Abstract
Identification of bacteria that produce carbohydrolytic enzymes is extremely important given the increased demand for these enzymes in many industries. Twenty lignocellulose-degrading bacterial isolates from Algerian compost and different soils were screened for their potential to produce different enzymes involved in biomass deconstruction. Based on 16S rRNA gene sequencing, the isolates belonged to Proteobacteria and Actinobacteria. Differences among species were reflected both as the presence/absence of enzymes or at the level of enzyme activity. Among the most active species, Bosea sp. FBZP-16 demonstrated cellulolytic activity on both amorphous cellulose (CMC) and complex lignocellulose (wheat straw) and was selected for whole-genomic sequencing. The genome sequencing revealed the presence of a complex enzymatic machinery required for organic matter decomposition. Analysis of the enzyme-encoding genes indicated that multiple genes for endoglucanase, xylanase, β-glucosidase and β-mannosidase are present in the genome with enzyme activities displayed by the bacterium, while other enzymes, such as certain cellobiohydrolases, were not detected at the genomic level. This indicates that a combination of functional screening of bacterial cultures with the use of genome-derived information is important for the prediction of potential enzyme production. These results provide insight into their possible exploitation for the production of fuels and chemicals derived from plant biomass.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For decades, enzymes have been widely used in industrial processes. The growing demand to replace traditional chemical processes, generating pollution and toxic wastes for the environment, led to develop biotechnological processes that already integrate the use of cellulases and xylanases in various industries: pulp and paper, textile, food or detergents. Another promising area for the industrial application of these enzymes is second generation biofuel production from lignocellulosic biomass (Song et al. 2014).
The hydrolysis of the lignocellulosic biomass uses a wide range of enzymes (known as CAZymes for carbohydrate-active enzymes; listed in http://www.cazy.org) acting together to degrade the robust carbohydrate polymer of the cell wall into simple sugars that may be easily fermented (Gao et al. 2014). Among them, the superfamily of glycoside hydrolases (GH) is an important group of enzymes required for the degradation of plant cell walls as well as other polysaccharides.
Cellulases and xylanases have a major role in degradation of lignocellulosic biomass (Amore et al. 2015). Lignocellulosic biomass can be exploited in this context resource for the production of biofuels as it is very abundant, inexpensive and production of such resources contributes to the reduction of the intensity of environmental pollution (Maki et al. 2009; Nandimath et al. 2016). The cellulose and hemicellulose from agricultural by-products account for two-thirds of lignocellulose and serve as substrates for the production of green solvents such as bioethanol effluents (Jung et al. 2015). Cellulases are typically represented by three types of extracellular enzymes: endoglucanases, which perform internal hydrolysis at 1,4-β-bonds; cellobiohydrolases I and II (CBHI and CBHII) which act synergistically from the reducing and non-reducing ends of cellulose, respectively, releasing cellobiose; which is degraded by the β-glucosidases releasing two glucose monomers (Salvachúa et al. 2013). As for hemicellulases, they belong to multiple GH that are specific for degradation of xylose-, mannose-, arabinose-, glucose- and galactose-containing polysaccharides (Větrovský et al. 2014).
Although almost all lignocellulolytic enzymes that are used today are from fungi such as Trichoderma spp. and Fusarium spp., their addition as hydrolytic commercial enzymes for ethanol production is very expensive. Enzymes from bacteria with the combination of using agro-industrial residues, such as wheat straw, can reduce significantly production costs (Woo et al. 2014; El-Naggar et al. 2014).
Algeria is currently facing very recent problems of decrease in the cost of fossil fuels, pollution and proliferation of all kinds of wastes: agricultural wastes, industrial wastes, forest residues and urban wastes. Most of these wastes are often removed by burning of the biomass. All these wastes are potential sources of lignocellulosic biomass. Our work is in this context through the isolation of local bacterial strains producing cellulases and hemicellulases for use at industrial level.
The present work faces one of key bottlenecks of the process: the cost of enzymatic pre-treatment. The use of bacterial strains isolated from Algeria and known to utilize (hemi)cellulose as carbon source could be very promising for the improvement of the production of bioethanol from low-cost lignocellulosic residues.
