Abstract
Background and aims
To evaluate the influence of soil type on the symbiosis between Mimosa spp. and rhizobia.
Methods
A greenhouse experiment was carried out with trap plants using seeds of six species of Mimosa and soils from three different locations in central Brazil: Posse, Brasília and Cavalcante. Plant dry biomass and number of nodules were measured after four months. Symbiotic bacteria were isolated from nodules and their molecular identification was performed. Three housekeeping genes (16S rRNA, recA and gyrB) plus the nodC and nifH symbiotic genes were used to determine the identity of the symbionts and to reconstruct the phylogenetic relationships among the isolated nitrogen-fixing bacteria.
Results
Rhizobia from the Betaproteobacterial genus Paraburkholderia (former Burkholderia) and the Alphaproteobacterial genus Rhizobium were isolated from different species of Mimosa. As in previous studies, the phylogenies of their symbiosis-essential genes, nodC and nifH, were broadly congruent with their core housekeeping genes (16S rRNA, recA and gyrB), which suggests limited or no horizontal gene transfer. Edaphic factors such as pH and fertility influenced the occurrence of these unrelated rhizobial types in the nodules on these Mimosa spp.
Conclusions
Mimosa species have the ability to associate with different types of rhizobia (α- and β-proteobacteria), suggesting low specificity between host and bacterium in experimental conditions. Soil factors such as pH, nitrogen and fertility seem to favour the predominance of certain types of rhizobia, thus influencing the establishment of symbiotic relationships.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leguminous plants are important for natural and agricultural ecosystems because of their ability to fix atmospheric N2 in nodules formed on their roots via symbioses with diazotrophic bacteria (rhizobia). In this association, which is critical for plant nutrition and global N cycling, rhizobia provide nitrogen in exchange for carbon compounds supplied by the plant (Sprent 2009; Robledo et al. 2010).
All bacteria currently known to nodulate leguminous plants belong to the group of gram-negative proteobacteria. Until the beginning of the XXI century, bacteria of the order Rhizobiales, within the class α-proteobacteria, were considered the only ones that nodulated Leguminosae. However, since 2001, several reports have demonstrated that members of the β-proteobacteria class also nodulate and fix nitrogen in association with these plants (Chen et al. 2001; Vandamme et al. 2002; Chen et al. 2003; Elliott et al. 2007a, b, 2009; Bournaud et al. 2013; Liu et al. 2014; Lemaire et al. 2015). In particular, studies carried out with the pantropical Mimosa genus have been used to demonstrate that bacteria from the β-proteobacteria play a key role in nitrogen fixation in association with leguminous plants (Chen et al. 2005a, b; Elliott et al. 2009; Gyaneshwar et al. 2011; Bontemps et al. 2010; Reis Junior et al. 2010; Lammel et al. 2013; Platero et al. 2016). Indeed, Mimosa is now used as a model for studies involving these symbiotic relationships in natural ecosystems.
Previous studies have shown that bacteria from the genus Paraburkholderia (former Burkholderia) are the main symbionts of Mimosa species in the central region of Brazil (Chen et al. 2005a; Elliott et al. 2007a; Bontemps et al. 2010; Reis Junior et al. 2010), French Guiana, Costa Rica, and in areas where such plants were introduced such as Asia and Australia (Chen et al. 2005b; Barrett and Parker 2006; Parker et al. 2007; Liu et al. 2012; Mishra et al. 2012; Gehlot et al. 2013; Melkonian et al. 2014). A division of the genus Burkholderia was recently proposed, involving the creation of a novel genus Paraburkholderia containing the primarily environmental and plant-associated species which broadly corresponds to the “Plant Beneficial and Environmental (PBE)” clade of Suárez-Moreno et al. (2012), and which separates them from Burkholderia sensu stricto, which encompasses some environmental strains, but is particularly known for its human clinical and phytopathogenic species (Sawana et al. 2014; Beukes et al. 2017). All known nodulating Burkholderia species are currently placed in the genus Paraburkholderia, the exception being B. symbiotica which is most likely to be placed in a new genus along with some other members of the “Transition Group I” defined by Estrada-de los Santos et al. (2016), such as B. caryophylli, B. soli and B. rhizoxinica.
In Uruguay, the main symbionts associated with Mimosa belong to the genus Cupriavidus (Platero et al. 2016), which is related to Paraburkholderia in the β-proteobacteria class. In Mexico, an important center of Mimosa diversity, species of this genus are predominantly associated with α-proteobacteria from the genera Rhizobium and Ensifer (Bontemps et al. 2016). In India, the genus Ensifer is also predominant in two endemic species of Mimosa (Gehlot et al. 2013).
The preference of Mimosa for association with some distinct groups of rhizobia in different regions may be related to soil characteristics, such as pH and fertility, which influence symbiont selection (Elliott et al. 2009; Garau et al. 2009; Thrall et al. 2011; Liu et al. 2012, 2014; Mishra et al. 2012; Lammel et al. 2015; Stopnisek et al. 2014; Lemaire et al. 2015). For example, sites with high levels of heavy metals (zinc, copper and nickel) in soil tend to favour symbiosis with Cupriavidus, as verified in Uruguay (Platero et al. 2016) and New Caledonia (Klonowska et al. 2012). In central Brazil, where soils are generally acidic, Paraburkholderia spp. are the main Mimosa symbionts (Bontemps et al. 2010; Reis Junior et al. 2010). On the other hand, the predominance of α-proteobacteria in Mimosa species endemic to Mexico may be related to the presence of more fertile and pH neutral soils (Bontemps et al. 2016).
Symbiotic interactions generally depend on the expression of lipo-chitoligosaccharides (LCOs) or “Nod factors” produced by bacteria in response to plant secreted flavonoids, proteins and polysaccharides from the root surface. Any changes in the structures of these Nod factors can alter the specificity between plants and bacteria (Wang et al. 2012). In addition, this specificity varies among legumes, as some species are associated only with bacteria from one group (α- or β-proteobacteria), or with a few species of rhizobia, while others, more promiscuous, are associated with several types of rhizobia (Peix et al. 2015, Dall'Agnol et al. 2016). Factors that can influence the choice of symbionts by a plant species include the composition and diversity of the microbial community, soil characteristics, such as pH, salinity, nitrogen and nutrient levels, and altitude (Thrall et al. 2011; Lemaire et al. 2015, 2016).
