of both Ensifer and Bradyrhizobium in the alkaline (RJ, HR, PB, PY) and neutral (MP) soils; while in acidic soils (ML) only Bradyrhizobium microsymbionts were isolated. Such soil pH-based distribution of Ensifer and Bradyrhizobium are similar to previous reports that suggests Ensifer as a predominant root nodule microsymbiont in alkaline soils while Bradyrhizobium predominates in acidic soils (Gehlot et al. 2012; Tak et al. 2016; Sankhla et al. 2017; Ojha et al. 2017; Rathi et al. 2018; Choudhary et al. 2020; Jorrin et al. 2021; Chouhan et al. 2022). Phenotypic variation, such as distinctive carbon utilization and IAR patterns was also observed among the tested Bradyrhizobium and Ensifer strains, with a few strains that could tolerate high salt concentrations (up to 3%) and high temperature (up to 48 °C) as reported earlier (Zhang et al. 1991; Tak et al. 2016; Sankhla et al. 2015, 2017; Rathi et al. 2017; Choudhary et al. 2017, 2018, 2020; Gaur et al. 2018; Chouhan et al. 2020, 2022).
4.2 Phylogenetic diversity and mosaicism of core and sym genes in D. cinerea-nodulating Ensifer and Bradyrhizobium strains
In comparison to 16S rRNA, phylogenies based on the protein coding recA gene have better resolution (Tak et al. 2016; Sankhla et al. 2017; Rathi et al. 2018; Chouhan et al. 2022). Accordingly, 20 bacterial strains isolated from different sampling sites were identified as species of Ensifer and Bradyrhizobium based on their recA genes. Of the five MLSA types formed in concatenated gene phylogeny three are novel types divergent from the described type strains. Based on multiple core gene loci Ensifer strains DC-RJ2 and DC-HR18 shared similarity with ‘E. aridi’ (strain LMR001 isolated from V. gummifera in North Africa Le Quéré et al. 2017), but differed in sym loci (nodA and nifH) suggesting these strains have typically Old World core genomes, but have evolved in terms of plasmid-borne loci. The strains DC-MP27, DC-PB24, DC-RJ12 and DC-RJ16 showed typical incongruence in core and sym loci. Based on core and nodA genes majority of D. cinerea- Ensifer strains were closely related with the symbionts of C. pumila and L. leucocephala (Rathi et al. 2018; Chouhan et al. 2022). Diverse Ensifer strains have been identified from D. cinerea trapped in PB and HR soils. From these same sampling sites [Mansa (PB) and Sirsa (HR)] Ensifer strains were trapped in root nodules of V. nilotica (Choudhary et al. 2020) and L. leucocephala (Chouhan et al. 2022) but the Ensifer genotypes isolated from these three host plants are divergent from each other. Symbiotic preferences for Ensifer strains by members of the Caesalpinioideae (including the Mimosoids), and the Papilionoideae have evolved differently. Some mimosoids such as L. leucocephala (Chouhan et al. 2022) and D. cinerea (this study) are able to establish symbiosis both with Papilionoid-derived Ensifer and Mimosoid-derived Ensifer types (Tak et al. 2016; Sankhla et al. 2017; Rathi et al. 2018; Tak and Gehlot 2019; Choudhary et al. 2020; Chouhan et al. 2022). Ensifer strains from the non-Mimosoid Caesalpinioideae species C. pumila shared similarity only with Papilionoid-derived Ensifer and did not associate with Mimosoid-derived Ensifer strains (Rathi et al. 2018) highlighting the complexity of host factors involved in the legume-rhizobia symbiosis. It is intriguing question if legume host have evolved to pair with multiple types of microsymbionts or if the microsymbionts have evolved to infect multiple host plants and hence define promiscuity. As reported by multiple studies (Sankhla et al. 2017; Rathi et al. 2018; Choudhary et al. 2020; Chouhan et al. 2022) the present study on D. cinerea-Ensifer reinforces the notion that Ensifer is the dominant root nodule microsymbiont of native and invasive legumes in alkaline soils of India, and that the enormous genetic diversity of Ensifer has been created through horizontal gene transfer (HGT) resulting in several mosaic combinations of core and sym genes (Tak et al. 2016; Sankhla et al. 2017; Andrews et al. 2018; Chouhan et al. 2022).
