Abstract
One of the important applications of computer-aided diagnosis is the detection of connective tissue disorders through automatic classification of antinuclear autoantibodies (ANAs). The recognition of ANAs is primarily done by analyzing indirect immunofluorescence (IIF) images of human epithelial type 2 (HEp-2) cells. In this regard, the paper introduces a novel approach for automatic classification of ANAs by staining pattern recognition of HEp-2 cell IIF images. Considering a set of HEp-2 cell images, the proposed method selects a set of relevant local texture descriptors for a pair of staining pattern classes, as well as identifies a set of important features corresponding to each relevant descriptor. The set of features for multiple classes is obtained from each of the important feature sets selected under various relevant local texture descriptors for all possible class-pairs. The relevance of a descriptor is evaluated based on the theory of rough hypercuboid approach, while the important feature set of a local descriptor is formed by reducing the impact of both noisy pixels present in an HEp-2 cell image and noisy HEp-2 cell images in a staining pattern class. Finally, the support vector machine is used to recognize one of the known staining patterns present in IIF images. The effectiveness of the proposed staining pattern recognition method, along with a comparison with related approaches, is illustrated on two benchmark databases of HEp-2 cell images using different classifiers and experimental set-up. The results show that the proposed approach performs significantly better than existing methods, with respect to both classification accuracy and F1 score, irrespective of the databases and classifiers used.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The main objective of computer-aided diagnosis (CAD) systems is to overcome the shortcomings of manual test procedure through proper automation. It helps to reduce both time and effort needed for manual analysis. Pattern recognition, machine learning and image processing techniques are widely used to develop the CAD systems, which make the analysis faster, easier and more reliable. In recent years, one of the important applications of CAD systems is the diagnosis of autoimmune disorders—a group of disorders where the immune system malfunctions and the tissues are attacked by autoantibodies. Connective tissue disease (CTD) is one such disorder, which can be predicted by the presence of antinuclear autoantibodies (ANAs) in the serum of patients. The ANAs behave as markers to detect the disorder and hence, play an important role in the diagnosis of many CTDs [2, 44].
The standard method used to predict the presence of ANAs is indirect immunofluorescence (IIF) [16], while human epithelial type 2 (HEp-2) cell is commonly used as cell substrate for IIF analysis of ANAs [47, 53]. The HEp-2 cells are tissue culture cells which have two components, namely, nucleus and cytoplasm. The nucleus of HEp-2 cells is quite large, providing better microscopic observation and greater sensitivity in the detection of antibodies. During the IIF test, fluorescent labelled secondary antibodies bind with different ANAs (primary antibodies) present in the patient’s serum. As a consequence, different staining patterns are formed both at nucleus and cytoplasm regions of the HEp-2 cells. If the nucleus of the cells is affected, staining patterns like Centromere and Nucleolar are formed, whereas Cytoplasmatic pattern, surrounding the nucleus, is formed when cytoplasm region is affected. Figure 1 presents specimen IIF images of the HEp-2 cells with different staining patterns. Several CTDs are mainly characterized by different staining patterns [1, 32, 47]. For example, Polymyositis and Sjogren’s Syndrome (SS) are primarily associated with Centromere (Fig. 1a) and Homogeneous (Fig. 1b) staining pattern classes, mixed CTDs and primary SS correspond to Speckled (Fig. 1d, e) staining patterns, while Cytoplasmatic (Fig. 1f) pattern is associated with Systemic Lupus Erythematosus disease. So, the main objective here is to recognize the known staining pattern classes of ANAs, by analyzing IIF images of HEp-2 cells, for the diagnosis of CTDs.
In last few years, some efforts have been made for the development of CAD systems to recognize staining patterns present in HEp-2 cell IIF images. Different shape-based, edge-based and texture-based features [24, 40, 50] can be used to capture the information of surface of the HEp-2 cells. Several global texture descriptors based on gray level co-occurrence matrix [20] are used in [9, 43, 45], while edge orientation histograms [39], histograms of oriented gradients [6], rotation-invariant Gabor features [3], and modified Zernike moments [41] are also found to provide important information of an HEp-2 cell image [9]. However, it has been shown in [9] that the local variations of intensity patterns are more effective than global information, obtained from the HEp-2 cell images, in differentiating various staining pattern classes. In this background, several local texture descriptors, namely, local binary pattern (LBP) [36], rotation invariant LBP (\(\hbox {LBP}^{\mathrm{ri}}\)) [37], rotation invariant uniform LBP (\(\hbox {LBP}^{\mathrm{riu2}}\)) [37], completed LBP [19], co-occurrence of adjacent LBPs (CoALBP) [35] and rotation-invariant co-occurrence of adjacent LBPs [34], have been used to characterize an HEp-2 cell image [9, 34], while the concept of gradient-oriented co-occurrence of LBPs has been introduced in [48]. For automatic recognition of mitotic cell, a combination of textural features obtained using LBPs and morphological features has been used in [14]. To capture local textural information, a modified version of uniform LBPs descriptor has been incorporated in this work. Wiliem et al. [54] have proposed a system for cell classification, comprising of nearest convex hull classifier and a dual-region codebook-based descriptor. In order to encode gradient and textural characteristics of the HEp-2 patterns, Theodorakopoulos et al. [48] have proposed a new descriptor, based on co-occurrence of uniform LBPs along directions dictated by the orientation of local gradient.
In general, a particular texture descriptor at a single scale is used to describe the characteristics of all the classes of HEp-2 cell images. However, the inherent texture patterns of HEp-2 cells are quite different from each other. The inter-class visual similarities, intra-class variations, presence of noise, and ambiguity in staining pattern class definition further enhance the difficulty in actual pattern recognition. So, it is expected that a specific descriptor considered under a particular scale may be important in differentiating a given pair of HEp-2 pattern classes, but may not be relevant in capturing the inherent characteristics of another pair of staining pattern classes. In this regard, the main contributions of this paper are as follows:
-
1.
development of a novel approach for the diagnosis of connective tissue disorders through automatic identification of ANAs, by staining pattern recognition of HEp-2 cell IIF images;
-
2.
formulation of the problem of descriptor selection to select a set of important local texture descriptors under appropriate scales for a pair of staining pattern classes, along with a set of relevant features corresponding to each selected descriptor;
-
3.
computation of the relevance of a descriptor, based on the theory of rough hypercuboid approach;
-
4.
evaluation of the importance of a feature set, corresponding to a relevant descriptor, by defining an energy function; and
-
5.
demonstrating the effectiveness of the proposed approach on benchmark HEp-2 cell image databases.
The proposed descriptor selection method integrates judiciously the theory of rough sets and the merits of local texture descriptors for HEp-2 cell staining pattern classification. It helps in finding both the dominant features representing the important characteristics of each HEp-2 cell image and relevant feature set for each of the HEp-2 cell staining pattern classes. The resultant feature set is independent of both noisy pixels in an HEp-2 cell image and noisy HEp-2 cell images in a staining pattern class. All the important features, selected under several relevant local descriptors for all pairs of classes, form the final feature set for multiple classes. Finally, the recognition of staining patterns present in IIF images is done by using support vector machine. The effectiveness of the proposed staining pattern recognition method, along with a comparison with related approaches, is demonstrated on two benchmark HEp-2 cell image databases using different classifiers and experimental set-up.