In the case of Mimosa, it is also possible that the plants of each country or continent may have coevolved with their respective symbionts over millions of years resulting in increased specificity. For example, after the ancestors of the main lineages of Mimosa from Brazil and Mexico diverged, their descendants may have coevolved with the rhizobia of the local rhizosphere, Paraburkholderia in the case of the Cerrado and Caatinga biomes in Brazil, and Rhizobium and Ensifer in the central highlands of Mexico (Bontemps et al. 2010; Moulin et al. 2015; Bontemps et al. 2016; Sprent et al. 2017), which may have resulted in high rhizobia host-specificity. This might be particularly relevant for endemic plant species, which are restricted to very particular sites and narrow environmental conditions, and might therefore tend to restrict their symbiotic ability to a very limited range of rhizobia.
Despite the growing knowledge on the symbionts associated with leguminous plants in several ecosystems, the rhizobia host-specificity in natural environments and how different types of soil can affect this interaction are still poorly understood. In this study, we conducted an experiment to compare the nodulation of six native Brazilian species of the genus Mimosa in three soil types from central Brazil with distinct physicochemical characteristics. We hypothesized that endemic host legume species are expected to exhibit greater specificity in relation to their association with rhizobia, while widely distributed species may associate with a wide variety of symbionts. Regarding soil characteristics, according to recent literature (e.g. Elliott et al. 2009; Bontemps et al. 2016), it is expected that more fertile soils with high pH would favour association with α-proteobacteria, whereas more acidic and nutrient poor soils would result in nodulation with β-proteobacteria.
The aims of the present study were to verify the influence of soil type on nodulation and the composition of associated rhizobial species. Three housekeeping genes (16S rRNA, recA and gyrB) plus the nodC and nifH symbiosis-essential genes were used to determine the identity of the symbionts and to reconstruct the phylogenetic relationships among the isolated nitrogen-fixing bacteria.
Materials and methods
Collecting sites and plant species
In order to perform the nodulation tests, seeds of Mimosa species and soil samples containing their associated rhizosphere from three different sites in central Brazil with contrasting edaphic and vegetation characteristics, were used (Table 1). Soil samples from each locality were collected (0 to 20 cm depth) and analysed according to the protocols of Embrapa (1997).
A deciduous seasonal forest predominates in the collection site at Posse, with soil derived from limestone with high fertility and pH tending to neutral. The Brasília site is a typical woody savanna (cerrado) in a deep clay Oxisol, with low fertility and low pH. The Cavalcante site comprises an open savanna shrubby vegetation (cerrado rupestre) on sandy-rocky soil with low fertility and low pH (Table 1). Soil analysis shows high cation exchange capacity with elevated levels of Ca, Mg, K, P, nitrogen and organic matter in Posse, whereas the Brasília and Cavalcante sites have lower levels of cations, nitrogen and organic matter, but high concentrations of Al (Table 2).
Mature seeds of two Mimosa species per site were collected in the field. Plant species used in the study (Table S1) vary according to the extent of their geographic distribution: M. xanthocentra is a ruderal species of wide distribution in South America; M. acutistipula occurs in the Cerrado and Caatinga regions in Brazil often associated with fertile soils; M. claussenii and M. radula are widely distributed within the Cerrado region; M. kalunga and the undescribed Mimosa sp. are endemic to the region of Cavalcante in northern Goiás (Barneby 1991; Simon et al. 2010). The identity of the so far unidentified Mimosa sp. could not be determined as it might represent a new taxon. These species were chosen because of their abundance in each locality. They represent different lineages distributed throughout the phylogeny of the genus, with M. claussenii, M. kalunga and the Mimosa sp. belonging to the same clade (Simon et al. 2011).
Experiments with trap plants
Experiments with trap plants (Bontemps et al. 2016; Mishra et al. 2012) were performed to evaluate nodulation and growth of the Mimosa species cultivated in three different soil types, including the original soil where the seeds were collected. Seeds of six species of Mimosa were used (Table S1): two from Posse (M. acutistipula and M. xanthocentra), two from Brasília (M. claussenii and M. radula) and two from Cavalcante (M. kalunga and Mimosa sp.). Seeds were immersed in 70% alcohol for 30 s and sodium hypochlorite solution (2.5% active chlorine) for five minutes for surface sterilization. Subsequently, seeds were placed in a 23 mm mesh sieve to be washed five times with sterile distilled water. The dormancy was broken by scratching the testa of the seeds under sterile conditions. The seeds were then placed in Petri dishes with moistened filter paper, and after ten days, seedlings were transferred to 300 ml pots filled with soil (no fertilizer added), and cultivated for four months. Seedlings cultivated in sterilized sand were used as a negative control. Nodules from four to seven plants of each species were used for isolating rhizobia.
For comparison of plant growth (dry biomass) and nodulation among different types of soil an analysis of variance (ANOVA) was conducted and significant differences between means were assessed by Duncan’s test at the 5% level of significance using the MSTAT-C software (Michigan State University). Means and standard errors were calculated from values of three replicates of each Mimosa spp. in each soil.
Rhizobia isolation
We randomly harvested one to six nodules per treatment (species/soil type), depending on their availability. Collected nodules were rehydrated in sterile distilled water for 2.5 h. Superficial sterilization of nodules was performed by soaking them in ethanol (95%) for 30 s, followed by immersion in 2.5% sodium hypochlorite solution for five minutes and then five washes in sterile water. After this procedure, 500 μl of sterile saline solution was added to 2 ml Eppendorf tubes and the nodules were crushed in this solution with sterile forceps. The resulting solution was serially diluted to 10−3 in sterile saline solution. Thereafter, 100 μl of the last two dilutions (10−2 and 10−3) were plated on Medium 79 of Fred and Waskman (1928), otherwise known as YMA medium, with Congo red (Vincent 1970), using two replicates for each dilution. The plates were incubated for two to seven days at 30 °C and, after this, individual colonies were collected to obtain a pure culture of each bacterial isolate. Some nodules did not allow for the isolation of potentially symbiotic bacteria.
DNA extraction, amplification and sequencing
Bacteria isolated from nodules formed during the experiment with trap plants were cultured in YMA solid medium with Congo red for 72 h at 28 °C. Thereafter, a purified colony was transferred into YMA liquid medium for 24 h at 28 °C. Subsequently, bacterial DNA was extracted using Pure Link Genomic DNA Kits (Invitrogen), following manufacturer’s instructions.
The extracted DNA from each isolate was used as template for PCR reactions and the sequencing of five genes: 16S rRNA, recA, gyrB, nodC and nifH, which are widely used in phylogenetic studies with symbiotic bacteria (e.g. Peix et al. 2015). The PCR products were generated using Dr. Max DNA polymerase (MGMED, Co.) and sequenced in both directions (Macrogen, Korea). Primers used in gene amplification are listed in Table S2. The sequences obtained (GenBank accession numbers MG182874-MG183123) were compared to sequences deposited in the GenBank database (Table S3).
