Abstract
Many soil Actinobacteria are potent producers of extracellular enzymes decomposing lignocellulose. Four strains of Actinobacteria with a high potential to hydrolyse cellulose and hemicellulose were identified among environmental isolates. The strains were grown on raw lignocellulosic substrates (olive pomace, oat flakes, sawdust, and wheat straw) under submerged fermentation in a laboratory scale. Modified Melin Norkrans Medium amended with raw lignocellulosic substrates as carbon sources (0.5%) was used to enhance lignocellulosic biomass decomposition. Three strains belonged to the genus Streptomyces and one strain to the genus Mycobacterium. Annotation of genomes showed high proportion of genes encoding for carbohydrate-active enzymes in Streptomyces sp. GESEQ-4 (537, i.e. 6% of 8404 genes), Streptomyces sp. GESEQ-13 (351 (5%) of 6705 genes), Streptomyces sp. GESEQ-35 (608 (6%) of 9788 genes), and Mycolicibacterium fortuitum subsp. fortuitum GESEQ-9 (222 (3%) of 6405 genes). These included plant cell wall-degrading enzymes belonging to the families GH1, GH2, GH3, GH5, GH6, GH9, GH10, GH12, GH16, GH26, GH30, GH39, GH48, GH51, and GH74, of which GH1, GH2, GH3, GH5, GH6, and GH16 were found in all four genomes. Assays for cellulose and hemicellulose degrading extracellular enzymes confirmed the ability of the isolates to decompose cellulose and hemicellulose. The highest endo-cleaving enzyme activities were produced by the strain Steptomyces sp. GESEQ-4 DSM 106287. Our results provide new perspectives into the enzymatic array by which the Actinobacteria break down complex lignocellulosic biomass. It is crucial to assess the genome to determine enzyme function as well as the enzyme families responsible for the degradation process in Actinobacteria. The potential degradation functions for these actinobacterial strains were validated by testing their cellulolytic and hemicellulolytic activities with various lignocellulosic substrates.
Graphic Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Statement of Novelty
We confirm that this work is original and has neither been published elsewhere nor is currently under consideration for publication. In this contribution, we address actinobacterial strains as enzyme producers involved in the decomposition of cellulose and hemicellulose. We sequenced their genomes and identified their corresponding genes encoding enzymes. Since the decomposition of lignocellulosic biomass is a complex process, we have exploited genome sequencing and enzyme assays to reveal the presence of a complex enzymatic machinery required for raw lignocellulose material.
Introduction
The photosynthetically derived polymers cellulose and hemicellulose represent the most abundant renewable resources on Earth. These materials are derived from fixed carbon dioxide and they form the main building blocks of lignocellulosic biomass [1]. Agriculture, forestry, industry, and urban waste are promising clean and sustainable sources of lignocellulose. In recent years, increasing attention has been devoted to the production of bioethanol from low-cost lignocellulose contained in agri-food residues (zero waste energy) using enzymes to access the energy embedded in the C-H-bonded molecules of cellulose and hemicellulose polymers [2].
Although many Actinobacteria are as efficient at degrading lignocellulosic biomass as fungi [3, 4], their enzymatic systems are so far underrepresented in literature [5]. Both groups share almost the same morphological features including hyphal growth and spore formation [2] which allows them to proliferate in bulky substrate such as dead plant biomass. Actinobacteria have been observed in a broad spectrum of habitats which correlates with their metabolic diversity in decomposing various organic components [6].
Carbohydrate-active enzymes (CAZymes) include glycoside hydrolases (GHs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), glycosyltransferases (GTs), auxiliary activities (AAs) and non-catalytic, carbohydrate-binding modules (CBMs) [7]. Within each group, enzymes are distinguished into families on the basis of the amino acid sequence in the CAZy database (for Carbohydrate Active enZymes) (http://www.cazy.org/) [8]. GHs are enzymes responsible for cleaving glycosidic bonds and are often associated to CBMs that facilitate specific substrate binding. CBMs are typical to aerobic bacteria and fungi, and their role may ensure the efficient processing of the polymeric substrate compared to cellulosome systems of anaerobic microbial degraders of cellulose [9]. Cellulose breakdown can be either partial, resulting in the formation of cellodextrins, which are oligosaccharides formed of two or more glucose monomers, or complete, resulting in separated glucose molecules [10].