2 Proposed method for HEp-2 cell staining pattern classification
During the development of a CAD system, a specific texture descriptor at a fixed scale is generally used to describe the intrinsic characteristics of all HEp-2 cell images, irrespective of their staining pattern classes. Let us consider the scatter plots of HEp-2 cell images corresponding to MIVIA database [15], presented in Fig. 2. In each plot, feature values, corresponding to the HEp-2 cell images belonging to a particular pair of classes, are computed by considering a specific descriptor under a fixed scale. The x-axis of each plot represents the most relevant feature, while the y-axis denotes the feature which provides the maximum dependency or joint relevance when considered along with the most relevant feature at x-axis. From Fig. 2a, b, it can be seen that some of the HEp-2 cell images of Homogeneous and Coarse Speckled classes, considered under \(\hbox {LBP}^{\mathrm{ri}}\) at scale 1 (\({\mathcal {S}}_1\)), overlap with each other, while they are almost separable under the same descriptor but at a different scale (\({\mathcal {S}}_4\): scale 4). It establishes that the HEp-2 cell images, which are not well-separated under a specific scale, can be properly classified if a different scale is considered keeping the descriptor fixed. However, Fig. 2c depicts that the cell images of another class-pair, namely, {Homogeneous, Centromere}, are poorly classified if same descriptor (\(\hbox {LBP}^{\mathrm{ri}}\)) under the same scale (\({\mathcal {S}}_4\)) is considered, while Fig. 2d shows that they become more distinguishable if a different descriptor (CoALBP) at a different scale (\({\mathcal {S}}_2\)) is considered. So, it can be concluded that the information obtained from a particular modality may found to be relevant in differentiating samples from a specific class-pair, but may not be significant in categorizing samples from another pair of classes. In the proposed method, a modality refers to a particular local texture descriptor considered under a specific scale.
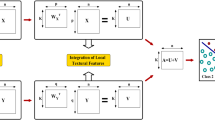
Hence, the proposed method selects important features from the relevant modalities for each pair of classes, and finally forms the resultant feature set for multiple staining pattern classes. Figure 3 depicts the block diagram of the proposed method for HEp-2 cell staining pattern classification. At first, the histogram of feature values corresponding to each modality is obtained for each of the input HEp-2 cell images. Then, class specific feature set is formed for each of the HEp-2 cell staining pattern classes under each modality. Thereafter, the class-pair specific feature sets are obtained for each modality, from each of the class specific feature sets, for all possible class-pairs. The relevance of each class-pair specific feature set under a particular modality is evaluated using the concept of rough hypercuboid approach. After selecting a set of most relevant feature sets for each class-pair, the final feature set for multiple staining pattern classes is formed, which is further used for training the support vector machine. Each of these steps is elaborated next one by one.
2.1 Generation of class specific feature set
Let \(X=\{x_1,\ldots ,x_k,\ldots ,x_n\}\) be a set of n training HEp-2 cell images, where each image \(x_k \in {\mathfrak {R}}^m\) is represented by a set \(F=\{f_1,\ldots ,f_j,\ldots ,f_m\}\) of m features. So, the set \(h_k=\{h_{k1}, \ldots , h_{kj}, \ldots , h_{km}\}\), containing m feature values, describes the image \(x_k\), where \(h_{kj}=x_k(f_j)\) is the value of the feature \(f_j\) corresponding to the image \(x_k\). In the proposed research work, four local descriptors, namely, LBP [38], \(\hbox {LBP}^{\mathrm{ri}}\) [37], \(\hbox {LBP}^{\mathrm{riu2}}\) [37], and CoALBP [35], are considered. Hence, \(h_k\) represents the normalized histogram of \(x_k\), corresponding to one of the four texture descriptors. Let, \({\tilde{h}}_k=\{{\tilde{h}}_{k1},\ldots , {\tilde{h}}_{kj},\ldots ,{\tilde{h}}_{km}\}\) be the sorted \(h_k\), in descending order, such that \({\tilde{h}}_{k1} \ge \cdots \ge {\tilde{h}}_{kj} \ge \cdots \ge {\tilde{h}}_{km}\), where the set \({I}_k=\{{I}_{k1}, \ldots ,{I}_{kj},\ldots ,{I}_{km}\}\) preserves the corresponding feature index of \({\tilde{h}}_{k}\). Let us consider that each of the HEp-2 cell images of X belongs to one of the c staining pattern classes \(\{\omega _{1},\ldots , \omega _{i},\ldots ,\omega _{c}\}\).
The textural properties of a cell image \(x_k\) can be primarily described in terms of the feature values of the set \(h_k\). In the proposed work, it is assumed that all the feature values of \(h_k\) do not contribute equally in representing the intrinsic textural patterns of the image \(x_k\). Indeed, a subset \(V_k\) of the feature set F can precisely capture the significant characteristics of \(x_k\), which is termed as the dominant feature set of \(x_k\). However, different images have their own characteristics, which can be represented by different dominant feature sets. In order to identify the dominant features of each cell image \(x_k\) from the corresponding \(h_k\), the following function is defined:
where \({{\vartheta }}(x_k,q)\in [0,1]\) and \({{\vartheta }}(x_k,m)=1, \forall x_k \in X\). Here, \(\vartheta\) is known as the energy function. It depicts the amount of total energy, present in the sorted normalized histogram \({\tilde{h}}_k\), preserved by the first q features of \(x_k\). Hence, the energy of \(x_k\), computed from \({\tilde{h}}_k\), expresses the relevant information about the cell image \(x_k\).
In order to achieve a pre-specified fraction of energy \(\vartheta _{given}\) from the sorted histogram \({\tilde{h}}_{k}\), the required number of dominant features for the sample \(x_{k}\) can be computed as follows:
So, the average number of dominant features for the entire set of samples X is given by
while the set of dominant features \(V_k\), corresponding to the sample \(x_k\), is defined as
Hence, the set \(V_k \subseteq F\) contains only the dominant features of \(x_k\), which can sufficiently represent the important characteristics of the image \(x_k\).
Generally, the samples belonging to a particular class are expected to exhibit similar characteristics, which can be reflected in the corresponding dominant feature sets of the samples. In other words, the samples from same class will have similar sets of dominant features, while the samples belonging to different classes will have different sets of dominant features. However, if a noisy sample is present in a class \(\omega _i\), the dominant feature set of that sample may contain certain features which do not provide any important characteristics of the class \(\omega _i\). Presence of such features in the dominant set of the sample may delude classification of the samples of \(\omega _i\). So, it is necessary to identify those features from the dominant sets of samples that represent significant properties of a class to which the samples belong. To address the above mentioned problem, the probability of occurrence \(p(f_{j}|\omega _{i})\) of a feature \(f_j\) in the dominant sets of samples of a particular class \(\omega _i\) is computed as follows:
Here, \(|\omega _{i}|\) denotes the number of samples belonging to the class \(\omega _{i}\). So, the features with low probability of occurrence values in a particular class \(\omega _i\) can be assumed to occur inadvertently in the dominant sets of samples belonging to \(\omega _i\) and hence, can be discarded from the set without much loss of information. In order to discard those features, a threshold parameter \(\delta\) is used here, and class specific feature set \({\mathcal {N}}(\omega _i)\) for the class \(\omega _i\) is formed, based on the value of \(\delta\), as follows:
So, the set \({\mathcal {N}}(\omega _i)\) represents the important characteristics specific to the class \(\omega _i\). A feature \(f_j\) will only be present in the set \({\mathcal {N}}(\omega _i)\) if it is dominant in most of the samples of \(\omega _i\) as well as bears significant amount of information regarding the characteristics of class \(\omega _i\).
2.2 Pairwise class specific and final feature sets
Let, the pairwise class specific feature set be denoted by \({\mathcal {N}} (\{\omega _{i},\omega _{r} \})\), corresponding to the pair of classes \(\{\omega _{i}, \omega _{r} \}\). So, \({\mathcal {N}}(\{\omega _ {i},\omega _{r} \})\) should contain information that is relevant for both the classes \(\omega _{i}\) and \(\omega _{r}\). Let, \({\mathcal {N}}(\omega _{i})\) and \({\mathcal {N}}(\omega _{r})\) be the class specific feature sets, computed using (7), corresponding to the classes \(\omega _{i}\) and \(\omega _{r}\), respectively. Then, \({\mathcal {N}}(\{\omega _{i} ,\omega _{r} \})\) is defined in terms of \({\mathcal {N}}(\omega _{i})\) and \({\mathcal {N}} (\omega _{r})\) as follows:
Hence, \({\mathcal {N}}(\{\omega _{i},\omega _{r}\})\) contains the features that represent significant characteristics, common to the particular pair of classes \(\omega _{i}\) and \(\omega _{r}\).