The concatenated phylogeny of Bradyrhizobium provided clarity in the taxonomic positions of strains that were dispersed in Bradyrhizobium Mega Clade-I and II forming several novel MLSA types. Strains DC-RJ13 and DC-MP30 were divergent from B. yuanmingense based on their core genomes but shared similarity with strains reported from C. pumila and L. leucocephala (Rathi et al. 2018; Chouhan et al. 2022). Strains with such a genetic make-up are predominant in the alkaline soils of arid and semi-arid regions of Thar Desert (RJ) and other states of India, and even in Pakistan (Appunu et al. 2009; Gehlot et al. 2012; Choudhary et al. 2017; Rathi et al. 2017, 2018; Sankhla et al. 2018; Tak and Gehlot 2019; Chouhan et al. 2020, 2022; Jorrin et al. 2021; Hakim et al. 2021). The D. cinerea-nodulating Bradyrhizobium MLSA types Ia and Ib harbored completely divergent nodA genes whereby DC-RJ13 retained its position with other Bradyrhizobium strains reported from alkaline RJ soils, but strain DC-MP30 clustered with other MP strains to form a novel sym-type clade. This is the first incidence in which we have found B. yuanmingense-type strains showing incongruence in their core and sym gene phylogenies. Strains DC-PY35 and DC-PY37 were trapped by D. cinerea grown in PY soils collected from the rhizosphere of L. leucocephala. Both strains (DC-PY35 and DC-PY37) are symbionts that are apparently specific to D. cinerea on the basis of their core genes and nodA loci. Interestingly, the exotic legume L. leucocephala was nodulated only by strains of Ensifer in these same soils and did not select Bradyrhizobium (Chouhan et al. 2022). Moreover, at the pan-India level sampling/rhizobia trap experiments with L. leucocephala the Bradyrhizobium strains were trapped only from MP soils while in the case of D. cinerea Bradyrhizobium strains were trapped from sampling sites covering RJ, PY (alkaline), MP (neutral) and ML (acidic) soils. Therefore, the host genotype clearly plays a role in the selection of rhizobia from the same soil. Fields et al. (2023) in a recent study on clover reported the influence of host selection and local growth conditions on diversity and composition of Rhizobium within clover nodules. Strains DC-ML38 and DC-ML39 isolated from acidic soils of ML formed novel lineages in Mega Clade-II. Remarkably L. leucocephala failed to nodulate in acidic ML soils collected from NEHU Campus, Shillong (Chouhan et al. 2022) but D. cinerea effectively picked up Bradyrhizobium suggesting a more elaborated promiscuity of this invasive mimosoid. As reported in Rathi et al. (2018) our results also suggests that a significant diversity of Bradyrhizobium has evolved in the acidic soils of ML which has a large pool of novel Bradyrhizobium strains carrying a mosaic of novel combinations of core and sym genes.