Nowadays, there has been substantial progress in developing the approaches for enzyme discovery and the isolation of bacteria capable of producing these enzymes (Baldrian and López-Mondéjar 2014). The advent of genomics and metagenomics sequencing efforts provide pool of genes encoding enzymes that make these efforts possible. Application of genomic techniques has significantly advanced our understanding of the genetic potential of microorganisms.
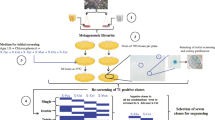
The aim of this work was to isolate new bacterial strains producing efficient cellulose and hemicelluloses-degrading enzymes and to identify the relationship between genes encoding enzymes and activity of secreted enzymes in pure bacterial cultures. In the present work, we report the results of different methods employed for an effective screening of carbohydrate-degrading enzymes produced by various bacterial strains isolated from Algerian ecosystems with widely differing characteristics. We have successfully isolated cellulose-degrading bacterial strains and selected one efficient cellulose-degrading bacterium producing different types of cellulases/hemicellulases for whole genome sequencing.
Materials and methods
Study sites and sampling
A total of 10 samples of locally-produced compost generated from plant wastes (Biocompost, Bejaia) were collected in May 2014 from two 10 kg packets and 20 soil cores (45 mm diameter) that were distinct with respect to soil and site characteristics were collected in August 2014 from four areas in Batna city located in the east of Algeria. Soil samples were collected in five defined plots (10 m2, approximately 100 m from each other) at each sampling site. After collection, the samples were stored at 4 °C and analysed within 24 h after collection.
The soils and compost were air-dried to determine the soil physicochemical properties. Soil pH was determined using a glass electrode (sensION + PH3, France) after shaking a suspension of 1:2.5 w/v soil/water ratio for 30 min (Pochon and Tradieux 1962). Conductivity was determined using pH/Conductivity Meter (Eutech Instruments con 510, Singapore) in a solution of 1:5 w/v soil/water ratio (U.S.S.L.S. 1954). Gravimetric water content (GWC) was determined by drying at 105 °C for 48 h for estimation of loss of organic matter (Grossman and Reinsch 2002). Organic matter content was determined by mass loss on ignition for 16 h at 550 °C in a muffle furnace (Nabertherm, Germany) (Girard et al. 2011). Air dried soil samples were sieved to 2 mm, ground, and analysed for total C and N, using an Elemental analyser (Elementar Vario EL III, Elementar, Hanau, Germany) in an external laboratory at the Research Institute for Soil and Water Conservation (RISWC) Prague-Zbraslav, Czech Republic. C content was measured using sulfochromic oxidation, and N content was estimated by the Kjeldahl method (Bremner 1960).
Temperature during sampling was mesothermal ranging between 27 and 30 °C. Samples were characterized by high content of organic matter, neutral and slightly alkaline pH (Table 1). The chemical properties of soils and compost differed, with the forest soil containing more organic matter, but lower nutrients (C and N) than compost and exhibiting slightly but higher pH. The conductivity was noted higher for the forest soil and compost; this translates into a richness of mineral salts. The results of the GWC % reflects well the semi-arid regions of the sampling. The carbon and nitrogen (C/N) ratio was higher in the compost (Table 1).
Isolation of bacteria
For the isolation of the bacteria, using the conventional dilution plate technique, 10 g of each sample were suspended in 100 ml of sterile distilled water in 250 ml Erlenmeyer flask, shaken at 150 rpm, at room temperature (25 °C) for 20 min, further diluting to 10−5. About 100 µL of the suspension was spread on the surface of five different isolation media in Petri dishes in duplicate, using a sterile glass spreader and incubated under aerobic conditions at 28 °C for 4 weeks. In order to isolate and explore the diversity of bacteria from the targeted sites, different isolation media were used: Starch Casein Agar, Kuster’s Agar (KUA) medium (Kuster and Wiliams 1964), Glucose-Yeast extract-Malt (GLM) (Kitouni et al. 2005), (ISP2 Agar) (Shirling and Gottlieb 1966) and humic acid vitamin agar (HV agar) (Hayakawa and Nonomura 1987). The pH of mediums was adjusted to 7.2–7.4 before autoclaving at 121 °C for 20 min. After sterilization, the mediums were supplemented with nystatine (50 µg/mL) to prevent the growth of fungi (Williams and Davis 1965). The isolated colonies were purified, maintained on agar slants and in 25% (v/v) glycerol at −20 °C for further study.