Considering a set of t number of modalities \({\mathcal {M}}=\{{\mathcal {M}}_1,\ldots , {\mathcal {M}}_p,\ldots ,{\mathcal {M}}_t\}\), the proposed method computes the relevance \(\varGamma _p (\{\omega _i,\omega _r \})\) of each pairwise class specific feature set \({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\})\), corresponding to the pair of classes \(\{\omega _{i},\omega _{r}\}\) under the pth modality \({\mathcal {M}}_p\). The theory of hypercuboid equivalence partition matrix of rough hypercuboid approach [27] is employed to compute the relevance, which is illustrated in Sect. 2.4. The relevance of the feature set \({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\})\) represents the relevance of the pth modality \({\mathcal {M}}_p\), with respect to the class-pair \(\{\omega _{i},\omega _{r}\}\).
After computing the relevance of each pairwise class specific feature set \({{\mathcal {N}}}_p (\{\omega _{i},\omega _{r}\})\) corresponding to t modalities for each class-pair \(\{\omega _i,\omega _r \}\), \({\tilde{t}}\) most relevant feature sets \(\{{{\mathcal {N}}}_p (\{\omega _{i},\omega _{r}\})\}\) are chosen, and the set \(\tilde{{\mathcal {N}}}_{ir}= \{{{\mathcal {N}}}_p(\{\omega _{i},\omega _{r}\})\}\) is formed for the pair of classes \(\omega _{i}\) and \(\omega _{r}\). Finally, the feature set \({\mathcal {D}}\) for all possible pairs of classes is defined as
The support vector machine (SVM) is used to train and predict the staining patterns present in IIF images of HEp-2 cells, based on the final feature set \({\mathcal {D}}\).
2.3 Novelty of the proposed approach
In this regard, it should be noted that the method due to Liao et al. [26], termed as dominant LBP (DLBP), defines the most frequently occurring local textural patterns as dominant patterns of an image. The method considers the occurrence values of dominant patterns to form the feature vector of an image, without considering the information of pattern types. In effect, a particular dimension of the feature vector may correspond to different patterns for different images, while two different dimensions of two feature vectors may indicate same pattern type. So, different texture images are represented by occurrence values of different sets of patterns. As DLBP method disregards the pattern type information while generating feature vector, it fails to account the discriminative properties of individual patterns, which are essential to classify images properly.
Guo et al. [18], on the other hand, proposed a three-layered model to learn the most discriminative patterns, from the original set of patterns, for representing a texture image. At the first layer, the most frequently occurring patterns, which occupy a certain percentage of total occurrence, are identified for all the training images. The set of such patterns is considered to reflect the important textural properties of an image and termed as the dominant pattern set of that image. As opposed to the DLBP method, this method encodes the pattern types, together with the pattern occurrences, for computing the dominant pattern set. Since the dominant pattern sets may vary greatly for different images of the same class, the discriminative pattern set for each class is formed at the second layer by taking the intersection of dominant pattern sets over all the training images belonging to the same class. At the last layer, union of all the discriminative pattern sets, corresponding to all the classes, forms the final feature set. As the discriminative pattern set of a class \(\omega _i\) is formed with the features having probability of occurrence 1 with respect to dominant pattern sets of all images belonging to \(\omega _i\), the set is exactly same with the class specific feature set \({\mathcal {N}}(\omega _i)\), introduced in the proposed method, with \(\delta =1\) as in (7). It can be assumed that a pattern, which bears important characteristics of a class \(\omega _i\), will be dominant in all samples of \(\omega _i\). However, if a noisy sample is present in the class \(\omega _i\), then the pattern may not be dominant in the noisy sample, and hence will not be selected as a discriminative pattern of class \(\omega _i\) if we set \(\delta =1\). So, the discriminative pattern set, obtained using the method of Guo et al. [18], is comparatively smaller in size, but has less discriminative information.
In order to have a compact representation of class specific feature set \({\mathcal {N}}(\omega _i)\) for the class \(\omega _i\), the proposed method computes the probability of occurrence of a feature in the dominant feature sets of samples belonging to \(\omega _i\) and the class specific feature set \({\mathcal {N}}(\omega _i)\) is formed with features which are dominant in most of the samples of \(\omega _i\) as mentioned in (7). Also, in the proposed method, different modalities are considered for differentiating different pairs of classes, based on the relevance of the modalities, while the methods of Liao et al. [26] and Guo et al. [18] consider only the information of a fixed set of modalities to distinguish the samples belonging to different classes.
2.4 Computation of relevance
Let \({{\mathbb {U}}}=\{x_1,\ldots ,x_k, \ldots ,x_n\}\) be the finite set of n samples or objects, \({{\mathbb {C}}}=\{f_1,\ldots ,f_j, \ldots ,f_m\}\) be the condition attribute set in \({{\mathbb {U}}}\), and \({{{\mathbb {D}}}}\) be the decision attribute set, where \({\mathbb U}/{{{\mathbb {D}}}}=\{\omega _1,\ldots ,\omega _i,\ldots , \omega _c\}\) denotes the c equivalence classes or information granules of \({{\mathbb {U}}}\) generated by the equivalence relation induced from the decision attribute set \({{{\mathbb {D}}}}\). Given a set \(\omega _i \subseteq {{\mathbb {U}}}\), in general, it may not be possible to describe \(\omega _i\) precisely in the approximation space \(<{\mathbb U},{{\mathbb {C}}} \cup {\mathbb {D}}>\). The class \(\omega _i\) can be approximated using the equivalence relation induced from each condition attribute or feature \(f_j \in {\mathbb {C}}\) by constructing the ith hypercuboid equivalence partition of \({\mathbb {U}}\) corresponding to \(\omega _i\). So, for c classes, c-hypercuboid equivalence partitions of the universe can be obtained and arrayed as \((c \times n)\) matrix \({\mathbb {H}}(f_j)=[{\mu }_{ik}(f_j)]\). The matrix \({\mathbb {H}}(f_j)\) is termed as the hypercuboid equivalence partition matrix of the feature \(f_j\), where each \(\mu _{ik}(f_j)\) can be obtained as follows [27]:
Here, \(\mu _{ik}(f_j)\) represents the membership of object \(x_k\) in the ith equivalence partition, based on the equivalence relation induced by \(f_j\). The interval \([{\mathcal {L}}_{ij},{\mathcal {U}}_{ij}]\) is the value range of the feature \(f_j\) with respect to the class \(\omega _{i}\). It is spanned by the objects with class label \(\omega _{i}\). So, the feature value \(x_k(f_j)\) of each object \(x_k\) with class label \(\omega _{i}\), corresponding to the feature \(f_j\), falls within the interval \([{\mathcal {L}}_{ij},{\mathcal {U}}_{ij}]\), which implies that an equivalence partition is nonempty.
Each row of \({\mathbb {H}}(f_j)\) is a hypercuboid equivalence partition corresponding to a particular class, induced by the feature \(f_j\). The ith hypercuboid equivalence partition, corresponding to the class \(\omega _i\), is denoted as:
Here, ‘+’ represents the union set operation. Each class \(\omega _i\) can be approximated using only the information contained within the feature \(f_j\) by constructing the A-lower and A-upper approximations of \(\omega _i\) [27]:
where equivalence relation A is induced from \(f_j\).
The hypercuboid equivalence partition matrix \({\mathbb {H}}({\mathbb {C}})\), induced by the condition attribute set \({\mathbb {C}}\), can be computed as [27]:
Hence, the equivalence partition \(\mu _i({\mathbb {C}})\) represents a hypercuboid in m-dimensional Euclidean space, where each dimension corresponds to a certain feature. The m-dimensional hypercuboid is defined as the Cartesian product of m orthogonal intervals at m respective dimensions, corresponding to class \(\omega _i\). As each hypercuboid corresponds to a particular class, c hypercuboids will be formed for c given classes. Figure 4 presents an example of two hypercuboids, corresponding to two HEp-2 cell staining pattern classes, namely, Centromere and Homogeneous. Each hypercuboid encompasses the objects, that is, HEp-2 cell images, belonging to a particular class, considering two features selected under CoALBP at scale \({\mathcal {S}}_2\).