This study aims to relate the enzymatic activity and lignocellulose hydrolysis in the isolates of actinobacterial lignocellulose degraders during their growth on various lignocellulose raw materials and to compare the diversity of CAZymes within their genomes.
Material and Methods
Actinobacterial Strains and Culture Conditions
Four (hemi)cellulolytic Actinobacteria strains were isolated from soil and compost, GESEQ-4, isolated on Starch Casein Agar [11] from an agricultural soil (N 35° 30′ 46.9″, E 6° 15′ 31.812″) on August 8th, 2014, GESEQ-9 isolated on Kuster’s Agar [11] and GESEQ-13 on humic acid vitamin agar [12], from a compost supplied by Biocompost (Algeria) on April 5th, 2014, and GESEQ-35 isolated on Kuster’s Agar [11] from a forest soil (N 35° 34′ 19.81″, E 6° 11′ 40.634″) on August 8th, 2014. The inocula were grown for seven days in 250 mL flasks containing 50 mL (g L−1): glucose, 4; malt extract, 10; yeast extract, 4; (pH 7.2) at 25 °C. Four distinct substrates were tested as carbon sources: wheat straw, olive pomace, oat flakes, and sawdust. The residues were milled to a fine powder using an Analytical Mill (IKA-A 11 Basic Mill, Germany). A total of 2.0 g of each substrate was added to 100 mL of Modified Melin Norkrans Medium (MNM) consisting of (g L−1): NH4H2PO4, 2, KH2PO4, 0.6, MgSO4.7H2O, 0.5, K2HPO4, 0.4 and 10 mL L−1 of trace elements solution containing (g L−1): CaCl2⋅2H2O, 7.40; FeSO4⋅7H2O, 1.20; ZnSO4⋅7H2O, 0.66; MnSO4⋅4H2O, 0.5; CoCl2⋅6H2O, 0.1 at pH 7.0 followed by sterilization at 121 °C for 15 min. The cultures were incubated at 25 °C in the dark for 21 days. A 25 mL aliquot of hydrolysed biomass was centrifuged at 4 °C at 6000x g for 20 min (Hermle, Z 323K, Germany). The strains were deposited at the DSMZ, as GESEQ-4 (= DSM 106287), GESEQ-9 (= DSM 107958), GESEQ-13 (DSM 105759) and GESEQ-35 (= DSM 105785).
Enzyme Assays of Cellulases and Hemicellulases
The activities of cellulases (endoglucanase, cellobiohydralase and β-glucosidase) and hemicellulases (endoxylanase, β-xylosidase, α-glucuronidase, α-glucosidase, β-galactosidase, β-mannosidase and α-arabinosidase) were measured after 21 days of incubation as described previously [13, 14]. Following the protocols of the substrate supplier (Megazyme, Ireland), endo-cleaving enzymes were quantified using azo-dyed carboxymethyl cellulose and birchwood xylan. The samples were collected after centrifugation of the culture broth at 4000×g for 20 min (HERMLE Z 324, Germany) to obtain the culture supernatant which is tested for enzymatic activity. A reaction mixture containing 0.15 mL of substrate and 0.15 mL of sample was prepared. After 60 min incubation at 40 °C, the reaction was stopped by adding 0.75 mL ethanol followed by 10 s of shaking and spinning down precipitate by 10 min of centrifugation at 1000x g. The enzyme activity was measured at 595 nm based on standard curves correlating dye and reducing sugar release. The activity of cellulases and hemicellulases were determined by measuring the fluorescence of 4-methylumbelliferone (MUF)-linked substrates, as described previously [13]. Fluorescence was measured after 5 and 125 min using a 96-well microplate incubated at 40 °C. Enzyme activity is defined as the amount of enzyme that releases 1 nmol of reducing sugars per minute. All measurements were conducted in triplicate. Statistical analysis was performed using the R software package (version 3.6.2; https://www.r-project.org/). Kruskal–Wallis test with Bonferroni correction was performed for pairwise comparisons of all possible combinations of categories related to enzyme activity with different carbon sources.