Semiquantitative CMCase assay
All isolates were screened for the ability to cleave amorphous cellulose on minimal medium agar with 1% of carboxymethylcellulose as the only carbon and energy source (CLM), containing (g/L): CMC, 10.0; NaNO3, 1.2; KH2PO4, 3.0; K2HPO4, 6.0; MgSO·7H2O, 0.2; CaCl2, 0.05; MnSO4·7H2O, 0.01; ZnSO4·7H2O, 0.001; pH 7.0 (El-Naggar et al. 2014). The pure cultures of bacteria were individually spot inoculated at the centre of CMC agar plates using Petri dishes (90 mm, 20 mL/plate) and incubated at 28 °C to allow for the production of cellulases. After 5 days of incubation, the agar plates were stained with aqueous solution of 0.1% (w/v) Congo red. The dye was decanted after 30 min and the plates were washed with 1 M NaCl for counterstaining the plates and make the zone visible and clear. To indicate the CMCase activity, the diameters of clear zones surrounding bacterial colonies were measured with dial callipers. Positive isolates were tested again for confirmation. Any indication of clearing was considered as a positive result.
Colorimetric assay using 3,5-dinitrosalicylic acid reagent
Endoglucanase and endoxylanase activities were quantitatively measured by determining the amount of reducing sugars from soluble CMC and xylan, respectively, using the 3,5-dinitrosalicyclic acid reagent (DNS) method (Miller 1959). The growth of bacteria was performed into 250 ml plugged and shaked Erlenmeyer flasks, each containing 50 ml of the CLM medium. Only those strains that had previously been selected using the Congo red test, displaying strong and very strong CMCase digestion halo, were evaluated in this step. The hydrolysis of CMC and beechwood xylan was quantified by using the DNS reagent to estimate the amount of reducing sugars released. After 1 week of incubation at 28 °C, the cultures were centrifuged and the resulting culture supernatants were used as crude enzyme solution for estimation of reducing sugars (i.e. glucose for CMCase and xylose for Xylanase). The reaction mixture composed of 250 µl of the crude enzyme and 250 µl of 1% (w/v) substrate solution prepared in 50 mM phosphate buffer (pH 7.0) were incubated at 50 °C and the reaction was stopped by adding 1.5 ml of DNS reagent. Then the treated samples were boiled in boiling bath for 5 min and after cooling on ice for colour stabilization. Appropriate enzyme and substrate controls were also included in the assay. Absorbance was measured at 540 nm using UVmini-1240 spectrophotometer (Shimadzu, Japan). The enzyme activities were calculated by estimating the amounts of liberated reducing sugars (glucose and xylose equivalents) against a glucose and xylose standard curve, respectively. Enzyme activity is expressed as U/mL, i.e., μmol of glucose or xylose released per ml per min.
Enzyme assays on wheat straw as growth resource
The strains selected from the second quantitative screening were further tested for the production of various extracellular enzymes in order to evaluate their ability to grow on wheat straw as natural substrate, and to produce extracellular enzymes that participate in the degradation of cellulose, hemicellulose and other polysaccharides. Bacterial strains were cultivated in liquid medium containing 0.5% of air-dried, milled wheat straw into 250-mL flasks for 14 days at 25 °C without agitation (three replicates). The cultivation liquid was collected the first two weeks and the centrifuged supernatant of fermented broth was used as crude enzyme extract for determination of enzymatic activities.
Endocellulase and endoxylanase assays
The activities of endo-1,4-β-glucanase (endocellulase) and endo-1,4-β-xylanase (endoxylanase) were measured using azo-xylan and azo-cellulose as substrates, respectively, using the protocol of the supplier (Megazyme, Bray, Ireland), and determined by referring to a standard curve as described previously (Baldrian 2009). The reaction mixture contained 0.15 ml of substrate and 0.15 ml sample. The reaction mixture was incubated at 40 °C for 60 min and the reaction was stopped by adding 0.75 ml of ethanol followed by 10 s vortexing and 10 min centrifugation. The amount of released dye was measured at 595 nm under UV–VIS spectrophotometer (Lambda 11, Perkin-Elmer) and the enzyme activity was calculated according to standard curves correlating the dye release with the release of reducing sugars. One unit of enzyme activity was defined as the amount of enzyme releasing 1 nmol of reducing sugars per min.