Phylogenetic analyses
To characterize the isolates at taxonomic level, representative sequences of several bacterial strains were obtained from GenBank and aligned with the sequences generated in this work using ClustalW, imported into BioEdit 4.8.4 (Hall 1999) and manually corrected. Phylogenetic analysis based on the 16S rRNA, recA, gyrB, nodC and nifH genes was performed following a maximum likelihood analysis (ML) implemented by the RAxML-HPC v.8 program using the GTR-CAT nucleotide substitution model (Stamatakis 2006) from the CIPRES portal (Miller et al. 2010). To obtain support values, data sets were retested a thousand times using the bootstrap method (Felsenstein 1985). Individual phylogenetic trees were constructed with the sequences aligned for each of the previously cited genomic DNA regions, inferring genetic distance and similarity among the studied bacteria. Sequences from reference strains closest to the genera Paraburkholderia, Cupriavidus, Burkholderia, Rhizobium, Bradyrhizobium, Ensifer and Mesorhizobium were included in the phylograms of the five genes. Whenever possible, sequences from the same reference species were used for all five genes. In addition, a phylogeny based on concatenated sequences of 16S rDNA, recA and gyrB was generated.
Evaluation of nodulation capacity
Confirmation of nodulation capacity was verified by means of an authentication experiment. The species M. pudica L. was chosen as a “model host” because it is a fast-growing species and has an ability to nodulate with a wide range of rhizobia, mainly β-proteobacteria (Chen et al. 2005a; Bontemps et al. 2010; Mishra et al. 2012), but also some Rhizobium species (Elliott et al. 2009; Baraúna et al. 2016). Seeds were sterilized and their dormancy was broken as cited before. Subsequently these seeds were pre-germinated on a cotton tray soaked with sterile dH2O and incubated for 48 h at 27 °C.
Seedlings were tested in a perlite substrate according to Elliott et al. (2009). Glass tubes (50 ml volume) were half-filled with perlite and then autoclaved at 121 °C and 1 atm for 30 min. Sterilized Hoagland nutrient solution (Hoagland and Arnon 1938) without N was applied to the perlite until the saturation point. The seedlings were inoculated with one ml culture of the isolates grown in YMA liquid medium for 48 h. The possibility of cross-contamination was investigated using an uninoculated negative control randomly inserted among treatments. All plants remained in a growth chamber with controlled temperature (26 °C) and 16 h light / 8 h dark cycle. After one month the plants were collected and the presence / absence of nodules was recorded. Because the Rhizobium isolates did not nodulate effectively with M. pudica, we also used Siratro (Macroptilium atropurpureum Moc. & Sessé ex DC.) as an additional host, since this is a promiscuous papilionoid species with high capacity for nodulating with α-proteobacteria (Elliott et al. 2007b; Mishra et al. 2012). Comparisons between the vigour of inoculated and control plants, as well as observation of effective nodules, were used as qualitative evidence for the symbiotic capacity of rhizobial isolates.
Results
Plant biomass and number of nodules
All Mimosa species grew in their original and in the other tested soils (Fig. 1). M. acutistipula and M. xanthocentra, from Posse, produced higher biomass in their soil of origin compared to the biomass obtained in other soils (Table 3). Mimosa kalunga and Mimosa sp. from Cavalcante, and M. claussenii and M. radula from Brasília, also produced slightly more biomass in the soil from Posse, the most fertile of all soils, but these differences, with the exception of M. kalunga, were not statistically significant (Table 3).
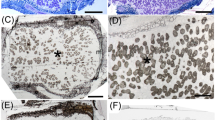
Plants of six Mimosa species (a M. acutistipula and detail of nodules, b M. xanthocentra, c M. radula, d Mimosa sp., e M. kalunga, f M. claussenii) used in the trap experiment with soils from three different sites in central Brazil (P: Posse, B: Brasília, C: Cavalcante) (S: sand - negative control). Scale bar = 5 cm
Most Mimosa species were able to nodulate in the three soil types, the exceptions being M. claussenii and M. xanthocentra, which did not nodulate in soils from Posse and Cavalcante, respectively. The species from Posse, M. acutistipula and M. xanthocentra, showed higher number of nodules in their original soil than the other species growing with this same substrate. These two species also had higher nodulation in their original soil when compared to their nodulation in soils from Brasília and Cavalcante. The other species, with the exception of M. claussenii, did not have a notable nodulation difference when the three soils were compared (Table 3). There was no nodulation on plants grown in the control with sterilized sand.
Identification of rhizobia using sequences of the housekeeping genes 16S rRNA, recA and gyrB
The 54 isolates obtained in this study were identified at least to genus level by comparing their 16S rRNA and recA gene sequences with those in the GenBank database using the BLASTN algorithm (Altschul et al. 1990). The BLASTN results classified them in the α- and β-classes of Proteobacteria and showed that they shared a high similarity (97% - 100%) with already known species of bacteria (data not shown). Nine isolates were assigned to the genus Rhizobium, while the other 45 isolates belonged to the genus Paraburkholderia. All bacteria isolated from nodules of plants growing on soils from Brasília and Cavalcante were identified as Paraburkholderia spp. On the other hand, all isolates from Posse grouped within Rhizobium, except isolate POS_MSP2 that grouped with Paraburkholderia (Table 4).
Individual phylogenies based on housekeeping genes (16S rRNA, recA and gyrB) were highly congruent, although the 16S rRNA tree was less resolved (Figs. S1-S3). A phylogeny based on the concatenation of these three genes (Fig. 2) separated the isolates between the α- and β- proteobacteria, and confirmed that all tested Mimosa species associated with bacteria from both classes, except for M. claussenii which did not nodulate with Rhizobium.
Maximum-likelihood tree based on concatenated 16S rRNA, recA and gyrB gene sequences (2571 bp) showing the phylogenetic relationship between Mimosa nodulating rhizobia isolates (this work) and reference strains (GenBank). Support values (1000 bootstrap replicates >50%) are shown. Scale bar in number of substitutions per site. Isolates code: Sites/Soils: POS = Posse (blue); BSB = Brasília (red); CAV = Cavalcante (green). Host plant: MAC = M. acutistipula; MCL = M. claussenii; MKA = M. kalunga; MRA = M. radula; MSP = Mimosa sp.; MXA = M. xanthocentra
The α-proteobacteria isolates grouped in a clade containing three Rhizobium strains from the study of Bontemps et al. (2016) and with R. altiplani, a newly-described species of Rhizobium associated with M. pudica in central Brazil (Baraúna et al. 2016) (Fig. 2). Isolates identified as β-proteobacteria grouped in Paraburkholderia, forming two distinct clades: one of them containing P. nodosa, and the other with P. tuberum and P. sprentiae. The first clade contains mostly isolates from Brasília and the second from Cavalcante (Figs. 2, S1-S3).