Genome Sequencing and Annotation
DNA was extracted from liquid axenic cultures using UltraClean Microbial DNA Isolation Kit (MoBio) according to manufacturer's protocol, with the modification that the cells were shaken in a lysis buffer on FastPrep 5 m/s for 2 × 30 s. Sanger sequencing to confirm the taxonomy of isolates was done in the Centre for DNA Sequencing, Prague, Czech Republic, using bacterial primer 1100R. The concentration of extracted DNA was measured on a Qubit 2.0 Fluorometer (Life Technologies, USA) using the Qubit dsDNA BR kit [15]. At least 500 ng of DNA was ligated using TruSeq Nano DNA Sample Preparation Kit and sequenced on Illumina MiSeq with 2 × 250 pair-end runs. Genome assembly was done using either Velvet (1.2.10) [16] or SPAdes (3.9.0) algorithm [17]. Genes were predicted and annotated on the RAST server [18]. GH families were annotated using dbCAN database (HMMdb 5.0 release) [19]. Additional genome annotation for CBM-enzyme couples identification was performed using Prokka gene prediction [20] followed by a search with dbCAN HMMs of CAZy families combined with a BLAST search with nucleotide sequences from the CAZy homepage (update 10/2020). The raw data, genome assembly files and 16S rRNA gene sequences are available with the accession numbers GESEQ-4 (PRJNA702041, JAFFSU000000000, MF150027), GESEQ-9 (PRJNA702043, JAFFSW000000000, MF150030), GESEQ-13 (PRJNA702042, JAFFSV000000000, MF150029) and GESEQ-35 (PRJNA702046, JAFFSX000000000, MF150031). To assess the phylogenetic placement of the cultivated strains, all RefSeq genomes of the genera Streptomyces (n = 2,143) and Mycolicibacterium (n = 303) were downloaded in February 2021 using NCBI-genome-download 0.3.0 script (https://github.com/kblin/ncbi-genome-180 download). GToTree v1.5.46 [21] together with Prodigal [22], HMMER3 [23], Muscle [24], trimAI [25], FastTree2 [26] and GNU Parallel [27] were used to infer phylogenetic trees of the two genera based on a set of 74 bacterial single-copy gene HMM profiles. Trees were visualized using iTOL [28].
Results and Discussion
(Hemi)cellulose hydrolysis by the Actinobacteria is highlighted by comparative genomics and supported by biochemical measurements of the extracellular enzymes produced into the culture liquid. Genomic information collected from Actinobacteria stresses their potential for lignocellulosic biomass decomposition. All the sequenced genomes of the strains have large genomes and a high G+C content (Table 1), which are common characteristics of Actinobacteria [29].
(Hemi)cellulolytic Potential of the Actinobacterial Strains on Biomass
Maximal endoglucanase production, 29.6 U mL−1, was observed when sawdust as a sole carbon source was used with the strain Streptomyces sp. GESEQ-4. This strain also exhibited the highest arabinosidase activity, 249 U mL−1, on oat flakes (Fig. 1). Sawdust induced the highest release of endoxylanase, 29.2 U mL−1, in Streptomyces sp, GESEQ-35. The highest activities of cellobiohydrolase (258 U mL−1), β-glucosidase (994 U mL−1), β-xylosidase (626 U mL−1), β-galactosidase (1692 U mL−1), and α-glucosidase (2106 U mL−1) were obtained on wheat straw with strain GESEQ-13. Wheat straw induced the highest activities of β-mannosidase (55.0 U mL−1) and α-glucuronidase (163 U mL−1) with strain GESEQ-35.
For strain GESEQ-4, the enzymatic activity of β-xylosidase activity was higher in wheat straw compared to olive pomace (p = 0.039) and cellobiohydrolase in wheat straw versus oat flakes (p = 0.019). For strain GESEQ-13 the activities of α-glucosidase (p = 0.013), arabinosidase (p = 0.012), β-galactosidase (p = 0.013), 1-4-β-glucosidase (p = 0.013), β-xylosidase (p = 0.013), and cellobiohydrolase (p = 0.019) were significantly different between wheat straw versus oat flakes. For strain GESEQ-35 the activities of α-d-Glucuronidase (p = 0.039), β-galactosidase (p = 0.013), 1–4-β-glucosidase (p = 0.019) and cellobiohydrolase (p = 0.019) were significantly different between wheat straw and olive pomace, whereas the activity of endoxylanase was significantly different between wheat straw and sawdust (p = 0.039). For strain GESEQ-9 the activity of α-glucosidase differed significantly between olive pomace and sawdust (p = 0.013), and the activity of 1-4-β-glucosidase differed significantly between wheat straw and oat flakes (p = 0.039).