Fluorometric assays using 4-methylumbelliferone (MUF)-linked substrates
The activities of the extracellular enzymes were measured using 4-methylumbelliferyl (MUF) linked substrates listed in Table 2. The assay is based on measurement of fluorescence of released 4-methylumbelliferyl (MUF) from the substrates upon enzymatic cleavage.
The assay was performed with 200 µL substrate and 40 µL sample, as described previously (Baldrian 2009). The reaction mixtures were incubated at 40 °C for 120 min in the dark. Substrates (in DMSO) were combined with the three technical replicates of the extracts in a 96-well multiwell plate. For the background fluorescence measurement, 4-methylumbelliferol standards were used to correct the fluorescence quenching. Fluorescence was determined on the Infinite microplate reader (TECAN, Austria) from 5 to 125 min at 355 nm excitation and 460 nm emission wavelength. Enzyme activities were determined from the fluorescence units using a standard calibration curve of methylumbelliferone (MUF) and expressed as rates of MUF production (nmol min−1 ml−1).
16S rRNA gene sequencing and phylogenetic analysis
Isolates were genotyped using the 16S rRNA gene sequence. A small amount of cell biomass from a single colony of each strain growing on ISP-2 agar plates was picked up with a sterile toothpick and resuspended in 20 µL of Ultra pure water and was added to 25 µL of the PCR reaction as template (“colony PCR”). When colony PCR failed, ArchivePure DNA Purification Kit for Yeast & Gram-positive bacteria was used to extract and purify DNA from isolates following the manufacturer’s instructions, and then PCR was performed.
The 16S rRNA gene was PCR amplified using the forward primer 27f (5′-AGAGTTT GATCCTGGCTCAG-3′), the reverse primer 1492r (5′ GGTTACCTTGTTACGACTT-3′) and 16S universal Eubacterial primers: EUB530F (5′-GTGCCAGCMGCNGCGG-3′) and EUB1100R (5′-GGGTTNCGNTCGTTG-3′). All PCR reactions were performed on a Mastercycler personal thermocycler (Eppendorf, Hamburg, Germany). Each 50 μL PCR reaction contained 5 μL of 10× polymerase buffer, 3 μL of 10 mg mL−1 purified Bovine Serum Albumin, 2 µl of each primer, 1 µl of PCR Nucleotide Mix (10 mM) and 1.5 µl of polymerase (2 U μL−1; Pfu DNA Polymerase: DyNAzyme II DNA polymerase, 1:24). When using the primers 27f/1492r, the PCR amplification protocol was conducted as follow: a 94 °C denaturation step for 2 min, then 35 cycles were performed as follows: 94 °C for 30 s, 55 °C for 1 min and 72 °C for 1 min 30 s, followed by an additional 10 min at 72 °C for 10 min to allow final extension. With the primers EUB530F/EUB1100R, the conditions for thermal cycling were as follows: 94 °C for 5 min, 35 cycles of 94 °C for 1 min, 62 °C for 1 min, 72 °C for 1 min, and 72 °C for 10 min. At the end of the cycling, the reaction mixture was cooled at 4 °C. To confirm the successful amplification, the PCR products were detected by agarose gel electrophoresis and were visualized by UV fluorescence after ethidium bromide staining. The Sanger sequencing reaction was performed by the Macrogen Company (Amsterdam, Netherlands). Quality of the 16S rRNA gene sequences of the bacterial isolates used in this study has been manually evaluated using SEED 1.2.1 (Větrovský and Baldrian 2013). The obtained sequences were identified through BLAST against GenBank nucleotide sequence database. Sequences were deposited in the GenBank database and their accession numbers are reported in Table 3.
Whole genome sequencing and annotation
Genome of Bosea sp. FBZP-16 was sequenced using the Illumina MiSeq platform (Illumina, Inc., CA, USA) at the C4SYS facility of the Institute of Microbiology of the Czech Academy of Sciences. The sequencing libraries were prepared from extracted DNA using the TruSeq DNA PCR-Free Library Preparation Kit (Illumina, Inc., CA, USA) and pooled for sequencing via a paired-end run (2 × 251 bp). This run yielded 605398 paired-end reads, representing approximately 50-fold coverage.