Phylogenetic analysis of the symbiotic nodC and nifH genes
Sequences of the 54 isolates were obtained for nodC, but nifH sequences were obtained for only 46 of them due to problems with PCR amplification. The tree topologies based on these two genes involved in nodulation (nodC) and nitrogen fixation (nifH) were similar to that of the other housekeeping genes, with the isolates also distributed within their respective α- and β-rhizobial groups (Figs. 3 and 4).
Maximum-likelihood tree based on the nodC gene sequences (610 bp) showing the phylogenetic relationship between Mimosa nodulating rhizobia isolates (this work) and reference strains (GenBank). Support values (1000 bootstrap replicates >50%) are shown. Scale bar in number of substitutions per site. Isolates code: Sites/Soils: POS = Posse (blue); BSB = Brasília (red); CAV = Cavalcante (green). Host plant: MAC = M. acutistipula; MCL = M. claussenii; MKA = M. kalunga; MRA = M. radula; MSP = Mimosa sp.; MXA = M. xanthocentra
Maximum-likelihood tree based on the nifH gene sequences (300 bp) showing the phylogenetic relationship between Mimosa nodulating rhizobia isolates (this work) and reference strains (GenBank). Support values (1000 bootstrap replicates >50%) are shown. Scale bar in number of substitutions per site. Isolates code: Sites/Soils: POS = Posse (blue); BSB = Brasília (red); CAV = Cavalcante (green). Host plant: MAC = M. acutistipula; MCL = M. claussenii; MKA = M. kalunga; MRA = M. radula; MSP = Mimosa sp.; MXA = M. xanthocentra
In the nodC and nifH phylogenies the isolates that grouped with α-proteobacteria were contained in a clade closest to Rhizobium sp. strain JPY479 which was isolated from M. xanthocentra by Bontemps et al. (2010). In general, the isolates identified as β-proteobacteria were grouped into two nodC clades, one closest to P. tuberum sv. mimosae and another to P. nodosa. For nifH two clades were also formed, one closest to P. mimosarum and P. nodosa and the other with strain JPY306 (Bontemps et al. 2010), which was subsequently placed in P. tuberum sv. mimosae by Mishra et al. (2012) along with several other strains from central Brazil.
Evaluation of nodulation capacity
The nodulation ability of all rhizobia isolated in the trap experiment was tested in M. pudica, a species known to be promiscuous. In this experiment all rhizobia confirmed their nodulation capacity, but the nine Rhizobium isolates (all obtained from Posse soil) formed small nodules with white coloration in their interior indicating ineffective nodulation. On the other hand, five of these Rhizobium isolates were tested on the alternative host Siratro and showed effective nodulation of it (Fig. S4). The 20 isolates from Cavalcante and the 24 isolates from Brasília soil, all belonging to the genus Paraburkholderia, resulted in effective nodulation in M. pudica, with nodules showing pinkish-red coloration in their interior, a sign of the presence of the symbiosis-essential protein leghemoglobin (Table 4). The strains of Paraburkholderia that nodulated M. pudica and those of Rhizobium that nodulated Siratro resulted in healthier (green leaves) and more vigorous plants compared to non-inoculated controls (which had no nodules and reduced growth; Fig. S4); taken together, these data provide compelling qualitative evidence for the symbiotic capacity of most of the isolates from this study.
Discussion
We observed a clear difference in the development of some species of Mimosa grown in different types of soil from central Brazil. Plants growing in the soil from Posse, which was the most fertile, tended to produce more biomass, which was statistically significant for the local species M. acutistipula and M. xanthocentra, as well as M. kalunga (Cavalcante endemic). Neither M. acutistipula nor M. xanthocentra performed well in the lower fertility Brasília and Cavalcante soils, even in the presence of potential symbiotic associations, suggesting that these plants are not well adapted to poor soils. On the other hand, most species native to Brasília and Cavalcante did not perform better when grown in richer soil (Posse), suggesting that they are better adapted to low fertility soils. Regarding the number of nodules, the mimosas did not show differences related to the soil types, except for the species from Posse, M. acutistipula and M. xanthocentra, which produced more nodules when growing in their original soil. Most likely, this large number of nodules found in an environment with high N and organic matter content may be related to the adaptation of both symbiotic partners (plant and rhizobia) to this environment. There are several studies supporting the concept that biological nitrogen fixation (BNF) is often (but not invariably) more active where the N supply is low (Vitousek et al. 2013). On the other hand, even in environments considered rich in N, BNF may be important to compensate for losses of this nutrient, thus avoiding its depletion in these soils (Pons et al. 2007).
The analysis of the 16S rRNA, recA and gyrB genes demonstrated that α- and β-rhizobia were found in this study. Overall, the rhizobia found here are in accordance with the high incidence of both α- and β-proteobacteria in the soil microbiota as detected in metagenomics studies carried out in different sites across the Cerrado region (Quirino et al. 2009; Araújo et al. 2012; Castro et al. 2016).
Rhizobium strains that predominated in Posse were recovered in our phylogenetic analyses as closely related to R. altiplani and to other Rhizobium strains isolated from Mimosa species. R. altiplani was recorded associated with M. pudica growing in an anthropogenic neutral to alkaline soil (pH 7.7) in the Distrito Federal in central Brazil (Baraúna et al. 2016). Isolates obtained from nodules of Mimosa spp. growing in relatively fertile soils with neutral-alkaline pH in Mexico (Bontemps et al. 2016) also grouped within the clade that contains our Rhizobium isolates and R. altiplani (Fig. 2). These observations reinforce the preference of Rhizobium isolates for less acidic and more fertile soils.
The isolates of Paraburkholderia obtained in our experiment were closest to P. nodosa, P. tuberum, and P. sprentiae, and hence were similar to rhizobia isolated from other studies that sampled nodules from Mimosa in different countries, including many localities in Brazil (Chen et al. 2005a, 2006, 2007). For example, several strains of P. nodosa and P. tuberum-like bacteria, together with P. mimosarum, P. diazotrophica, and other unidentified Paraburkholderia isolates were obtained from different sites in the Cerrado and Caatinga biomes in central and north-eastern Brazil (Bontemps et al. 2010) where acidic soils with low amounts of available nutrients occur, such as those found in the Brasília and Cavalcante sites in the present study.
Paraburkholderia nodosa has M. bimucronata and M. scabrella, from south Brazil as its original hosts (Chen et al. 2005a, 2007; Lammel et al. 2013), but it is also capable of nodulating with many different Mimosa species (Bontemps et al. 2010), and is very widespread in South America. Indeed, P. nodosa was also the main species found in nodules of common bean (Phaseolus vulgaris) when it was used as trap plants, showing its ability to associate with other legumes in Cerrado soils (Dall'Agnol et al. 2016).