Genome Sequencing and Annotation
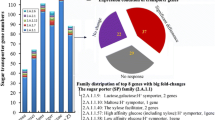
CAZyme identification in the genomes was used to describe the potential of the Actinobacteria to degrade cellulose and hemicelluloses. A set of genes encoding enzymes involved in lignocellulose degradation were predicted by annotating the CAZyme profiles (Table 2). Figure 2 comprises all the CAZymes identified in the genomes. With a larger genome size of over 10 Mbp, Streptomyces sp. GESEQ-35 encodes the highest number of CAZymes (608) and GHs (262). In general, Streptomyces strains showed better enzyme production, which was also confirmed by higher number of GHs as well as more pronounced visual growth on biomass (Fig. 3). Main cellulolytic families GH1, GH3, GH5, GH6 were detected in all the four actinobacterial genomes. GH9, GH12 and GH48 were identified in Streptomyces strains but not in Mycolicibacterium fortuitum subsp. fortuitum GESEQ-9. Cellulose degradation through lytic polysaccharide monooxygenases (LPMOs, AA10) and chitin (AA10 and GH18) is also predicted in the Streptomyces strains. Hemicellulolytic enzymes GH2, GH16 were annotated in the four genomes. Xylanases/xyloglucanases (GH10), mannanases (GH26), α-glucanases (GH31), (hemi)cellulases (GH36, GH43, GH51, GH74, and GH95) were not identified in Mycolicibacterium fortuitum subsp. fortuitum GESEQ-9. Endoxylanase (GH11) and hemicellulases (GH39) were identified in Streptomyces sp. GESEQ-4 and Streptomyces sp. GESEQ-13. (Hemi)cellulases (GH30) were identified in Streptomyces sp. GESEQ-4 and Streptomyces sp. GESEQ-35. GH54 with α-L-arabinosidase and/or β-xylosidase activity was detected in Streptomyces sp. GESEQ-4. α-1,4-galactosidase (GH27) and α-fucosidase (GH29) were encoded in both Streptomyces sp. GESEQ-4 and Streptomyces sp. GESEQ-35. Polygalacturonases (GH28), β-1,4-galactosidase (GH35 and GH42), α-1,4-galactosidase (GH36) and α-fucosidase (GH95) were encoded in the three Streptomyces strains. α-1,4-galactosidase or α-amylase (GH57) was only identified in Mycolicibacterium fortuitum subsp. fortuitum GESEQ-9 and α-1,4-galactosidase (GH97) in Streptomyces sp. GESEQ-13. Xylan esterases (CE1, CE2, CE3, CE4, and CE7) were identified in all actinobacterial strains, except for CE2 missing in Streptomyces sp. GESEQ-35 and Mycolicibacterium fortuitum subsp. fortuitum GESEQ-9. Although, the enzymatic degradation of other biomass compounds, including lignocellulosic biomass (lignin and pectin) and chitin was not verified by enzyme assays measurements, CAZyme families known in their biotransformation were detected at the genome level. In that respect, vanillyl-alcohol oxidase (AA4) and benzoquinone reductase (AA6) associated with lignin degradation were identified among the strains.
There are very few studies concerning bacterial degradation of lignin [3], however, recently discovered enzymes from bacteria such as Streptomyces sp. have been useful for delignification [30, 31]. Families involved in pectin degradation PL1, PL3 and PL9 and pectin methylesterase (CE8) were predicted except for GESEQ-9. Although the pectin content in plant biomass is typically lower than that of cellulose and hemicellulose, interest of pectin degradation has grown recently. Pectinases have also emerged a class of enzymes with particular interest in the degradation of pectin, along with their role in degradation of other polysaccharides [32]. α-1,4-glucosidase, α-rhamnosidase, β-amylase, or α-amylase (GH13), glucoamylase (GH15), chitinase (GH23) and α-1,4-glucosidase (GH63) were encoded in the four studied strains, while α-rhamnosidases (GH2 and GH78), were detected only in the Streptomyces strains, and α-rhamnosidase (GH106) in GESEQ-4 and GESEQ-35. In addition to lignocellulosic decomposing enzymes, GH annotation also showed the capacity of the strains to produce several other enzymes, such as amylases, proteases, chitinases, and pectinases.