However, in real-life data analysis, uncertainty arises due to overlapping class boundaries. So, every two hypercuboids may intersect with each other. The intersection of two hypercuboids forms an implicit hypercuboid (shaded region of Fig. 4), which encloses the misclassified objects that belong to more than one equivalence partitions with respect to a set of attributes. The relevance of the set of condition attributes or features with respect to the decision attribute set depends on the cardinality of the implicit hypercuboid. It can be observed that the relevance increases with the decrease in the cardinality of implicit hypercuboids.
In the current study, the relevance of pairwise class specific feature set \({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\})\), corresponding to the pair of classes \(\{\omega _{i},\omega _{r}\}\), is to be computed. In this case, \(c=2\) as only a particular class-pair \(\{\omega _i,\omega _r\}\) is considered at a time. So, \(n=|\omega _i|+|\omega _r|\), that is, the samples belonging to \(\{\omega _{i},\omega _{r}\}\) constitute the universe \({\mathbb {U}}\) and the condition attribute set \({\mathbb {C}}\) is given by \({\mathbb {C}}={\mathcal {N}}_p (\{\omega _{i},\omega _{r}\})\). As the number of equivalence partitions reduces to 2, the relevance of the feature set \({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\})\) with respect to the class labels of samples from \(\{\omega _{i},\omega _{r}\}\) is redefined from the hypercuboid equivalence partition matrix \({\mathbb {H}}({\mathcal {N}}_p(\{\omega _{i}, \omega _{r}\}))\) as follows:
Therefore, the relevance is obtained as the XOR output of two equivalence partitions corresponding to the pair of classes. If an object belongs to both the equivalence partitions, then the corresponding XOR value becomes zero. This validates the fact that the object is enclosed by the implicit hypercuboid, formed at the intersection of two equivalence partitions, and thus, cannot be classified correctly using the equivalence relation induced by the feature set. However, if the object belongs to exactly one of the two possible partitions, then XOR operation produces an output 1, demonstrating that the object outside the implicit hypercuboid contributes in the computation of relevance of the feature set. Thus, the relevance measure proposed in (16) significantly reduces the computational complexity of the earlier measure reported in [27]. From (16), it can be seen that if \(\varGamma ({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\}))=1\), then no implicit hypercuboid exists and both the classes \(\omega _{i}\) and \(\omega _{r}\) can be defined precisely using the knowledge of \({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\})\). On the other hand, if \(\varGamma ({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\}))=0\), then both of them cannot be defined using the information of \({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\})\). However, if \(\varGamma ({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\})) \in (0,1)\), then \(\omega _{i}\) and \(\omega _{r}\) can be approximated using the feature set \({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\})\).
The pairwise class specific feature set \({\mathcal {N}}_p(\{\omega _{i}, \omega _{r}\})\) may consists of redundant features that do not provide any significant information in classifying the samples of class \(\omega _i\) from that of \(\omega _r\). So, prior to computing the relevance \(\varGamma ({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\}))\) of the feature set \({\mathcal {N}}_p(\{\omega _{i},\omega _{r}\})\), insignificant and irrelevant features are discarded from the set, based on maximum relevance-maximum significance criterion reported in [29]. The basic steps of the proposed method to select the feature set \({\mathcal {D}}\) for HEp-2 cell classification are outlined in Algorithm 1.

3 Experimental results and discussions
The performance of the proposed method is extensively analyzed and corresponding results are presented in this section. Several local texture descriptors, namely, local binary patterns (LBP) [38], rotation invariant LBPs (\(\hbox {LBP}^{\mathrm{ri}}\)) [37], rotation invariant uniform LBPs (\(\hbox {LBP}^{\mathrm{riu2}}\)) [37] and co-occurrence among adjacent LBPs (CoALBP) [35], computed at different scales, such as scale 1 (\({\mathcal {S}}_1\)), scale 2 (\({\mathcal {S}}_2\)), scale 3 (\({\mathcal {S}}_3\)), scale 4 (\({\mathcal {S}}_4\)), concatenation of \({\mathcal {S}}_1\), \({\mathcal {S}}_2\) and \({\mathcal {S}}_3\) (\({\mathcal {S}}_{123}\)), concatenation of \({\mathcal {S}}_1\), \({\mathcal {S}}_2\) and \({\mathcal {S}}_4\) (\({\mathcal {S}}_{124}\)), are used in the current study to demonstrate the efficacy of the proposed method in HEp-2 cell classification. For CoALBP, 4-neighborhood is considered, while 8-neighborhood is considered for other local descriptors. The performance of the proposed method is extensively compared with that of dominant LBP (DLBP) [26], discriminative features for texture description (DFTD) [18], and several multimodal data integration methods. Different classifiers such as SVM [51] with linear, polynomial, and radial basis function (RBF) kernels, extreme learning machine (ELM) [23], restricted Boltzmann machine (RBM) [21], discriminative RBM (DRBM) [25], fuzzy RBM (FRBM) [13], random forest (RF) [4] and K-nearest neighbors (KNN) algorithm [10] are also used for evaluating the performance of the proposed approach.
Both training–testing and tenfold cross-validation (CV) methods are performed to validate the performance of the proposed method as well as the existing approaches. Results obtained from training–testing are pictorially presented using bar graphs and performance of tenfold CV is studied through box-and-whisker plots and tables of mean, median, standard deviation and p-values computed using paired-t (one-tailed) and Wilcoxon signed-rank (one-tailed) tests, with 95% confidence level. In box-and-whisker plots, median is represented by central line of the box, upper and lower boundaries depict upper and lower quartiles, respectively. Whiskers are drawn from mean to three standard deviations, so that extreme points can also be included. The outliers are plotted with ‘+’, individually.
3.1 Description of data sets
Two HEp-2 cell image databases, namely, MIVIA data set [15] (ICPR 2012 HEp-2 cell classification contest data set) and SNP HEp-2 database [54], are considered to validate the performance of various approaches. The images of two databases were captured at different assay and microscopic configurations. The MIVIA database contains images of 1455 cells obtained from 28 slides, out of which 721 cells are used as the training set while the rest of 734 cells comprise the test set. The entire set is divided into six staining pattern classes, namely, Centromere, Homogeneous, Nucleolar, Coarse Speckled, Fine Speckled and Cytoplasmic. On the other hand, the SNP HEp-2 database contains 1806 labelled cells, which is partitioned into training and test sets with 869 and 937 cells, respectively. Each of the cells belongs to one of the five staining pattern classes, namely, Centromere, Homogeneous, Nucleolar, Coarse Speckled and Fine Speckled. Both the MIVIA and SNP HEp-2 cell data sets are split into ten distinct folds for cross-validation, where the cell images are almost equally distributed with respect to each of the classes.
3.2 Importance of local texture descriptors
In this section, the performance of different texture descriptors is analyzed in characterizing the staining patterns of HEp-2 cell images. The corresponding results on MIVIA and SNP databases are reported in Table 1 for training–testing. Several global texture descriptors such as discrete cosine transform (DCT) [49], fast Fourier transform (FFT) [33], rotation invariant Gabor features [3], gray level co-occurrence matrix (GLCM) [20] and histograms of oriented gradients (HOG) [6] are used to represent the inherent textural properties of HEp-2 cell images. Similarly, various local texture descriptors, namely, LBP [38], \(\hbox {LBP}^{\mathrm{ri}}\) [37], \(\hbox {LBP}^{\mathrm{riu2}}\) [37] and CoALBP [35] have been used to characterize the HEp-2 staining pattern classes. From the results reported in Table 1, it can be observed that local variations of intensity patterns are proved to be more effective in differentiating staining pattern classes, than the global information obtained from the HEp-2 cell images. This observation also resembles the results reported in [9]. Hence, local texture descriptors are considered for further analysis in the current study.
3.3 Illustrative example
In order to illustrate the proposed method, let us consider HEp-2 cell images from training and test sets of MIVIA database [15], where the staining pattern classes of 734 test cell images need to be predicted based on the information of final feature set \({\mathcal {D}}\), obtained from the 721 training cell images. Six staining pattern classes are represented by the set \(\{\omega _1,\ldots ,\omega _i,\ldots ,\omega _6 \}\), where each \(\omega _i\) corresponds to one of the six possible classes, namely, Centromere, Homogeneous, Nucleolar, Coarse Speckled, Fine Speckled and Cytoplasmic, in the specified order. In this example, the parameters \(\vartheta _{given}\) and \(\delta\) are considered as 0.9 and 0.1, respectively.