Along with P. nodosa, bacteria related to P. tuberum are among the most commonly found β-rhizobia associated with different Mimosa species in Brazil and elsewhere in South America and Mexico (Bontemps et al. 2010, 2016; Mishra et al. 2012; Lammel et al. 2013). Closely related bacteria belonging to P. tuberum sensu stricto were originally isolated from the papilionoid legume Aspalathus carnosa L. in South Africa (Vandamme et al. 2002), and the type strain of this species (STM678T) is capable of nodulating many native South African papilionoid legumes (Elliott et al. 2007b; Beukes et al. 2013; Lemaire et al. 2015, 2016). Indeed, it has been proposed that P. tuberum has two symbiovars (sv. mimosae and sv. papilionoideae) in terms of host, geographical distribution and nod gene phylogeny. The sv. papilionoideae strains from South Africa cannot nodulate with Mimosa (Elliott et al. 2007b; Mishra et al. 2012; Estrada-de los Santos et al. 2016; Lemaire et al. 2016; de Meyer et al. 2016).
Paraburkholderia sprentiae was originally isolated from root nodules of Lebeckia ambigua E. Mey. growing in the Western Cape of South Africa; it is most closely related to P. tuberum (De Meyer et al. 2013), and it has not previously been isolated from any legume outside South Africa. It should be noted, however, that the large species complex represented by P. tuberum and P. sprentiae is currently being revised (Venter, Steenkamp, James & de Meyer, unpublished), and it is likely that the Neotropical “B. tuberum - B. sprentiae” will be allocated to a new and separate species which preferentially nodulates Mimosa and related mimosoids.
The nodC gene was sequenced for all isolates, indicating their capacity for nodulation. Probably owing to PCR amplification problems, we were unable to obtain amplicons of the nifH gene for some of the isolates. This does not imply that such isolates are ineffective symbionts that are not able to fix N2, since unsuccessful PCR reactions may be attributed to different causes, such as primer mismatch or poor DNA quality (Howieson et al. 2013).
The strains that grouped with α-proteobacteria in the nodC and nifH phylogenies were closely related to Rhizobium sp. strain JPY479. This strain was originally isolated from nodules of M. xanthocentra in Mato Grosso – Brazil, and it is one of only two α-proteobacterial strains isolated from Mimosa spp. in the study of Bontemps et al. (2010). According to Bontemps et al. (2010) the nifH and nodC genes of these strains were related to previously described α-proteobacterial symbionts of Mimosa isolated elsewhere; they are close to R. tropici and group with Rhizobium strains commonly isolated from Mexican Mimosa species (Bontemps et al. 2016).
The authentication experiment confirmed that all isolates tested were able to nodulate with M. pudica. Although both Paraburkholderia and Rhizobium induced the formation of nodules in this host, the nodules formed by Rhizobium were small and did not appear to be effective. Mimosa pudica is known to be a trap plant suitable for β-rhizobia, but the same is not generally true for α-rhizobium, and this may have influenced the effectiveness of nodulation on M. pudica by the α-rhizobial strains (Chen et al. 2005a; Elliott et al. 2009; Bontemps et al. 2010, 2016; Mishra et al. 2012; Klonowska et al. 2012; Gehlot et al. 2013; Melkonian et al. 2014). On the other hand, five out of the nine Rhizobium isolates were tested and effectively nodulated the alternative host Siratro, which is a promiscuous papilionoid species, confirming that the Rhizobium strains obtained here are genuine legume symbionts.
Our study revealed that the predominance of certain rhizobia in Mimosa nodules depends on the soil properties. The most acidic and less fertile soils (Brasília and Cavalcante) favoured the association of Mimosa with Paraburkholderia, while the soil with pH close to neutral and with higher fertility (Posse) led to the association with Rhizobium. Only one Paraburkholderia strain (POS_MSP2) was isolated in Posse.
Previous studies have demonstrated that Mimosa species tend to associate with α-proteobacteria when growing in soils with neutral/alkaline pH, such as those from Posse. For example, mimosas in India, where soil pH ranged from 7.8 to 8.2 (Gehlot et al. 2013), and in Mexico from 6.5 to 7.8 (Bontemps et al. 2016), were found in association with α-proteobacteria (Ensifer and Rhizobium). Interestingly, reports of Rhizobium associated with Mimosa in Brazilian soils, and in the Cerrado in particular, are very rare (Bontemps et al. 2010; Reis Junior et al. 2010). In fact, most host species used in our experiment have already been reported as nodulating with Paraburkolderia (Bontemps et al. 2010; Reis Junior et al. 2010), but the only one that has been previously found nodulating with Rhizobium was M. xanthocentra (Bontemps et al. 2010). One possible reason for such high prevalence of Paraburkholderia is that previous studies have mainly sampled in sites of low fertility acidic soils, which are more typical in the Cerrado region, the main centre of Mimosa diversity (Simon and Proença 2000). For the very few previous records of Rhizobium isolation from Mimosa carried out in this biome, there is no specific information on soil characteristics, except for that of R. altiplani, which was isolated from M. pudica growing in an alkaline soil substrate in a disturbed area close to Brasília (Baraúna et al. 2016). It is important to consider that several patches of highly-fertile limestone soil associated with seasonally dry forests, such as those found in Posse, occur in central Brazil. Therefore, it is likely that in such localities a range of Rhizobium strains or even other genera from the α-proteobacteria, as well as less acid-tolerant β-proteobacteria like Cupriavidus, would be prevalent.
In addition to environmental factors, such as soil fertility and pH, nitrogen concentration also seems to play a role in rhizobial dominance. In a competition experiment between rhizobia strains, when Mimosa is growing in substrate with low concentration of N, the nodulation by Paraburkholderia stands out in relation to Rhizobium. This situation is only reversed when there is an increase of N concentration in the environment (Elliott et al. 2009; Suárez-Moreno et al. 2012). Our results support these findings since soils from Posse, where Rhizobium predominates, have six-fold higher concentrations of nitrogen than in Brasília or Cavalcante (Table 2).
Although Paraburkholderia is not restricted to acidic soils, species of this genus have acquired mechanisms to tolerate acidity that enabled them to grow on soils where many other rhizobia groups cannot survive well, which would result in competitive advantages (Stopnisek et al. 2014). A predominance of Paraburkholderia was also found in some legume genera in the Cape region of South Africa, where poor and acidic soils seem to favour the association of Paraburkholderia with endemic legumes (Garau et al. 2009; Lemaire et al. 2015, 2016).