Genomics, along with genome sequencing, is a powerful tool for exploring lignocellulose enzymes. To complement genomic analysis, a biochemical analysis of the enzymes in question is required to determine their catalytic efficiency on natural substrates for CAZymes [33, 34]. Annotations on CAZyme show that the isolated Actinobacteria strains contain a broad range of enzymes designed to break down both cellulose and hemicellulose. Enzymes of the CAZy family are in accordance with the hydrolysis profile of the studied actinobacterial strains. We have found that GESEQ-4 and GESEQ-35 seem to be the dominant contributors to the numbers of genes encoding enzymes in lignocellulose degradation, while GESEQ-9 and GESEQ-13 appear to be less generalistic degraders. Phylogenetic clustering in the context of whole genus shows strain GESEQ-9 to be closely related with Mycolicibacterium fortuitum (Fig. 4). Streptomyces strains GESEQ-35 and GESEQ-4 cluster together in a separate clade from GESEQ-13: these strains are not closely related to identified Streptomyces species, which have higher numbers of sequenced genomes.
Phylogenetic trees based on 74 bacterial single-copy genes for the genus Streptomyces (A, n = 2,143 RefSeq genomes) and Mycolicibacterium (B, n = 303 RefSeq genomes). Positions of strains cultivated within this study are denoted as circles. Related taxa with multiple sequenced genomes are described with species names
Despite belonging to the same genus of Streptomyces, the types and numbers of enzyme families can differ drastically between bacterial strains and species [35]. In this way, some bacterial strains might possess a more specialized set of enzymes. Therefore, it is important to consider the sequences of closely related species to expand the number of valuable carbohydrate-producing bacteria that are efficient at degrading lignocellulose. In the study of Berlemont et al., Streptomyces strains have been shown to be able to utilize variable carbon sources, while Mycobacterium strains with few GHs do have a decreased capacity for polysaccharide deconstruction because they depend on their hosts for resources [36, 37]. Streptomyces biocatalytic enzymes have been studied extensively, to the point where they are now commercially available [4]. However, the relative contributions of the various enzymes responsible for modifying plant cell walls are not yet fully appreciated. A reliable model has not yet been developed to explain the evolutionary development of actinobacterial enzyme machinery [38]. These enzymes typically include multifunctional domains, each composed of two or more CBMs. Indeed, CBM2, CBM32, CBM48 and CBM50 have been annotated in the four genomes. The genomes comprised many couples of GH, AA, CE, PL with CBMs (Supplementary Material). However, most of the other CBMs such as CBM4 and CBM6 were only encoded in Streptomyces genomes. Some CBMs were strain specific, namely CBM14 in the Mycobacterium strain, CBM30 in GESEQ-35 and CBM46 in GESEQ-4. CBM3 and CBM11 were only detected in GESEQ-13 and GESEQ-35. CBMs do not possess their own catalytic activity, but they are still highly important because they bind the catalytic domain of the enzyme to cellulose and enhance enzyme–substrate interaction [7, 39].
Conclusion
Genome sequencing indicates that the strains contain strong candidates for biotechnologically important cellulose-degrading enzymes such as GH3 and GH12. As a result, actinobacterial strains studied here can be added to a list of microorganisms that convert biomass to sugars, as seen from an industrial perspective. Genomic studies of Actinobacteria also hold the promise enabling bacterial strains to be engineered to generate novel enzymes and cocktail enzymes to degrade lignocellulosic biomass.