4.3 Symbiotically efficient Ensifer and Bradyrhizobium harbor diverse sym genes
Ten Ensifer strains phylogenetically resolved into four nodA types of which three types (T-i, ii, iii) were identical or closely related to the Indian ‘E. aridi’-type of strains but were divergent to the African ‘E. aridi’ strain LMR001 (Tak et al. 2016; Rathi et al. 2018; Tak and Gehlot 2019; Rocha et al. 2020; Chouhan et al. 2022) while one type (T-iv) grouped within an exclusively Indian mimosoid-nodA clade. Incongruence was observed in the phylogenetic positioning of D. cinerea-Ensifer strains in core and nodA phylogenies with a few genetically diverse strains harboring a common nodA. Most Ensifer strains from Indian mimosoid legumes harbor novel nodA genes closely related to E. arboris and some harbor an E. kostiensis-type (Sankhla et al. 2017; Choudhary et al. 2017, 2018, 2020; Chouhan et al. 2022). Remarkably, the ‘E. aridi’nodA-type has not been previously encountered in Ensifer symbionts of native Mimosoid legumes, including species of Mimosa (Gehlot et al. 2013), Vachellia (Sankhla et al. 2017; Choudhary et al. 2017, 2020) and Senegalia (Choudhary et al. 2018), but Ensifer symbionts of the two exotic mimosoids (D. cinerea and L. leucocephala) and native Prosopis cineraria (Gehlot et al. 2016) have acquired this dominant and widely distributed nodA gene. The exotic legumes D. cinerea and L. leucocephala are nodulated by both (i) Ensifer with Mimosoid-nodA type and (ii) Ensifer strains that nodulate several Papilionoids (Alysicarpus vaginalis, Rhynchosia aurea, Crotalaria burhia and Tephrosia spp.) harboring novel Papilionoid-nodA genotypes (Rathi et al. 2017; Tak et al. 2016; Sankhla et al. 2018; Tak and Gehlot 2019). Bradyrhizobium strains from PY (DC-PY35 and DC-PY36) and MP (DC-MP30, DC-MP31 and DC-MP32) clustered to form novel clades divergent to B. agreste (isolated from Glycine clandestina, Australia) (Klepa et al. 2021). A similar clustering was observed in strains originating from acidic soils of ML (Ojha et al. 2017; Rathi et al. 2018). Strains DC-RJ11 and DC-RJ13 from alkaline soils of RJ were divergent to B. yuanmingense in both core and sym gene phylogenies, and shared close similarities with many strains reported from several Papilionoids (A. vaginalis, R. aurea, C. burhia and Tephrosia spp.), Caesalpinioideae-Mimosoids (V. leucophloea and L. leucocephala) and non-Mimosoid Caesalpinioideae (C. pumila) from different states (RJ, MP, Tamil Nadu, Uttarakhand) of India (Choudhary et al. 2017; Rathi et al. 2017, 2018; Sankhla et al. 2018; Tak and Gehlot 2019; Chouhan et al. 2022).
The symbiotic performance of Ensifer strains harboring different nodA types on the crop legume V. radiata was comparable as all nodulated and improved the biomass of inoculated plants in comparison to N-control plants, as did the Bradyrhizobium strains. Similar results have been reported by Tak et al. (2013, 2016), Gehlot et al. (2016), Le Quéré et al. (2017) and Rathi et al. (2018) whereby Ensifer strains harboring the ‘E. aridi’-type of nodA effectively nodulated V. radiata whereas Ensifer strains harboring the “standard” Indian mimosoid type of nodA fail to nodulate Vigna spp. as reported by Sankhla et al. (2017), Choudhary et al. (2017, 2018, 2020) and Chouhan et al. (2022), suggesting different host preferences based on the rhizobial sym type.
5 Conclusion
Dichrostachys cinerea is native to Africa, but in India it grows well and is nodulated in alkaline and acidic soils. Diverse strains of Ensifer and Bradyrhizobium occupy its root nodules in equal proportion depending on the local soil type. The MLSA of strains from D. cinerea is the first documentation of the phylogenetic diversity of its microsymbionts from India and elsewhere. The preferences in terms of selecting microsymbionts by D. cinerea is similar to that previously reported in India for native and exotic Caesalpinioideae, such as the invasive L. leucocephala (Chouhan et al. 2022) and the native C. pumila (Rathi et al. 2018) in that it is very promiscuous effectively nodulating with diverse strains of Ensifer (mainly Old World) and Bradyrhizobium (from both Mega clades). The symbiotaxonomy of Ensifer strains also differed as they cross-nodulated the crop V. radiata, which previously reported Indian mimosoid-nodulating Ensifer strains failed to achieve. Strains with such broad host range expanding from Mimosoids to legume crops are ideal candidates for inoculum preparation to improve soil fertility. Moreover, since D. cinerea nodulates and fix N in association with a broad diversity of rhizobia in alkaline soils and acidic soils of India, provided proper controls of its invasiveness are implemented, its cultivation can be advocated to increase soil fertility in both kinds of soil environment.