Contrary to our expectations we did not find pronounced host specificity, since most Mimosa species tested were able to nodulate with bacteria from different classes. Most species nodulated with bacteria of both the genera Paraburkholderia and Rhizobium. The exception was M. claussenii which appears not to associate with Rhizobium, as evidenced by the fact that it was the only species that failed to nodulate in the soil of Posse wherein a predominance of Rhizobium was found. Interestingly, even among the two endemic species, M. kalunga and Mimosa sp., which are found only in the Cavalcante region, no specificity in terms of α and β-rhizobia was observed. These results partly contradict expectations about symbiotic relationships between rhizobia and endemic Mimosa spp. (Bontemps et al. 2010), supporting the view that these plants are more promiscuous than previously thought (Thrall et al. 2011).
This lack of specificity between plant species and bacteria may be related to the presence of different nodulation genes in the genome of several rhizobial strains, allowing a response to different types of flavonoids secreted by the plant, resulting in bacterial infection in different legume species (Peix et al. 2015; Sprent et al. 2017). From a host perspective, even habitat specialists and narrow endemic plant species seem to maintain their ability to communicate and associate with various types of rhizobia, even with those outside their restricted habitat of occurrence. In this case, the capacity of symbiotic association with different types of rhizobia remains dormant in the host genome, even after many generations of coevolution and strong habitat/symbiont specialization. This was observed, for example, with the symbionts of native and endemic Mimosa species in Mexico: although their dominant symbionts were α-rhizobia, species belonging to recently diverged clades (1–3 my old) with close relatives in South America had retained their ability to nodulate with Paraburkholderia whereas species belonging to clades which had evolved in Mexico for up to 20 my had lost that ability (Bontemps et al. 2016).
Lemaire et al. (2015, 2016) surveying symbiotic relationships in a range of South African native legumes found that the plant genera investigated showed large variation in symbiotic specificity. Amphithalea and Podalyria were exclusively nodulated by Paraburkolderia, whereas Argyrolobium, Otholobium and Psoralea were associated only with α-proteobacteria (Mesorhizobium and Rhizobium). In contrast, Aspalathus and Indigofera were associated with a wide diversity of rhizobia including both α- and β-proteobacteria. This large variation in symbiotic preference within these groups may be related to environmental conditions, such as pH, elevation, geology and biogeography (Lemaire et al. 2015, 2016), as previously proposed in other studies (Mishra et al. 2012; Bournaud et al. 2013; Howieson et al. 2013; Gehlot et al. 2013).
Our results indicate that different Mimosa species have the ability to associate with different types of rhizobia (α- and β-proteobacteria), suggesting low specificity between host and bacterium. Even endemic plant species, when grown on soils from different localities, were able to nodulate with a range of bacteria present in non-native soil rhizospheres. Soil factors such as pH, nitrogen, organic matter and fertility seem to favour the predominance of certain types of rhizobia, irrespective of host identity, thus influencing the establishment of symbiotic relationships.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Araújo JF, de Castro AP, Costa MM, Togawa RC, Junior GJ, Quirino BF, Bustamante MMC, Williamson L, Handelsman J, Krüger RH (2012) Characterization of soil bacterial assemblies in Brazilian savanna-like vegetation reveals acido bacteria dominance. Microb Ecol 64:760–770
Baraúna AC, Rouws LFM, Simoes-Araujo JL, dos Reis Junior FB, Iannetta PP, Maluk M, Goi SR, Reis VM, James EK, Zilli JE (2016) Rhizobium altiplani sp. nov. isolated from effective nodules on Mimosa pudica growing in untypically alkaline soil in Central Brazil. Int J Syst Evol Microbiol 66:1–7
Barneby RC (1991) Sensitivae Censitae: a description of the genus Mimosa Linnaeus (Mimosaceae) in the new world. Mem N Y Bot Gard 65:1–835
Barrett CF, Parker MA (2006) Coexistence of Burkholderia, Cupriavidus and Rhizobium sp. nodule bacteria on two Mimosa species in Costa Rica. Appl Environ Microbiol 72:1198–1206
Beukes CW, Venter SN, Law IJ, Phalane FL, Steenkamp ET (2013) South African papilonoid legumes are nodulated by diverse Burkholderia with unique nodulation and nitrogen-fixation loci. PLoS One 8:e68406
Beukes C, Palmer M, Manyaka P, Chan WY, Avontuur J, Zyl E, Huntemann M, Clum A, Pillay M, Palaniappan K, Varghese N, Mikhailova N, Stamatis D, Reddy TBK, Daum C, Shapiro N, Markowitz V, Ivanova N, Kyrpides N, Woyke T, Blom J, Whitman WB, Venter SN, Steenkamp ET (2017) Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front Microbiol. https://doi.org/10.3389/fmicb.2017.01154
Bontemps C, Elliott GN, Simon MF, dos Reis Junior FB, Gross E, Lawton RC, Neto NE, Loureiro MF, de Faria SM, Sprent JI, James EK, Young JPW (2010) Burkholderia species are ancient symbionts of legumes. Mol Ecol 19:44–52
Bontemps C, Rogel MA, Wiechmann A, Mussabekova A, Moody S, Simon MF, Moulin L, Elliott GN, Lacercat-Didier L, Da Silva C, Grether R, Camargo-Ricalde SL, Chen W, Sprent JI, Martínez-Romero E, Young JPW, James EK (2016) Endemic Mimosa species from Mexico prefer alphaproteobacterial rhizobial symbionts. New Phytol 209:319–333
Bournaud C, de Faria SM, dos Santos JMF, Tisseyre P, Silva M, Chaintreuil C, Gross E, James EK, Prin Y, Moulin L (2013) Burkholderia species are the most common and preferred nodulating symbionts of the Piptadenia group (tribe Mimoseae). PLoS One 8:e63478
Castro AP, Silva MRSS, Quirino BF, Bustamante MMC, Krüger RH (2016) Microbial diversity in Cerrado biome (Neotropical savanna) soils. PLoS One 11:e0148785
Chen WM, Laevens S, Lee TM, Coenye T, De Vos P, Mergeay M, Vandamme P (2001) Ralstonia taiwanensis sp. nov isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int J Syst Evol Microbiol 51:1729–1735
Chen WM, James EK, Prescott AR, Kierans M, Sprent JI (2003) Nodulation of Mimosa spp. by the β-proteobacterium Ralstonia taiwanensis. Mol Plant-Microbe Interact 16:1051–1061