References
Ahmed, A.A.Q., Babalola, O.O., McKay, T.: Cellulase- and xylanase-producing bacterial isolates with the ability to saccharify wheat straw and their potential use in the production of pharmaceuticals and chemicals from lignocellulosic materials. Waste Biomass Valorization 9, 765–775 (2018). https://doi.org/10.1007/s12649-017-9849-5
Leo, V. V., Asem, D., Zothanpuia, Singh, B.P.: Actinobacteria: A highly potent source for holocellulose degrading enzymes. In: Singh, B.P., Gupta, V.K., Passari, A.K. (eds.) New and Future Developments in Microbial Biotechnology and Bioengineering: Actinobacteria: Diversity and Biotechnological Applications, pp. 191–205. Elsevier, Amsterdam (2018)
Větrovský, T., Steffen, K.T., Baldrian, P.: Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil Actinobacteria. PLoS ONE 9, e89108 (2014). https://doi.org/10.1371/journal.pone.0089108
López-Mondéjar, R., Algora, C., Baldrian, P.: Lignocellulolytic systems of soil bacteria: a vast and diverse toolbox for biotechnological conversion processes. Biotechnol. Adv. 37(6), 107374 (2019)
Kameshwar, S., Qin, W.: Recent developments in using advanced sequencing technologies for the genomic studies of lignin and cellulose degrading microorganisms. Int J Biol Sci. 12(2), 156 (2016). https://doi.org/10.7150/ijbs.13537
Anteneh, Y.S., Franco, C.M.M.: Whole cell actinobacteria as biocatalysts, www.frontiersin.org, (2019). https://doi.org/10.3389/fmicb.2019.00077
Blackman, L.M., Cullerne, D.P., Hardham, A.R.: Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the Phytophthora parasitica genome. BMC Genomics (2014). https://doi.org/10.1186/1471-2164-15-785
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P.M., Henrissat, B.: The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. (2014). https://doi.org/10.1093/nar/gkt1178
Anderson, I., Abt, B., Lykidis, A., Klenk, H.-P., Kyrpides, N., Ivanova, N.: Genomics of aerobic cellulose utilization systems in Actinobacteria. PLoS ONE 7, e39331 (2012). https://doi.org/10.1371/journal.pone.0039331
Cui, J., Mai, G., Wang, Z., Liu, Q., Zhou, Y., Ma, Y., Liu, C.: Metagenomic insights into a cellulose-rich niche reveal microbial cooperation in cellulose degradation. Front. Microbiol. (2019). https://doi.org/10.3389/fmicb.2019.00618
Küster, E., Williams, S.T.: Selection of media for isolation of streptomycetes. Nature 202, 928–929 (1964). https://doi.org/10.1038/202928a0
Hayakawa, M., Nonomura, H.: Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509 (1987). https://doi.org/10.1016/0385-6380(87)90108-7
Houfani, A.A., Větrovský, T., Baldrian, P., Benallaoua, S.: Efficient screening of potential cellulases and hemicellulases produced by Bosea sp. FBZP-16 using the combination of enzyme assays and genome analysis. World J. Microbiol. Biotechnol. 33, 1–14 (2017). https://doi.org/10.1007/s11274-016-2198-x
Baldrian, P.: Microbial enzyme-catalyzed processes in soils and their analysis. Plant Soil Environ. 55, 370–378 (2009). https://doi.org/10.1007/s11104-008-9731-0
Lane, D.J.: 16S/23S rRNA sequencing. In: Stackebrandt, E.G., Goodfellow, M. (eds.) Nucleic Acid Techniques in Bacterial Systematics, pp. 115–175. Wiley, New York (1991)
Zerbino, D.R., Birney, E.: Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008). https://doi.org/10.1101/gr.074492.107
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A.A., Dvorkin, M., Kulikov, A.S., Lesin, V.M., Nikolenko, S.I., Pham, S.O.N., Prjibelski, A.D., Pyshkin, A. V, Sirotkin, A. V, Vyahhi, N., Tesler, G., Alekseyev, M.A.X.A., Pevzner, P.A.: SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). https://doi.org/10.1089/cmb.2012.0021
Meyer, F., Paarmann, D., D’Souza, M., Olson, R., Glass, E., Kubal, M., Paczian, T., Rodriguez, A., Stevens, R., Wilke, A., Wilkening, J., Edwards, R.: The metagenomics RAST server: a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 9, 1–8 (2008). https://doi.org/10.1002/9781118010518.ch37
Yin, Y., Mao, X., Yang, J., Chen, X., Mao, F., Xu, Y.: DbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 40, 445–451 (2012). https://doi.org/10.1093/nar/gks479
Seemann, T.: Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069 (2014). https://doi.org/10.1093/bioinformatics/btu153
Lee, M.D.: GToTree: a user-friendly workflow for phylogenomics. Bioinformatics 35, 4162–4164 (2019). https://doi.org/10.1093/bioinformatics/btz188