References
Andrews M, De Meyer S, James EK, Stępkowski T, Hodge S, Simon MF, Young JPW (2018) Horizontal transfer of symbiosis genes within and between rhizobial genera: occurrence and importance. Genes 9(7):321. https://doi.org/10.3390/genes9070321
Appunu C, N’zoue A, Moulin L, Depret G, Laguerre G (2009) Vigna mungo, V. radiata and V. unguiculata plants sampled in different agronomical-ecological-climatic regions of India are nodulated by Bradyrhizobium yuanmingense. Syst Appl Microbiol 32:460–470. https://doi.org/10.1016/j.syapm.2009.05.005
Bhandari MM (1990) Flora of the Indian desert. MPS Repros, Jodhpur, p 135
Cheng HR, Jiang N (2006) Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnol Lett 28(1):55–59. https://doi.org/10.1007/s10529-005-4688-z
Choudhary S, Meghwal RR, Sankhla IS, Tak N, Gehlot HS (2017) Molecular characterization and phylogeny of novel diverse nitrogen fixing microsymbionts associated with Vachellia (Acacia) leucophloea in arid and semi-arid regions of Rajasthan. Indian Forester 143(3):266–278. https://doi.org/10.36808/if/2017/v143i3/113663
Choudhary S, Tak N, Gehlot HS (2018) Phylogeny and genetic diversity assessment of Ensifer strains nodulating Senegalia (Acacia) senegal (L.) Britton. in arid regions of Western Rajasthan, India. Microbiology. 87(1):127–142. https://doi.org/10.1134/S0026261718010058
Choudhary S, Tak N, Bissa G, Chouhan B, Choudhary P, Sprent JI, James EK, Gehlot HS (2020) The widely distributed legume tree Vachellia (Acacia) nilotica subsp. indica is nodulated by genetically diverse Ensifer strains in India. Symbiosis 80(1):15–31. https://doi.org/10.1007/s13199-019-00658-8
Chouhan B, Tak N, Gehlot HS (2020) Phenotypic characterization and molecular identification of N2 fixing symbiotic rhizobia of Dichrostachys cinerea from arid and semi-arid soils of Rajasthan, India. Plant Archives. 20(2):5899–5906
Chouhan B, Tak N, Bissa G, Adhikari D, Barik SK, Sprent JI, James EK, Jha S, Gehlot HS (2022) Evolution of novel strains of Ensifer nodulating the invasive legume Leucaena leucocephala (Lam.) de Wit in different climatic regions of India through lateral gene transfer, FEMS Microbiol Ecol 98(9): fiac086. https://doi.org/10.1093/femsec/fiac086
Coates-Palgrave K (1988) Trees of Southern Africa. Struik publishers, Cape Town
de Faria SM, Ringelberg JJ, Gross E, Koenen EJM, Cardoso D, Ametsitsi GKD, Akomatey J, Maluk M, Tak N, Gehlot HS, Wright KM, Teaumroong N, Songwattana P, De Lima HC, Prin Y, Zartmann C, Sprent JI, Ardley J, Hughes CE, James EK (2022) The innovation of the symbiosome has enhanced the evolutionary stability of nitrogen fixation in legumes. New Phytol 235:2365–2377. https://doi.org/10.1111/nph.18321
Fields B, Moeskjaer S, Deakin WJ, Moffat EK, Roulund N, Andersen SU, Young JPW, Friman VP (2023) Rhizobium nodule diversity and composition are influenced by clover host selection and local growth conditions. Mol Ecol. https://doi.org/10.1111/mec.17028.10.1111/mec.17028
Gaur S, Tak N, Rathi S, Choudhary S, Gehlot HS (2018) Identification and molecular characterization of root nodule microsymbiont of Trigonella foenum-graecum L growing in different soils from Western Rajasthan, India. J Environ Biol 39:684–692. https://doi.org/10.22438/jeb/39/5/MRN-709
Gehlot HS, Panwar D, Tak N, Tak A, Sankhla IS, Poonar N, Parihar R, Shekhawat NS, Kumar M, Tiwari R, Ardley J, James EK, Sprent JI (2012) Nodulation of legumes from the Thar desert of India and molecular characterization of their rhizobia. Plant Soil 357(1):227–243. https://doi.org/10.1007/s11104-012-1143-5