Based on the assumption that all the features of a particular modality are not dominantly present in a sample, it is required to identify the set of dominant features for each sample of the training set using (2), which preserves 90% of the total energy present in the histogram of the sample. The average number of dominant features (\({\overline{d}}\)) obtained for each modality is tabulated in Table 2, which indicates that in case of modalities with large number of features, only a small portion of the feature set carries important information.
After obtaining the dominant feature set for each sample, the probability of occurrence of a feature in the dominant sets of samples of a particular class is computed. The class specific feature set is formed for each class with only the features having probability of occurrence value greater than or equal to 0.1 in that class. Table 3 reports the cardinality of class specific feature set for two classes \(\omega _1\) (Centromere) and \(\omega _2\) (Homogeneous). From the given six classes of MIVIA data set, \({6 \atopwithdelims ()2}\) or 15 pairwise class specific feature sets are obtained for each modality. For each pair of classes, the irrelevant and insignificant features are eliminated from the pairwise class specific feature set using the maximum relevance-maximum significance criterion [29], and finally the relevance of the reduced class specific feature set is computed using (16). Table 3 presents an instance of cardinalities of initial and reduced pairwise class specific feature sets, along with the relevance of the reduced set computed for each of the 15 modalities, corresponding to the class-pair \(\{\omega _1,\omega _2 \}\).
From Table 3, it can be observed that the irrelevant and insignificant features can be effectively removed using maximum relevance-maximum significance criterion. The relevance values of different modalities suggest that all the local descriptors and scales are not effective in differentiating samples belonging to two classes \(\omega _1\) and \(\omega _2\). For example, LBP at \({\mathcal {S}}_4\) is more relevant than LBP computed at other scales to classify samples of \(\omega _1\) from that of \(\omega _2\). However, CoALBP at \({\mathcal {S}}_2\) is found to be the best modality for classifying samples belonging to \(\{\omega _1,\omega _2\}\). Three most relevant modalities, along with the corresponding number of selected features for each class-pair, are arrayed in Table 4, where J(K) represents the Jth most relevant modality with K number of pairwise class specific features. For example, based on the relevance values reported in Table 3, CoALBP at \({\mathcal {S}}_2\) having 11 features, CoALBP at \({\mathcal {S}}_1\) with 10 features, and LBP at \({\mathcal {S}}_4\) with 12 features are selected to form the set \(\tilde{{\mathcal {N}}}_{ir}=\{{{\mathcal {N}}}_p (\{\omega _i,\omega _r\}) \}\), corresponding to the class-pair \(\{\omega _1,\omega _2\}\). From both Tables 3 and 4, it is also clear that the spatial information, obtained using CoALBP, is more important to differentiate \(\omega _1\) and \(\omega _2\) than the rotation invariance property of both \(\hbox {LBP}^{\mathrm{ri}}\) and \(\hbox {LBP}^{\mathrm{riu2}}\). In fact, the co-occurrence of local textural features is an essential information required to discriminate Centromere class (\(\omega _1\)) from the rest of the staining pattern classes, while both rotation invariance and uniformity properties of local descriptors are found to be irrelevant. However, the rotation invariance characteristic plays a significant role, as highlighted in Table 4, for classifying the samples of class-pairs \(\{\omega _2,\omega _4\}\), \(\{\omega _2,\omega _6\}\), \(\{\omega _3,\omega _4\}\), \(\{\omega _3,\omega _5\}\), \(\{\omega _3,\omega _6\}\), \(\{\omega _4,\omega _5\}\), and \(\{\omega _5,\omega _6\}\), where either \(\hbox {LBP}^{\mathrm{ri}}\) or \(\hbox {LBP}^{\mathrm{riu2}}\) is selected as the most relevant modality.
For each class-pair, three most relevant modalities are selected, based on their relevance values. The final feature set \({\mathcal {D}}\), consisting of 277 features, is obtained by taking the union of all the selected feature sets corresponding to all class-pairs. The SVM with linear kernel is used for training HEp-2 cell images, based on the final feature set \({\mathcal {D}}\). It provides 100% classification accuracy on training samples, while 63.90% accuracy on test data set.
3.4 Effectiveness of energy and threshold
As defined in (1), energy \(\vartheta\) of an image, which represents the important information specific to a particular image, is quantified in terms of the feature occurrences in the image. On the other hand, threshold \(\delta\), as mentioned in (7), signifies the contribution of a feature in representing the characteristics of a class. Based on the values of \(\vartheta\) and \(\delta\), the class specific feature set \({\mathcal {N}}(\omega _i)\), corresponding to the class \(\omega _i\), is formed. Hence, it is important to find out the optimum values of both \(\vartheta\) and \(\delta\) such that an accurate representation of the class \(\omega _i\) can be obtained through \({\mathcal {N}}(\omega _i)\).
Figure 5 depicts the probability of occurrence values, in sorted order, of features present in the dominant feature sets of samples belonging to a particular class. While top row of Fig. 5 presents it for six staining pattern classes of MIVIA database, considering LBP at \({\mathcal {S}}_1\), first three graphs of bottom row report the same for Centromere class at \({\mathcal {S}}_2\), \({\mathcal {S}}_3\), and \({\mathcal {S}}_4\) for LBP, and last three graphs depict it for Centromere class considering \(\hbox {LBP}^{\mathrm{ri}}\), \(\hbox {LBP}^{\mathrm{riu2}}\) and CoALBP at \({\mathcal {S}}_1\). Results are reported for different values of energy \(\vartheta\), ranging in between 0.75 and 1.0. From the occurrence curves, it can be observed that as \(\vartheta\) increases, the area under the corresponding curve strictly increases, which is evident from the definition of \(\vartheta\). While low energy provides the dominant feature sets of samples to a restrictive representation, high energy signifies more information captured from the images and thus, more features are included in the dominant feature sets of the samples, leading to a descriptive representation of the sets. Although it seems that samples from the same class will have similar sets of dominant features, but in real-life problems, samples belonging to the same class exhibit a large degree of variations among themselves due to incompleteness in class definitions, overlapping characteristics of class boundaries, and presence of outlier and noise. Hence, the samples from same class are described with different sets of dominant features. As a consequence, probability of occurrence values of features in the dominant feature sets vary to a great extent. The features with high probability of occurrence values reflect common properties of the samples with respect to the class to which they belong. On the other hand, the features with relatively low probability of occurrence values signify the special properties of certain images, which are essential to distinguish the images properly.
Let us consider the occurrence curve for Centromere class considering LBP at \({\mathcal {S}}_1\) (first graph of top row of Fig. 5). Around \(\vartheta =0.8\), there is a sharp fall in the probability of occurrence curve, which implies that the dominant feature sets of samples corresponding to Centromere mainly contain the features which signify common characteristics of the samples, without considering the special properties of certain samples. For \(\vartheta =1.0\), the entire feature set gets selected as the dominant feature set for each sample of Centromere, which contradicts the concept of dominant feature set itself. The probability of occurrence graph for \(\vartheta =0.9\) reflects the presence of both common features with high probability of occurrence values and unique features with comparatively low occurrence values in the dominant feature sets of samples of the class. Hence, it is evident that low \(\delta\) value tends to complement the dominant feature sets of samples of Centromere, obtained for high \(\vartheta\) value. Now, in the proposed method, the class specific feature set \({\mathcal {N}}(\omega _i)\) corresponding to a particular class \(\omega _i\) is defined to contain those features which bear important information of most of the samples of \(\omega _i\) as well as provide significant characteristics of the class \(\omega _i\). So, \({\mathcal {N}}({\mathrm{Centromere}})\) can be appropriately formed with a high \(\vartheta\) value and a low \(\delta\) value. The inferences, drawn from the occurrence curves of LBP computed at \({\mathcal {S}}_1\) for the samples of Centromere, are equally true irrespective of the descriptors, scales and classes, which is evident from rest of the curves presented in Fig. 5. Thus, in the current study, the values of \(\vartheta\) and \(\delta\) are chosen to be 0.9 and 0.1, respectively, which imply that a particular class is represented by a set of features, which preserves 90% of total energy by at least 10% of the samples belonging to that class. The value of \(\vartheta =0.9\) also complies with that of the method proposed by Guo et al. [18].