Hyatt, D., Chen, G.L., LoCascio, P.F., Land, M.L., Larimer, F.W., Hauser, L.J.: Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 11, 119 (2010). https://doi.org/10.1186/1471-2105-11-119
Eddy, S.R.: Accelerated profile HMM searches. PLoS Comput. Biol. 7, 1002195 (2011). https://doi.org/10.1371/journal.pcbi.1002195
Edgar, R.C.: MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). https://doi.org/10.1093/nar/gkh340
Capella-Gutierrez, S., Silla-Martinez, J.M., Gabaldon, T.: trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). https://doi.org/10.1093/bioinformatics/btp348
Price, M.N., Dehal, P.S., Arkin, A.P.: FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010). https://doi.org/10.1371/journal.pone.0009490
Tange, O.: GNU Parallel 2018. (2018). https://doi.org/10.5281/ZENODO.1146014
Letunic, I., Bork, P.: Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478 (2011). https://doi.org/10.1093/nar/gkr201
Lacombe-Harvey, M.-È., Brzezinski, R., Beaulieu, C.: Chitinolytic functions in actinobacteria: ecology, enzymes, and evolution. Appl. Microbiol. Biotechnol. 102, 7219–7230 (2018). https://doi.org/10.1007/s00253-018-9149-4
Sharma, H.K., Xu, C., Qin, W.: Biological Pretreatment of Lignocellulosic Biomass for Biofuels and Bioproducts: An Overview. https://www.bp.com/ (2019)
Riyadi, F.A., Tahir, A.A., Yusof, N., Sabri, N.S.A., Noor, M.J.M.M., Akhir, F.N.M.D., Othman, N., Zakaria, Z., Hara, H.: Enzymatic and genetic characterization of lignin depolymerization by Streptomyces sp. S6 isolated from a tropical environment. Sci. Rep. 10, 1–9 (2020). https://doi.org/10.1038/s41598-020-64817-4
Shrestha, S., Kognou, A.L.M., Zhang, J., Qin, W.: Different facets of lignocellulosic biomass including pectin and its perspectives. Waste Biomass Valorization 12, 4805–4823 (2020). https://doi.org/10.1007/s12649-020-01305-w
Aakko, J., Pietilä, S., Toivonen, R., Rokka, A., Mokkala, K., Laitinen, K., Elo, L., Hänninen, A.: A carbohydrate-active enzyme (CAZy) profile links successful metabolic specialization of Prevotella to its abundance in gut microbiota. Sci. Rep. 10, 12411 (2020). https://doi.org/10.1038/s41598-020-69241-2
Boutard, M., Cerisy, T., Nogue, P.-Y., Alberti, A., Weissenbach, J., Salanoubat, M., Tolonen, A.C.: Functional diversity of carbohydrate-active enzymes enabling a bacterium to ferment plant biomass. PLoS Genet. 10, e1004773 (2014). https://doi.org/10.1371/journal.pgen.1004773
Lladó Fernández, S., Větrovský, T., Baldrian, P.: The concept of operational taxonomic units revisited: genomes of bacteria that are regarded as closely related are often highly dissimilar. Folia Microbiol. (Praha) 64, 19–23 (2019). https://doi.org/10.1007/s12223-018-0627-y
Berlemont, R., Martiny, A.C.: Genomic potential for polysaccharide deconstruction in bacteria. Appl. Environ. Microbiol. 81, 1513–1519 (2015). https://doi.org/10.1128/AEM.03718-14
Berlemont, R., Martiny, A.C.: Phylogenetic distribution of potential cellulases in bacteria. Appl. Environ. Microbiol. 79, 1545–1554 (2013). https://doi.org/10.1128/AEM.03305-12
Ventura, M., Canchaya, C., Tauch, A., Chandra, G., Fitzgerald, G.F., Chater, K.F., van Sinderen, D.: Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71, 495–548 (2007). https://doi.org/10.1128/mmbr.00005-07
Malik, A., Kim, Y.R., Jang, I.H., Hwang, S., Oh, D.C., Kim, S.B.: Genome-based analysis for the bioactive potential of Streptomyces yeochonensis CN732, an acidophilic filamentous soil actinobacterium. BMC Genomics 21, 118 (2020). https://doi.org/10.1186/s12864-020-6468-5
Acknowledgements
The authors acknowledge the financial support by the Algerian Ministry of Higher Education and Scientific Research and the General Direction for Scientific Research and Technological Development (Algeria). The authors acknowledge Lamia Medouni–Haroune (Laboratoire de Microbiologie Appliquée, Université de Bejaia) and Karel Švec (Laboratory of Fungal Genetics and Metabolism, Institute of Microbiology of the Czech Academy of Sciences) for providing olive pomace and oat flakes, respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Houfani, A.A., Tláskal, V., Baldrian, P. et al. Actinobacterial Strains as Genomic Candidates for Characterization of Genes Encoding Enzymes in Bioconversion of Lignocellulose. Waste Biomass Valor 13, 1523–1534 (2022). https://doi.org/10.1007/s12649-021-01595-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12649-021-01595-8