Gehlot HS, Tak N, Kaushik M, Mitra S, Chen WM, Poweleit N, Panwar D, Poonar N, Parihar R, Tak A, Sankhla IS, Ojha A, Rao SR, Simon MF, dos Reis Junior FB, Perigolo N, Tripathi AK, Sprent JI, Young JPW, James EK, Gyaneshwar P (2013) An invasive Mimosa in India does not adopt the symbionts of its native relatives. Ann Bot 112(1):179–196. https://doi.org/10.1093/aob/mct112
Gehlot HS, Ardley J, Tak N, Tian R, Poonar N, Meghwal RR, Rathi S, Tiwari R, Adnawani W, Seshadri R, Reddy TBK, Pati A, Woyke T, Pillay M, Markowitz V, Baeshen MN, Baeshen NA, Ivanova N, Kyrpides N, Reeve W (2016) High-quality permanent draft genome sequence of Ensifer sp. PC2, isolated from a nitrogen-fixing root nodule of the legume tree (Khejri) native to the Thar Desert of India. Stand Genomic Sci 11:43. https://doi.org/10.1186/s40793-016-0157-7
Hakim S, Imran A, Mirza MS (2021) Phylogenetic diversity analysis reveals Bradyrhizobium yuanmingense and Ensifer aridi as major symbionts of mung bean (Vigna radiata L.) in Pakistan. Braz J Microbiol 52:311–324. https://doi.org/10.1007/s42770-020-00397-9
Heuzé V, Tran G, Giger-Reverdin S (2015) Sicklebush (Dichrostachys cinerea). Feedipedia, a programme by INRA, CIRAD, AFZ and FAO
Howieson JG, Dilworth MJ (2016) Working with rhizobia. Canberra: Australian centre for international agricultural research
Jorrin B, Maluk M, Atoliya N, Kumar SC, Chalasani D, Tkacz A, Singh P, Basu A, Pullabhotla SV, Kumar M, Mohanty SR (2021) Genomic diversity of pigeon pea (Cajanus cajan L Millsp.) endosymbionts in India and selection of potential strains for use as agricultural inoculants. Front Plant Sci 12:680981
Klepa MS, Ferraz Helene LC, O’Hara G, Hungria M (2021) Bradyrhizobium agreste sp nov, Bradyrhizobium glycinis sp. Nov. and Bradyrhizobium diversitatis sp. nov, isolated from a biodiversity hotspot of the genus Glycine in Western Australia. Int J Syst Evol Microbiol 71(3):004742. https://doi.org/10.1099/2Fijsem.0.004742
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Le Quéré A, Tak N, Gehlot HS, Lavire C, Meyer T, Chapulliot D, Rathi S, Sakrouhi I, Rocha G, Rohmer M, Severac D, Filali-Maltouf A, Munive JA (2017) Genomic characterization of Ensifer aridi, a proposed new species of nitrogen-fixing rhizobium recovered from Asian. African and American Deserts BMC Genom 18(1):1–24. https://doi.org/10.1186/s12864-016-3447-y
LPWG: Legume Phylogeny Working Group (2017) A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66(1):44–77. https://doi.org/10.12705/661.3
Moulin L, Klonowska A, Caroline B, Booth K, Vriezen JA, Melkonian R, James EK, Young JP, Bena G, Hauser L, Land M, Kyrpides N, Bruce D, Chain P, Copeland A, Pitluck S, Woyke T, Lizotte-Waniewski M, Bristow J, Riley M (2014) Complete Genome sequence of Burkholderia phymatum STM815T, a broad host range and efficient nitrogen-fixing symbiont of Mimosa species. Stand Genomic Sci 9(3):763–774. https://doi.org/10.4056/sigs.4861021
Nielsen MO, Reinoso-Pérez M, Sorensen M, Hansen HO, Gustafsson J (2013) Eco-friendly alternatives for control and use of invasive plants in agroforestry systems: The case of Marabú (Dichrostachys cinerea) in Cuba
Ojha A, Tak N, Rathi S, Chouhan B, Rao SR, Barik SK, Joshi SR, Sprent JS, James EK, Gehlot HS (2017) Molecular characterization of novel Bradyrhizobium strains nodulating Eriosema chinense and Flemingia vestita, important unexplored native legumes of the sub-Himalayan region (Meghalaya) of India. Syst Appl Microbiol 40(6):334–344. https://doi.org/10.1016/j.syapm.2017.06.003