Figure 6 presents the variation of classification accuracy with respect to both energy \(\vartheta\) and threshold \(\delta\), considering different kernels of SVM, namely, linear, polynomial, and radial basis function. Results are reported for \(\vartheta \in [0.75,1.00]\) with a gap of 0.05 and \(\delta \in [0.0,0.5]\) with a gap of 0.1. Both training–testing and tenfold CV are performed, considering both MIVIA and SNP databases. From all the results reported in Fig. 6, it can be seen that the classification accuracy of the proposed method increases with the increase in energy \(\vartheta\), while it decreases in increase of threshold \(\delta\). The proposed method attains better performance at \(\vartheta =0.90\) and \(\delta =0.1\), irrespective of the experimental set-up, classifiers, and data sets used.
3.5 Relevance of support vector machine
In the current study, the SVM with linear kernel is employed to evaluate the performance of the proposed texture descriptor selection method. In order to establish the importance of using SVM with linear kernel for classification, extensive experiments are carried out and corresponding results are reported in Table 5 with respect to different kernels of the SVM. Table 5 compares the classification accuracy (ACC) obtained using the proposed method at \(\vartheta =0.90\) and \(\delta =0.1\) with that of maximum accuracy (Best) obtained from all possible combinations of \(\vartheta\) and \(\delta\). In Table 5, both highest accuracy and lowest difference between best and obtained accuracy (\(\varDelta =\) Best − ACC) are marked in bold. Finally, the quality of each kernel is evaluated by the following measure:
As both lower value of \(\varDelta\) and higher value of ACC are desirable, the lower value of \(\varTheta\) indicates better classifier. From the results reported in Table 5, it can be seen that the SVM with linear kernel outperforms the SVM with other kernels for both training–testing and tenfold CV, on both MIVIA and SNP databases. Also, the accuracy obtained at \(\vartheta =0.90\) and \(\delta =0.1\) is comparable with the best accuracy in this case.
In order to assess the performance of the proposed method, different classifiers are employed to obtain the classification accuracy for both training–testing and tenfold CV using the feature set \({\mathcal {D}}\), obtained in (9). In this experiment, the accuracy obtained using SVM with linear kernel is compared with that of ELM [23], RBM [21], DRBM [25], FRBM [13], RF [4] and KNN [10], and the corresponding results are outlined in Table 6 for both MIVIA and SNP databases. The features extracted by the RBM are classified using SVM with linear kernel, while the value of K, for the KNN algorithm, is considered as 5 through extensive experimentation by varying it from 1 to 9.
From the results reported in Table 6, it can be noted that both RBM and proposed method with SVM perform better than other classifiers, in classifying staining patterns present in HEp-2 cell images of two data sets. Both ELM and FRBM also achieve satisfactory classification results, whereas DRBM, RF and KNN fail to identify the staining patterns of HEp-2 cell images. Since HEp-2 cell pattern classes are imbalanced in nature and histogram of textural patterns is considered as feature vector in the current study, the RF does not proved to be efficient in classifying staining pattern classes. However, if global features like DCT and FFT are considered to construct the RF classifier, its classification accuracy is increased to 25.61% and 29.16%, respectively, for MIVIA data set and 24.87% and 25.19%, respectively, for SNP database, in case of training–testing. In brief, the SVM with linear kernel exhibits better performance with reference to rest of the classifiers, irrespective of experimental set-up and data sets used. Hence, in the proposed work, the SVM with linear kernel is used to classify HEp-2 cell staining pattern classes.
3.6 Importance of rough hypercuboid approach
The proposed method assumes that the modalities with high relevance values are effective in differentiating the samples of class \(\omega _i\) from that of class \(\omega _r\). The relevance of a modality is computed using (16), based on hypercuboid equivalence partition matrix of rough hypercuboid approach. Based on the relevance measure \(\varGamma\), three pairwise class specific feature sets, corresponding to best three modalities, are considered to form the set \(\tilde{{\mathcal {N}}}_{ir}\) for each pair of classes \(\{\omega _i,\omega _r\}\). In order to establish the importance of \(\varGamma\) measure of rough hypercuboid approach, the relevance of pairwise class specific feature set, corresponding to each modality and each pair of classes \(\{\omega _i,\omega _r\}\), is computed and presented in Fig. 7a. The SVM with linear kernel is used to train the samples of \(\{\omega _i,\omega _r\}\), based on the pairwise class specific feature set corresponding to that modality. Finally, the SVM is used to predict the staining pattern classes of the samples belonging to the test set of each {\(\omega _i,\omega _r\)}. The classification accuracy on test data is presented in Fig. 7b. From Fig. 7a, b, it is evident that in most of the cases, a high \(\varGamma\) value corresponds to a high test accuracy, which indicates that \(\varGamma\) and accuracy values are highly correlated with each other. The values of both \(\varGamma\) and classification accuracy, obtained for different modalities corresponding to two representative class-pairs {Centromere, Cytoplasmic} and {Homogeneous, Cytoplasmic}, are plotted in Fig. 7c, d, respectively. The Pearson correlation coefficient \(\rho\) for the two aforementioned class-pairs are found to be 0.95 and 0.94, respectively.
In order to discard irrelevant and insignificant features from each pairwise class specific feature set, Step 6(ii) of the proposed algorithm uses maximum relevance-maximum significance (MRMS) criterion of feature selection [29]. Using the above criterion, the proposed method reduces the cardinality of feature set drastically, keeping the value of \(\varGamma\) unchanged. Table 7 establishes the importance of using MRMS criterion in the proposed method. The means and standard deviations of the cardinalities corresponding to initial and reduced pairwise class specific feature sets are reported in Table 7, which are computed considering all possible class-pairs and different scales of a particular descriptor. All the results indicate that MRMS criterion reduces the dimensions of feature sets to a great extent, especially for the descriptors with large number of features. Also, statistical significance analysis, based on both paired-t test and Wilcoxon signed rank test, reveals that the cardinality of reduced pairwise class specific feature set, corresponding to a particular descriptor, is significantly lower as compared to that of initially obtained feature set, irrespective of the descriptors used.
To compute the relevance of a class-pair specific feature set, several other feature evaluation measures such as class separability (CS) index [10], Davies-Bouldin (DB) index [7], and Dunn index [11] can be used. In order to establish the importance of rough hypercuboid (RH) approach over CS, DB and Dunn indices, extensive experiment is carried out on two data sets and corresponding results are reported in Table 8. From the results reported in Table 8, it can be seen that the proposed method using \(\varGamma\) measure, based on rough hypercuboid approach, performs better in most of the cases for both MIVIA and SNP databases, irrespective of experimental set-up used. It establishes the effectiveness of \(\varGamma\) measure, computed based on the concept of rough hypercuboid approach, in selecting the relevant modalities corresponding to each pair of classes.
3.7 Significance of class-pair specific modalities
In the proposed method, a modality refers to a specific local texture descriptor considered under a particular scale. Generally, a fixed set of modalities for all the classes is considered by the existing approaches for HEp-2 cell classification, while the proposed method selects class-pair specific modalities during the analysis of HEp-2 cell images. To establish the effectiveness of class-pair specific modalities over uniform modalities for all the classes, extensive experiment is carried out on two HEp-2 cell image databases, considering fifteen modalities corresponding to LBP, \(\hbox {LBP}^{\mathrm{ri}}\), \(\hbox {LBP}^{\mathrm{riu2}}\), and CoALBP, along with their four scales. Figures 8 and 9 and Table 9 present the comparative performance analysis between existing approaches and proposed method, considering both single and multiple modalities for two HEp-2 cell image databases. From the results reported in top row of Fig. 8, it can be seen that the proposed method attains highest classification accuracy of training–testing in all the cases, irrespective of the data sets, descriptors, scales and number of modalities used. The results reported in top row of Fig. 9 also confirm that the proposed method achieves highest F1 score in all the cases, except for CoALBP at \({\mathcal {S}}_2\) on MIVIA dataset.