The sequence data were assembled using Velvet 1.2.10, generating 61 contigs (N 50 = 259,947 bp) that represented the Bosea sp. strain FBZP-16 genome. The gene calling and annotation for the genome was performed using The RAST Server 2.0 (Rapid Annotations using Subsystem Technology) (Aziz et al. 2008; Overbeek et al. 2014; Brettin et al. 2015), which predicted tRNAs, open reading frames (ORFs), and rRNAs. Carbohydrate-active enzymes (CAZymes) annotation from the whole genome was performed by analysing amino acid sequences with the web server dbCAN (http://csbl.bmb.uga.edu/dbCAN/ ) (Yin et al. 2012).
The extracted 16S rRNA from RNAmmer was queried using BLASTn against the NCBI nucleotide database. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession LNCQ00000000. The version described in this paper is version LNCQ01000000.
Genome to genome comparisons
Once the genome of strain Bosea sp. FBZP-16 was sequenced, we were able to compare it to the genomes of Bosea species available on NCBI databases. A tool developed by DSMZ called the genome to genome distance calculator (GGDC) reports the percent G+C difference between two given genomes (Meier-Kolthoff et al. 2014). Glycosyl hydrolases of these bacteria were also investigated by analysing amino acid sequences with the web server dbCAN (http://csbl.bmb.uga.edu/dbCAN/) (Yin et al. 2012).
Phylogenetic analyses
All evolutionary analyses were conducted in MEGA6 (Tamura et al. 2013). Maximum Likelihood method used for construction of evolutionary tree Fig. 5. was based on the Kimura 2-parameter model (Kimura 1980). The tree with the highest log likelihood (−2640.2828) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood (MCL) approach, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+G, parameter = 0.0500)]. The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 3.6230% sites). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 24 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 1341 positions in the final dataset.
Neighbor-Joining method was used for tree construction in Fig. 2,3,4 (Saitou and Nei 1987). The optimal tree with the sum of branch length = 1.05228348 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al. 2004) and are in the units of the number of base substitutions per site. The analysis involved 20 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1195 positions in the final dataset.
Results
Semiquantitative CMC assay
In the study, conventional culture techniques led to the recovery of 170 bacterial isolates from soil and compost samples collected in Algeria. The screening led to 115 isolates (68%) which showed endocellulase activity on agar plates. CMCase activities were evaluated according to the extent and intensity of hydrolytic clearing zones as weak (1–10 mm); moderate (11–20 mm), strong (21–30 mm) and very strong (>30 mm). These zones frequency were represented by 8.2, 16, 22.4 and 21.2% of isolates, respectively (Fig. 1a). Many colonies showed a good CMC-degrading activity since, after Congo Red staining and consistent CMC degradation halos were detected. Strain Bosea sp. FBZP-16 showed large clear zone diameter around its colonies (Fig. 1b). Among the bacterial strains grown on CMC-agar plates, several isolates were selected as efficient cellulase producers.
Colorimetric assay using 3.5-dinitrosalicylic acid assay
Bacterial strains that were found to be positive by CMC agar plate assay were grown in liquid medium, using a 1% xylan- and 1% CMC-containing medium. The CMC media appeared to lose viscosity over the course of bacterial growth., this may happen in response to endoglucanase activity. Comparison of the endogluconase and endoxylanase profiles (Fig. 2) indicated endoglucanase activities between 0.1 and 2.65 U min−1 mL−1 and endoxylanases activities ranging from 0.2 to 32 U min−1 mL−1 with Nocardia sp. FBZK-20 showing the highest endogluconase activity among all 20 cultures tested, while Streptomyces sp. ALTH-1 contained the highest levels of endoxylanase activity.