Pant P, Pant P (2017) Ecological restoration techniques for management of degraded, mined-out areas and the role played by rhizospheric microbial communities. In: Singh R, Kumar S (eds) Green technologies and environmental sustainability. Springer, Cham, pp 437–453. https://doi.org/10.1007/978-3-319-50654-8_19
Pires RC, Reis Junior FB, Zilli JE, Fischer D, Hofmann A, James EK, Simon MF (2018) Soil characteristics determine the rhizobia in association with different species of Mimosa in central Brazil. Plant Soil 423(1–2):411–428. https://doi.org/10.1007/s11104-017-3521-5
Pueppke SG, Broughton WJ (1999) Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol Plant Microbe Interact 12(4):293–318. https://doi.org/10.1094/MPMI.1999.12.4.293
Pule-Meulenberg F, Dakora FD (2009) Assessing the symbiotic dependency of grain and tree legumes on N2 fixation for their N nutrition in five agro-ecological zones of Botswana. Symbiosis 48(1):68–77. https://doi.org/10.1007/BF03179986
Rathi S, Tak N, Bissa G, Chouhan B, Ojha A, Adhikari D, Barik SK, Satyawada RR, Sprent JI, James EK, Gehlot HS (2018) Selection of Bradyrhizobium or Ensifer symbionts by the native Indian caesalpinioid legume Chamaecrista pumila depends on soil pH and other edaphic and climatic factors. FEMS Microbiol Ecol 94(11):fiy180. https://doi.org/10.1093/femsec/fiy180
Rathi S, Gaur S, Tak N, Tak A, Gehlot HS (2017) Genetically diverse root nodule bacteria associated with Alysicarpus vaginalis from alkaline soil of Rajasthan, India. Plant Archives 17:495–505
Richardson AE, Viccars LA, Watson JM, Gibson AH (1995) Differentiation of Rhizobium strains using the polymerase chain reaction with random and directed primers. Soil Biol Biochem 27(4–5):515–524. https://doi.org/10.1016/0038-0717(95)98626-Y
Rocha G, Le Quéré A, Medina A, Cuéllar A, Contreras JL, Carreño R, Bustillos R, Muñoz-Rojas J, Villegas MDC, Chaintreuil C, Dreyfus B, Munive JA (2020) Diversity and phenotypic analyses of salt- and heat-tolerant wild bean Phaseolus filiformis rhizobia native of a sand beach in Baja California and description of Ensifer aridi sp. nov. Arch Microbiol 202(2):309–322. https://doi.org/10.1007/s00203-019-01744-7
Sáez SJM, Alfayate JAE (2020) Sicklebush (Dichrostachys cinerea) as a Medicinal Plant. J Anim Prod 32 (3)
Sankhla IS, Tak N, Meghwal RR, Choudhary S, Tak A, Rathi S, Sprent JI, James EK, Gehlot HS (2017) Molecular characterization of nitrogen fixing microsymbionts from root nodules of Vachellia (Acacia) jacquemontii, a native legume from the Thar Desert of India. Plant Soil 410(1–2):21–40. https://doi.org/10.1007/s11104-016-2838-9
Sankhla IS, Meghwal RR, Tak N, Tak A and Gehlot HS (2015) Phenotypic and molecular characterization of microsymbionts associated with Crotalaria medicagenia, a native legume of the Indian Thar Desert. Plant Archives 15:1003–1010
Sankhla IS, Meghwal RR, Choudhary S, Rathi S, Tak N, Tak A and Gehlot HS (2018) Molecular characterization of microsymbionts associated with root nodules of Crotalaria burhia Buch.-Ham. ex Benth., a native keystone legume species from Thar Desert of India. Indian J Exp Biol 56:373–385
Somasegaran P, Hoben HJ (1994) Handbook for rhizobia: Methods in Legume Rhizobium Technology. Springer, Verlag New York, USA
Sprent JI, Ardley J, James EK (2017) Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol 215(1):40–56. https://doi.org/10.1111/nph.14474