For tenfold CV, the comparative performance analysis is reported in middle and bottom rows of Fig. 8 for classification accuracy and Fig. 9 for F1 score using box-and-whisker plots and in Table 9 based on mean, median, standard deviation and p-values computed through both paired-t test and Wilcoxon signed rank test. All the results reported here confirm that the proposed method provides higher mean and median values of both classification accuracy and F1 score than that of all other methods for the two data sets, except CoALBP at \({\mathcal {S}}_{124}\) for SNP database. Also, the proposed method attains significantly better accuracy as well as F1 score in 78 cases each, considering 95% confidence level, out of total 88 cases each, and better but not significant in 8 cases each. All the results establish the fact that both classification accuracy and F1 score can be increased significantly by considering class-pair specific modalities, instead of taking into account a fixed set of modalities for all the classes.
3.8 Comparative performance analysis
The performance of the proposed approach is compared with that of several existing HEp-2 cell classification methods due to Cordelli and Soda [5], Strandmark et al. [46], and Wiliem et al. [54]. Results are reported in Fig. 10, considering ICPR 2012 contest protocol of cell level classification for MIVIA database [15] and fivefold training–testing for SNP database [54]. From the results reported in Fig. 10, it can be seen that the proposed method performs better than all other approaches for MIVIA database. In this regard, it may be noted that some other techniques, participated in ICPR 2012 contest, such as the methods of Di Cataldo et al. [8], Ersoy et al. [12], Ghosh and Chaudhary [17], and Snell et al. [42] achieve classification accuracy of 48.5%, 49.2%, 59.8% and 44.6%, respectively, on MIVIA database, which are lower than 63.9% accuracy obtained by the proposed method. For SNP database [54], the proposed method provides better performance than the method of Cordelli and Soda [5] and comparable performance with respect to the method due to Strandmark et al. [46]. However, the performance of the method due to Wiliem et al. [54] on their SNP database [54] is significantly better than all other approaches, whereas it is very poor on MIVIA database as compared to that of the proposed method.
Finally, the performance of the proposed method is extensively compared with that of (1) two effective textural feature extraction methods, namely, dominant LBP (DLBP) [26] and discriminative features for texture description (DFTD) [18]; (2) four statistical multimodal data integration methods, namely, canonical correlation analysis (CCA) [22], regularized CCA (RCCA) [52], CuRSaR [28] and FaRoC [30]; and (3) a CCA based method for HEp-2 cell staining pattern recognition, called CanSuR [31]. In [31], the SVM with RBF kernel is considered to compute classification accuracy, while SVM with linear kernel is used for rest of the methods. The results, corresponding to both training–testing and tenfold CV, are reported in Fig. 11 and Table 10 with respect to accuracy and F1 score. The performance of the DLBP and DFTD is evaluated for both \({\mathcal {S}}_{123}\) and \({\mathcal {S}}_{124}\), while four multimodal data integration methods and CanSuR, as suggested in [31], consider \({\mathcal {S}}_{2}\) and \({\mathcal {S}}_{3}\) of \(\hbox {LBP}^{\mathrm{ri}}\), for modality integration.
All the results reported in top row of Fig. 11 confirm that the proposed method attains maximum classification accuracy as well as F1 score for both the data sets. From the results reported in middle and bottom rows of Fig. 11, corresponding to tenfold CV on MIVIA and SNP databases, respectively, it is seen that the proposed method provides highest values of mean and median in both the cases. The statistical significance analysis, reported in Table 10 based on both paired-t test and Wilcoxon signed rank test, reveals that the proposed algorithm achieves significantly lower p values in 68 cases out of total 72 cases, and better but not significant p values in remaining 4 cases, with respect to both classification accuracy and F1 score.
The better performance of the proposed method is achieved due to the following facts:
-
1.
the proposed method considers class-pair specific modalities for analyzing HEp-2 cell images, rather than considering a fixed set of modalities for all the staining pattern classes;
-
2.
the relevance of each modality is evaluated through hypercuboid equivalence partition matrix of rough hypercuboid approach, which can approximate the sample categories of staining patterns efficiently;
-
3.
a set of dominant features is extracted under each relevant modality, based on the probability of texture pattern occurrence; and
-
4.
the maximum relevance-maximum significance criterion, used in the proposed method, facilitates identification of a reduced set of discriminative features for HEp-2 cell classification.
In effect, the proposed method provides significantly better performance as compared to existing methods.
4 Conclusion
The main contribution of this paper lies in developing a methodology for the diagnosis of connective tissue diseases, by recognizing staining patterns present in HEp-2 cell IIF images. The proposed method judiciously integrates the merits of rough hypercuboid approach and local textural descriptors. The rough hypercuboid approach facilitates selection of pairwise class specific important modalities, while relevant and significant features under selected modalities are obtained, based on the probability of occurrence of a feature in a class and maximum relevance-maximum significance criterion used in feature selection. Finally, support vector machine with linear kernel is used to recognize one of the known staining patterns present in IIF images. The effectiveness of the proposed method, along with a comparison with related approaches, has been demonstrated on two benchmark HEp-2 cell image databases.
In order to address the shortcomings of manual test procedure, one can use the proposed system to automatically determine the patterns in given HEp-2 cell images of a specimen. It may allow performing a pre-selection of the cases to be examined, which will enable physician to focus attention only on most relevant cases.
References
Agmon-Levin N, Damoiseaux J, Kallenberg C, Sack U, Witte T, Herold M, Bossuyt X, Musset L, Cervera R, Plaza-Lopez A, Dias C, Sousa MJ, Radice A, Eriksson C, Hultgren O, Viander M, Khamashta M, Regenass S, Andrade LEC, Wiik A, Tincani A, Rönnelid J, Bloch DB, Fritzler MJ, Chan EKL, Garcia-De La Torre I, Konstantinov KN, Lahita R, Wilson M, Vainio O, Fabien N, Sinico RA, Meroni P, Shoenfeld Y (2014) International recommendations for the assessment of autoantibodies to cellular antigens referred to as anti-nuclear antibodies. Ann Rheum Dis 73(1):17–23
Agmon-Levin N, Shapira Y, Selmi C, Barzilai O, Ram M, Szyper-Kravitz M, Sella S, Katz BS, Youinou P, Renaudineau Y, Larida B, Invernizzi P, Gershwin ME, Shoenfeld Y (2010) A comprehensive evaluation of serum autoantibodies in primary biliary cirrhosis. J Autoimmun 34(1):55–58
Bianconi F, Fernandez A, Mancini A (2008) Assessment of rotation-invariant texture classification through gabor filters and discrete fourier transform. In: Proceedings of the 20th international congress on graphical engineering
Breiman Leo (2001) Random forests. Mach Learn 45(1):5–32
Cordelli E, Soda P (2011) Color to grayscale staining pattern representation in IIF. In: Proceedings of the 24th IEEE international symposium on computer-based medical systems, pp 1–6
Dalal N, Triggs B (2005) Histograms of oriented gradients for human detection. In: Proceedings of IEEE computer society conference on computer vision and pattern recognition, vol 1, pp 886–893
Davies DL, Bouldin DW (1979) A cluster separation measure. IEEE Trans Pattern Anal Mach Intell 1:224–227
Di Cataldo S, Bottino A, Ficarra E, Macii E (2012) Applying textural features to the classification of HEp-2 cell patterns in IIF images. In: Proceedings of the 21st international conference on pattern recognition, pp 3349–3352. IEEE