Neighbor-joining tree of bacterial strains with enzymatic activity rates. Organism labels in the phylogenetic tree are NCBI taxonomy at the genus level. Each strain is characterized by horizontally stacked bar graphs of enzymatic activities in a liquid medium with CMC as the sole carbon source: endoglucanase and endoxylanase in U mL−1, i.e., μmol of substrate released per ml per min
Enzyme assays on wheat straw as growth resource:
To further test the cellulolytic and hemicellulolytic capacity of the bacterial isolates, we performed growth assays on wheat straw to detect these activities in a natural substrate. Some isolates showed individual enzyme activity i.e. either cellulase or xylanase and rest showed the activity for both the enzymes. The result show that enzymatic activities may considerably differ among species. The production of endocellulase was lower than that of endoxylanase (Fig. 3).
Bosea sp. FBZP-16, isolated from a coniferous forest soil in Algeria, produced several biomass-degrading enzymes, including cellulases and hemicellulases. Indeed, the strain demonstrated the ability to produce all three glycoside hydrolase (GH) activities involved in complete enzymatic hydrolysis of cellulose. Also, Bosea sp. FBZP-16 had the highest activity of cellobiohydrolase, significantly higher than other isolates of the collection (Fig. 4). The highest β-glucosidase activity (1718,16 nmol min−1 mL−1) was produced by Streptomyces sp. FBZP-13.
Neighbor-joining tree of bacterial strains with enzyme activities involved in carbohydrate metabolism. Each strain is characterized by horizontally stacked bar graphs of enzymatic activities after cultivation on wheat straw expressed in nmol min−1 mL−1. Isolates have filled circles according to the width of the clearing zones around colonies obtained with Congo Red test for screening of positive CMC degradation. (Color figure online)
Sequencing of 16S rRNA genes of xylanolytic and cellulolytic isolates
Besides enzyme production profiling, our second attempt was to examine molecular systematics of the isolated cellulolytic bacteria. Sequencing of the 16S rRNA gene from isolates that displayed positive activity on CMC identified members of the Proteobacteria and Actinobacteria. Phylogenetic analysis of Proteobacteria isolates revealed bacteria closely related to that in the genera Bosea, Lysobacter, Pseudomonas, Achromobacter, Ralstonia and Leifsonia. All Actinobacteria isolates belong to the genus Streptomyces, expect one strain to the genus Nocardia. Data derived from BLAST showed that the newly isolated strains have high sequence similarities with different species (Table 3).
Bosea sp. FBZP-16 genome properties
The total combined contig size of the Bosea sp. FBZP-16 genome was 6,081,062 bp, consisting of 61 contigs and containing 5904 coding sequences (CDSs), of which 70.2% were assigned to known functional genes. The properties and the statistics of the genome are summarized in Table 4. Phylogenetic analysis of the full-length 16S rRNA gene classified strain FBZP-16 as a Bosea species (Fig. 5).
To further characterize the potential of Bosea sp. FBZP-16 to degrade cellulose, hemicellulose and other carbohydrates, we identified the predicted glycoside hydrolase (GH) families of CAZymes (Carbohydrate-Active enZYmes) encoded within the genome of the bacterium. The genome of Bosea sp. FBZP-16 contained 161 CAZymes, comprising 43 glycoside hydrolases (GHs), 58 glycosyl transferases (GTs), 4 polysaccharide lyases (PLs), 38 carbohydrate esterases (CEs), 15 auxiliary activity enzymes (AAs) and 3 carbohydrate-binding modules (CBMs), which indicated the high potential of Bosea sp. FBZP-16 to degrade plant cell wall material. Among these are genes from three groups of enzymes involved in cellulose deconstruction, which are: endo-1,4-β-glucanases from families GH5 and GH8 and 1,4-β glucosidases from families GH51 and GH3 (Table 5). Also, of note is the presence of genes from families GH2 and GH120, which may be involved in deconstructing hemicellulose. Arabinan degradation potential is provided by an arabino-furanosidase (GH3), which is known to degrade arabinoxylan hemicelluloses. Indeed, when grown on wheat straw, Bosea sp. FBZP-16 produced both endoxylanases and β-xylosidases. The genes contributing to the decomposition of other important polysaccharides like starch and chitin were identified as well.
This annotations was corroborated by enzyme assays with selected substrates. However, Bosea sp. FBZP-16 does not harbour the gene for cellobiohydrolase (i.e., the glycosyl hydrolase family GH6, 9 or 48), although a high cellobiohydrolase activity (1562 nmol min−1 mL−1) was detected during enzyme assays.