Sprent JI (2005) Nodulated legume trees. In: Werner D, Newton WE (eds) Nitrogen fixation in agriculture, forestry, ecology, and the environment. Nitrogen fixation: origins, applications and research progress. Springer, Dordrecht 4:113–141. https://doi.org/10.1007/1-4020-3544-6_7
Tak N, Gehlot HS, Kaushik M, Choudhary S, Tiwari R, Tian R, Hill Y, Bräu L, Goodwin L, Han J, Liolios K, Huntemann M, Palaniappan K, Pati A, Mavromatis K, Ivanova N, Markowitz V, Woyke T, Kyrpides N, Reeve W (2013) Genome sequence of Ensifer sp. TW10; a Tephrosia wallichii (Biyani) microsymbiont native to the Indian Thar Desert. Stand Genomic Sci 9:304–314. https://doi.org/10.4056/sigs.4598281
Tak N, Awasthi E, Bissa G, Meghwal RR, James EK, Sprent JS, Gehlot HS (2016) Multi locus sequence analysis and symbiotic characterization of novel Ensifer strains nodulating Tephrosia spp. in the Indian Thar Desert. Syst Appl Microbiol 39(8):534–545. https://doi.org/10.1016/j.syapm.2016.08.002
Tak N, Gehlot HS (2019) Diversity of nitrogen-fixing symbiotic rhizobia with special reference to Indian Thar Desert. In: Satyanarayana T, Das S, Johri B (eds.). Microbial diversity in ecosystem sustainability and biotechnological applications. Singapore Springer pp. 31–55. https://springerlink.bibliotecabuap.elogim.com/chapter/10.1007/978-981-13-8487-5_2
Tak N, Bissa G, Gehlot HS (2020) Methods for isolation and characterization of nitrogen-fixing legume-nodulating bacteria. In: Gupta KJ (ed) Nitrogen metabolism in plants: methods and protocols. Methods in molecular biology. 2057:119–143. https://doi.org/10.1007/978-1-4939-9790-9_12
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680. https://doi.org/10.1093/nar/22.22.4673
Trinick MJ (1980) Relationships amongst the fast-growing rhizobia of Lablab purpureus, Leucaena leucocephala, Mimosa spp., Acacia farnesiana and Sesbania grandiflora and their affinities with other rhizobial groups. J Appl Microbiol 49:39–53. https://doi.org/10.1111/j.1365-2672.1980.tb01042.x
Wakeling JL, Bond WJ (2007) Disturbance and the frequency of root suckering in an invasive savanna shrub, Dichrostachys cinerea. Afr J Range Forage Sci 24(2):73–76. https://doi.org/10.2989/AJRFS.2007.24.2.3.157
Yates RJ, Howieson JG, Nandasena KG, O’Hara GW (2004) Root-nodule bacteria from indigenous legumes in the north-west of Western Australia and their interaction with exotic legumes. Soil Biol Biochem 36(8):1319–1329. https://doi.org/10.1016/j.soilbio.2004.04.013
Zhang X, Harper R, Karsisto M, Lindström K (1991) Diversity of Rhizobium bacteria isolated from the root nodules of leguminous trees. Int J Syst Evol 41(1):104–113. https://doi.org/10.1099/00207713-41-1-104
Acknowledgements
Authors acknowledge UGC-NFSC, New Delhi, for financial assistance in the form of Junior and Senior Research Fellowship to BC. Authors also acknowledge Department of Biotechnology (DBT), New Delhi (BT/49/NE/2014; BT/PR24584/NER/95/762/2017); DST-FIST, New Delhi and University Grants Commission, New Delhi for the major equipment and instrumental facilities used in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
One sentence summary: The invasive mimosoid legume Dichrostachys cinerea is promiscuous and nodulates in alkaline and acidic soils of India.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chouhan, B., Tak, N., James, E.K. et al. The invasive mimosoid legume Dichrostachys cinerea (L.) Wight & Arn is nodulated by diverse strains of Ensifer and Bradyrhizobium in different agroclimatic regions of India. Symbiosis 92, 421–438 (2024). https://doi.org/10.1007/s13199-024-00983-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-024-00983-7