Di Cataldo S, Bottino A, Islam I, Vieira TF, Ficarra E (2014) Subclass discriminant analysis of morphological and textural features for HEp-2 staining pattern classification. Pattern Recognit 47(7):2389–2399
Duda RO, Hart PE (1973) Pattern classification and scene analysis. Wiley, Hoboken
Dunn JC (1974) A fuzzy relative of the ISODATA process and its use in detecting compact, well-separated clusters. J Cybern 3:32–57
Ersoy I, Bunyak F, Peng J, Palaniappan K (2012) HEp-2 cell classification in IIF images using shareboost. In: Proceedings of the 21st international conference on pattern recognition. IEEE, pp 3362–3365
Feng S, Chen CLP (2016) A fuzzy restricted boltzmann machine: novel learning algorithms based on the crisp possibilistic mean value of fuzzy numbers. IEEE Trans Fuzzy Syst 26(1):117–130
Foggia P, Percannella G, Soda P, Vento M (2010) Early experiences in mitotic cells recognition on HEp-2 slides. In: Proceedings of the 23rd IEEE international symposium on computer-based medical systems, pp 38–43
Foggia P, Percannella G, Soda P, Vento M (2013) Benchmarking HEp-2 cells classification methods. IEEE Trans Med Imaging 32(10):1878–1889
Friou GJ, Finch SC, Detre KD, Santarsiero C (1958) Interaction of nuclei and globulin from lupus erythematosis serum demonstrated with fluorescent antibody. J Immunol 80(4):324–329
Ghosh S, Chaudhary V (2012) Feature analysis for automatic classification of HEp-2 florescence patterns: computer-aided diagnosis of auto-immune diseases. In: Proceedings of the 21st international conference on pattern recognition. IEEE, pp 174–177
Guo Y, Zhao G, Pietikäinen M (2012) Discriminative features for texture description. Pattern Recognit 45(10):3834–3843
Guo Z, Zhang L, Zhang D (2010) A completed modeling of local binary pattern operator for texture classification. IEEE Trans Image Process 19(6):1657–1663
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern 3(6):610–621
Hinton GE, Osindero S, Teh Y-W (2006) A fast learning algorithm for deep belief nets. Neural Comput 18(7):1527–1554
Hotelling H (1936) Relations between two sets of variates. Biometrika 28(3/4):321–377
Huang G-B, Zhu Q-Y, Siew C-K (2006) Extreme learning machine: theory and applications. Neurocomputing 70(1):489–501
Karargyris A, Siegelman J, Tzortzis D, Jaeger S, Candemir S, Xue Z, Santosh KC, Vajda S, Antani S, Folio L, Thoma G (2016) Combination of texture and shape features to detect pulmonary abnormalities in digital chest X-rays. Int J Comput Assist Radiol Surg 11(1):99–106
Larochelle H, Mandel M, Pascanu R, Bengio Y (2012) Learning algorithms for the classification restricted boltzmann machine. J Mach Learn Res 13(Mar):643–669
Liao S, Law MWK, Chung ACS (2009) Dominant local binary patterns for texture classification. IEEE Trans Image Process 18(5):1107–1118
Maji P (2014) A rough hypercuboid approach for feature selection in approximation spaces. IEEE Trans Knowl Data Eng 26(1):16–29
Maji P, Mandal A (2017) Multimodal omics data integration using max relevance-max significance criterion. IEEE Trans Biomed Eng 64(8):1841–1851
Maji P, Paul S (2011) Rough set based maximum relevance-maximum significance criterion and gene selection from microarray data. Int J Approx Reason 52(3):408–426
Mandal A, Maji P (2018) FaRoC: fast and robust supervised canonical correlation analysis for multimodal omics data. IEEE Trans Cybern 48(4):1229–1241
Mandal A, Maji P (2019) CanSuR: a robust method for staining pattern recognition of HEp-2 cell IIF images. Neural Comput Appl. https://doi.org/10.1007/s00521-019-04108-w
Mariz HA, Sato EI, Barbosa SH, Rodrigues SH, Dellavance A, Andrade LE (2011) Pattern on the antinuclear antibody-HEp-2 test is a critical parameter for discriminating antinuclear antibody-positive healthy individuals and patients with autoimmune rheumatic diseases. Arthritis Rheum 63(1):191–200
Nixon M, Aguado SA (2012) Feature extraction and image processing for computer vision. Academic Press, Cambridge
Nosaka R, Fukui K (2014) HEp-2 cell classification using rotation invariant co-occurrence among local binary patterns. Pattern Recognit 47(7):2428–2436
Nosaka R, Ohkawa Y, Fukui K (2012) Feature extraction based on co-occurrence of adjacent local binary patterns. In: Proceedings of the advances in image and video technology, pp 82–91
Ojala T, Pietikainen M, Harwood D (1994) Performance evaluation of texture measures with classification based on Kullback discrimination of distributions. In: Proceedings of the 12th IAPR international conference on pattern recognition, conference a: computer vision and image processing, pp 582–585
Ojala T, Pietikainen M, Maenpaa T (2002) Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans Pattern Anal Mach Intell 24(7):971–987
Ojala T, Pietikainen M, Harwood D (1996) A comparative study of texture measures with classification based on feature distributions. Pattern Recognit 29(1):51–59
Pinheiro A (2009) Image descriptors based on the edge orientation. In: Proceedings of the 4th international workshop on semantic media adaptation and personalization, pp 73–78
Santosh KC, Antani S (2017) Automated chest X-ray screening: can lung region symmetry help detect pulmonary abnormalities? IEEE Trans Med Imaging 37(5):1168–1177
Sim D-G, Kim H-K, Park R-H (2004) Invariant texture retrieval using modified Zernike moments. Image Vis Comput 22:331–342
Snell V, Christmas W, Kittler J (2012) Texture and shape in fluorescence pattern identification for auto-immune disease diagnosis. In: Proceedings of the 21st international conference on pattern recognition. IEEE, pp 3750–3753
Snell V, Christmas W, Kittler J (2014) HEp-2 fluorescence pattern classification. Pattern Recognit 47(7):2338–2347
Solomon DH, Kavanaugh AJ, Schur PH (2002) Evidence-based guidelines for the use of immunologic tests: antinuclear antibody testing. Arthritis Rheum 47(4):434–444
Stoklasa R, Majtner T, Svoboda D (2014) Efficient k-NN based HEp-2 cells classifier. Pattern Recognit 47(7):2409–2418
Strandmark P, Ulén J, Kahl F (2012) HEp-2 staining pattern classification. In: Proceedings of the 21st international conference on pattern recognition. IEEE, pp 33–36
Tan EM (1989) Antinuclear antibodies: diagnostic markers for autoimmune diseases and probes for cell biology. Adv Immunol 44:93–151
Theodorakopoulos I, Kastaniotis D, Economou G, Fotopoulos S (2014) HEp-2 cells classification via sparse representation of textural features fused into dissimilarity space. Pattern Recognit 47(7):2367–2378
Unser M (1986) Local linear transforms for texture measurements. Signal Process 11(1):61–79
Vajda S, Karargyris A, Jaeger S, Santosh KC, Candemir S, Xue Z, Antani S, Thoma G (2018) Feature selection for automatic tuberculosis screening in frontal chest radiographs. J Med Syst 42(8):146
Vapnik V (1995) The nature of statistical learning theory. Springer, New York
Vinod HD (1976) Canonical ridge and econometrics of joint production. J Econom 4(2):147–166
Wiik AS (2005) Anti-nuclear autoantibodies: clinical utility for diagnosis, prognosis, monitoring, and planning of treatment strategy in systemic immunoinflammatory diseases. Scand J Rheumatol 34(4):260–268
Wiliem A, Wong Y, Sanderson C, Hobson P, Chen S, Lovell BC (2013) Classification of human epithelial type 2 cell indirect immunofluoresence images via codebook based descriptors. In: Proceedings of the IEEE workshop on applications of computer vision, pp 95–102
Acknowledgements
This publication is an outcome of the R&D work undertaken in the project under the Visvesvaraya Ph.D. Scheme of Ministry of Electronics and Information Technology, Government of India, being implemented by Digital India Corporation. The authors would like to thank Ankita Mandal of Indian Statistical Institute, Kolkata for her valuable experimental support.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kumar, D., Maji, P. Selection of relevant texture descriptors for recognition of HEp-2 cell staining patterns. Int. J. Mach. Learn. & Cyber. 11, 2127–2147 (2020). https://doi.org/10.1007/s13042-020-01106-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-020-01106-6