Chen WM, de Faria SM, Straliotto R, Pitard RM, Simoes-Araujo JL, Chou JH, Barrios E, Prescott AR, Elliott GN, Sprent JI, Young JPW, James EK (2005a) Proof that Burkholderia strains form effective symbioses with legumes: a study of novel Mimosa-nodulating strains from South America. Appl Environ Microbiol 71:7461–7471
Chen WM, James EU, Chou JH, Sheu SY, Yang SZ, Sprent JI (2005b) β-rhizobia from Mimosa pigra, a newly discovered invasive plant in Taiwan. New Phytol 168:661–675
Chen W-M, James EK, Coenye T, Chou J-H, Barrios E, de Faria SM, Elliott GN, Sheu S-H, Sprent JI, Vandamme P (2006) Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int J Syst Evol Microbiol 56:1847–1851
Chen WM, de Faria SM, James EK, Elliott GN, Lin KY, Chou JH, Sheu SY, Cnockaert M, Sprent JI, Vandamme P (2007) Burkholderia nodosa sp. nov., isolated from root nodules of the woody Brazilian legumes Mimosa bimucronata and Mimosa scabrella. Int J Syst Evol Microbiol 57:1055–1059
Dall'Agnol RF, Plotegher F, Souza RC, Mendes IC, dos Reis Junior FB, Béna G, Moulin L, Hungria M (2016) Paraburkholderia nodosa is the main N2-fixing species trapped by promiscuous common bean (Phaseolus vulgaris L.) in the Brazilian ‘Cerradão’. FEMS Microbiol Ecol 92: fiw108
De Meyer SE, Cnockaert M, Ardley JK, Maker G, Yates R, Howieson JG, Vandamme P (2013) Burkholderia sprentiae sp. nov., isolated from Lebeckia ambigua root nodules. Int J Syst Evol Microbiol 6:3950–3957
De Meyer SE, Briscoe L, Martínez-Hidalgo P, Agapakis CM, de-los Santos PE, Seshadri R, Reeve W, Weinstock G, O’Hara G, Howieson JG, Hirsch AM (2016) Symbiotic Burkholderia species show diverse arrangements of nif/fix and nod genes and lack typical high-affinity cytochrome cbb3 oxidase genes. Mol Plant-Microbe Interact 29:609–619
Elliott GN, Chen W-M, Chou J-H, Wang H-C, Sheu S-Y, Perin L, Reis VM, Moulin L, Simon MF, Bontemps C, Sutherland JM, Bessi R, de Faria SM, Trinick MJ, Prescott AR, Sprent JI, James EK (2007a) Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol 173:168–180
Elliott GN, Chen WM, Bontemps C, Chou JH, Young JPW, Sprent JI, James EK (2007b) Nodulation of Cyclopia spp. (Leguminosae, Papilionoideae) by Burkholderia tuberum. Ann Bot 100:1403–1411
Elliott GN, Chou J-H, Chen WM, Bloemberg GV, Bontemps C, Martínez-Romero E, Velázquez E, Young JPW, Sprent JI, James EK (2009) Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ Microbiol 11:762–778
Embrapa (1997) Manual de métodos de análise de solo. 2ª ed. Centro Nacional de Pesquisa de Solos, Rio de Janeiro, p 212
Estrada-de los Santos P, Rojas-Rojas FU, Tapia-Garcıa EY, Vásquez-Murrieta MS, Hirsch AM (2016) To split or not to split: an opinion on dividing the genus Burkholderia. Ann Microbiol 66:1303–1314
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fred EB, Waskman SA (1928) Yeast extract-manitol agar. Laboratory manual of general microbiology. McGraw-Hill, New York, p 145
Garau G, Yates R, Deiana P, Howieson JG (2009) Novel strains of nodulating Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soil. Soil Biol Biochem 41:125–134
Gehlot HS, Tak N, Kaushik M, Mitra S, Chen WM, Poweleit N, Panwar D, Poonar N, Parihar R, Tak A, Sankhla IU, Ojha A, Rao SR, Simon MF, Reis Junior FB, Perigolo N, Tripathi AK, Sprent JI, Young JPW, James EK, Gyaneshwar P (2013) An invasive Mimosa in India does not adopt the symbionts of its native relatives. An of Bot 112:179–196
Gyaneshwar P, Hirsch AM, Moulin L, Chen WM, Elliott GN, Bontemps C, Estrada-de Los Santos P, Gross E, Reis Junior FB, Sprent JI, Young JPW, James EK (2011) Legume-nodulating betaproteobacteria: diversity, host range and future prospects. Mol Plant-Microbe Interact 24:1276–1288
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95 ⁄ 98 ⁄ NT. Nucleic Acids Symp Ser 41:95–98
Hoagland DR, Arnon DI (1938) The water-culture method for growing plants without soil. California agricultural experimental station. Circ. n.347
Howieson JG, De Meyer SE, Vivas-Marfisi A, Ratnayake S, Ardley JK, Yates RJ (2013) Novel Burkholderia bacteria isolated from Lebeckia ambigua - a perennial suffrutescent legume of the fynbos. Soil Biol Biochem 60:55–64
Klonowska A, Chaintreuil C, Tisseyre P, Miché L, Melkonian R, Ducousso M, Laguerre G, Brunel B, Moulin L (2012) Biodiversity of Mimosa pudica rhizobial symbionts (Cupriavidus taiwanensis, Rhizobium mesoamericanum) in New Caledonia and their adaptation to heavy metal-rich soils. FEMS Microbiol Ecol 81:618–635
Lammel DR, Cruz LM, Carrer H, Cardoso EJBN (2013) Diversity and symbiotic effectiveness of beta-rhizobia isolated from sub-tropical legumes of a Brazilian Araucaria Forest. World J Microbiol Biotechnol 29:2335–2342
Lammel DR, Cruz LM, Mescolotti D, Stürmer SL, Cardoso EJBN (2015) Woody Mimosa species are nodulated by Burkholderia in ombrophylous forest soils and their symbioses are enhanced by arbuscular mycorrhizal fungi (AMF). Plant Soil 393:123–135
Lemaire B, Dlodlo O, Chimphango S, Stirton C, Schrire B, Boatwright JS, Honnay O, Smets E, Sprent J, James EK, Muasya AM (2015) Symbiotic diversity, specificity, and distribution of rhizobia in native legumes of the Core cape subregion (South Africa). FEMS Microbiol Ecol 91:1–17
Lemaire B, Chimphango S, Stirton C, Rafudeen S, Honnay O, Smets E, Chen W-M, Sprent J, James EK, Muasya AM (2016) Biogeographical patterns of legume-nodulating Paraburkholderia: from African Fynbos to continental scales. Appl Environ Microbiol 82:5099–5115
Liu XY, Wei S, Wang F, James EK, Guo XY, Zagar C, Xia LG, Dong X, Wang YP (2012) Burkholderia and Cupriavidus spp. are the preferred symbionts of Mimosa spp. in Southern China. FEMS Microbiol Ecol 80:417–426
Liu WYY, Ridgway HJ, James TK, James EK, Chen WM, Sprent JI, Young JPW, Andrews M (2014) Burkholderia sp. induces functional nodules on the South African invasive legume Dipogon lignosus (Phaseoleae) in New Zealand soils. Microb Ecol 68:542–555
Melkonian R, Moulin L, Béna G, Tisseyre P, Chaintreuil C, Heulin K, Rezkallah N, Klonowska A, Gonzalez S, Simon M, Chen W-M, James EK, Laguerre G (2014) The geographical patterns of symbiont diversity in the invasive legume Mimosa pudica can be explained by the competitiveness of its symbionts and by the host genotype. Environ Microbiol 16:2099–2111
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, LA pp 1–8.
Mishra RP, Tisseyre P, Melkonian R, Chaintreuil C, Miche L, Klonowska A, Gonzalez S, Bena G, Laguerre G, Moulin L (2012) Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol Ecol 79:487–503
Moulin L, James EK, Klonowska A, de Faria SM, Simon MF (2015) Phylogeny, diversity, geographical distribution, and host range of legume-nodulating betaproteobacteria: what is the role of plant taxonomy? In: de Bruijn F (ed) Biological nitrogen fixation, Wiley, Hoboken, vol 1
Parker MA, Wurtz A, Paynter Q (2007) Nodule symbiosis of invasive Mimosa pigra in Australia and in ancestral habitats: a comparative analysis. Biol Invasions 9:127–138
Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ (2015) Bacterial associations with legumes. Crit Rev Plant Sci 34:17–42
Platero R, James EK, Rios C, Iriarte A, Sandes L, Zabaleta M, Battistoni F, Fabiano E (2016) Novel Cupriavidus strains isolated from root nodules of native Uruguayan Mimosa species. Appl Environ Microbiol 82:3150–3164
Pons TL, Perreijn K, van Kessel C, Werger MJA (2007) Symbiotic nitrogen fixation in a tropical rainforest: 15N natural abundance measurements supported by experimental isotope enrichment. New Phytol 173:154–167
Quirino BF, Pappas GJ, Tagliaferro AC, Collevatti RG, Neto EL, da Silva MR, Bustamante MMC, Krüger RH (2009) Molecular phylogenetic diversity of bacteria associated with soil of the savanna-like Cerrado vegetation. Microbiol Res 164:59–70
Reis Junior FB, Simon MF, Gross E, Boddey RM, Elliott GN, Neto NE, Loureiro MF, Queiroz LP, Scotti MR, Chen WW, Norén A, Rubio MC, Faria SM, Bontemps C, Goi SR, Young JPW, Sprent JI, James EK (2010) Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol 186:934–946
Robledo M, García-Trigueros C, Rivera LP, Manyani H, Mateos PF, Meguías M (2010) El viaje de Rhizobium hacia una simbiosis eficiente. In: Guijo MM, Palma RR, MJD M, MJD I, García EG, PFM G, Barrios ML, Gonzáles BR, EJB G (eds) Fundamentos y aplicaciones agroambientales de las interaciones beneficiosas plantas-microorganismos. SEFIN, Spain, pp 141–170
Sawana A, Adeolu M, Gupta RS (2014) Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet 5:429
Simon MF, Proença C (2000) Phtytogeografic patterns of Mimosa (Mimosoideae, Leguminosae) in the Cerrado biome of Brazil: an indicator genus of high-altitude centers of endemism? Biol Conserv 96:279–296
Simon MF, Hughes CE, Harris AS (2010) Four new species of Mimosa (Leguminosae) from the central highlands of Brazil. Syst Bot 35:277–288
Simon MF, Grether R, Queiroz LP, Sarkinen TE, Dutra VF, Hughes CE (2011) The evolutionary history of Mimosa (Leguminosae): towards a phylogeny of the sensitive plants. Am J Bot 98:1201–1221
Sprent JI (2009) Legume nodulation. A global perspective. Wiley-Blackwell, Chichester
Sprent JI, Ardley JA, James EK (2017) Biogeography of nodulated legumes and their nitrogen fixing symbionts. New Phytol 215:40–56
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stopnisek N, Bodenhausen N, Frey B, Fierer N, Eberl L, Weisskopf L (2014) Genus-wide acid tolerance accounts for the biogeographical distribution of soil Burkholderia populations. Environ Microbio 16:1503–1512
Suárez-Moreno ZR, Caballero-Mellado J, Coutinho BG, Mendonca-Previato L, James EK, Venturi V (2012) Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb Ecol 63:249–266
Thrall PH, Laine A-L, Broadhurst LM, Bagnall DJ, Brockwell J (2011) Symbiotic effectiveness of rhizobial mutualists varies in interactions with native Australian legume genera. PLoS One 6:1–11
Vandamme P, Goris J, Chen WM, Vos P, Willems A (2002) Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst Appl Microbiol 25:507–512
Vincent JM (1970) A manual for the practical study of root-nodule bacteria. Oxford: Blackwell Scientific (International Biological Programme handbook, 15)
Vitousek PM, Menge DNL, Reed SC, Cleveland CC (2013) Biological nitrogen fixation: rates, patterns and ecological controls in terrestrial ecosystems. Phil Trans R Soc B 368:20130119
Wang D, Yang S, Tang F, Zhu H (2012) Symbiosis specificity in the legume-rhizobial mutualism. Cell Microbiol 14:334–342
Acknowledgements
We thank managers and authorities for permission to collect in areas under their care, and Aécio Amaral, Alessandra Fidelis, Lucas Rolim, Clodoaldo Alves de Souza, for field and laboratory assistance. Authors also acknowledge Ieda de Carvalho Mendes and Marco Pessoa Filho (Embrapa Cerrados) for revising and giving suggestions on the manuscript. RCP was supported by a Master’s scholarship from the Brazilian National Council for Scientific and Technological Development (CNPq), EKJ was funded by the CAPES/CNPq Ciência sem Fronteiras program, and MFS was supported by a CNPq productivity fellowship. This research was partially financed by Embrapa (02.13.08.001.00.00) and INCT - Plant-Growth Promoting Microorganisms for Agricultural Sustainability and Environmental Responsibility (CNPq 465133/2014-4, Fundação Araucária-STI, CAPES).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editor: Katharina Pawlowski
Electronic supplementary material
ESM 1
(PDF 326 kb)
Rights and permissions
About this article
Cite this article
de Castro Pires, R., dos Reis Junior, F.B., Zilli, J.E. et al. Soil characteristics determine the rhizobia in association with different species of Mimosa in central Brazil. Plant Soil 423, 411–428 (2018). https://doi.org/10.1007/s11104-017-3521-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-017-3521-5