In order to calibrate how much similarity to other Bosea sp. bacteria would be expected on average, the genome of Bosea sp. FBZP-16 was compared with those of 12 other published genomes of Bosea species using the NCBI genome databases (Table 6). The genome of Bosea sp. FBZP-16 (6.08 Mb) is smaller in size than those of Bosea sp. LC85 (6.56 Mb), Bosea sp. Root483D1 (6.61 Mb) and Bosea sp. WAO (6.13 Mb) but larger than in other strains of the same genus. Bosea sp. FBZP-16 has more predicted protein coding genes (5,859) in comparison with the most related strains but fewer genes than Bosea sp. LC85 and Bosea sp. Root483D1(5,967 and 6,049, respectively). Bosea sp. FBZP-16 has a similar G+C content than that of Bosea sp. Root381, a smaller G+C content (66.7%) than those of Bosea sp. 117, Bosea sp. Leaf344 and Bosea sp. WAO (68.2%, 68.2 and 66.8%, respectively) but higher than the rest of the strains.
The closer relatives of Bosea sp. FBZP-16 has a comparable numbers of GH families. Despite the global similarity in the total number of GHs, the number of members in individual GH families varies widely, other members of the genus Bosea contained between 26 and 50 families of glycosyl hydrolases. Within the so-far sequenced members of the Bosea genus, Bosea sp. FBZP-16 exhibits one of the highest GH densities (Fig. 6). Some GH families - e.g., GH3, GH13, GH15, GH23, GH99, GH105 - are common in the genomes, while other families, such as GH1, GH2, GH5, GH8, and GH113 were not detected in all the genomes (Fig. 7).
Predicted numbers of selected glycosyl hydrolases (GH) in the genome of Bosea sp. FBZP-16 and the genomes of other species of Bosea. On the left total number of GHs found in the genome; on the right gene content in GH families containing enzymes involved in the degradation of cellulose and hemicelluloses
Discussion
Carbohydrate active enzymes from bacteria are considered promising candidates for the preparation of novel enzymatic cocktails for the conversion of lignocellulosic biomasses into fermentable sugars. The biodiversity of Algerian soils and compost have been exploited for the isolation of new bacterial strains able to produce cellulolytic activities. All purified bacteria were screened for endo-cellulase production by Congo Red assay on CMC-containing plates. The clear zone formed following the hydrolysis of the CMC is due to the action of cellulases, because the Congo red dye remain attached to regions where β-(1,4)-linked D-glucopyranosyl units remains. However, a longer time for the reaction of the dye with the medium may increase the visibility of the zones of hydrolysis, while the diameter of the halo could be helpful in the selection of strains having a high cellulose degradation activity (Florencio et al. 2012). Bacteria residing in soils and compost are likely to produce a variety of hydrolytic enzymes. The use of wheat straw, a very abundant substrate for the production of industrial enzymes is an important way to reduce costs of the entire process (Bhalla et al. 2015). Some strains of bacteria are negative when growing on wheat straw, although being positive with CMC as a carbon source. This can be explained by the fact that CMC can be degraded by enzymes that are not necessarily active on the most recalcitrant forms of cellulose (Adams et al. 2011). The ability to degrade cellulose and hemicellulose is assigned to certain limited groups of bacteria (Baldrian and Šnajdr 2011; López-Mondéjar et al. 2016). Furthemore, the degradation of crystalline regions takes more time than the amorphous ones and some microorganisms are able to attack only amorphous cellulose (Baldrian and Šnajdr 2011). Enzyme performance can be reduced during lignocellulose hydrolysis by interaction with lignin or lignin–carbohydrate complex (LCC) (Berlin et al. 2006). Composition of polymers of hemicelluloses are typically branched and contain neutral and/or acidic side groups that render hemicelluloses amorphous or poorly crystalline (Baldrian and Šnajdr 2011). Bosea sp. FBZP-16 produced the highest cellobiohydrolase activity among the isolated strains, interesting fact as this class of cellulase represents the rate-limiting enzyme in the decomposition of cellulose (Vetrovsky and Baldrian 2014). However, during its genomic analysis, no GH family containing cellobiohydrolases was detected. While this may be surprising, the observations that microbes can perform certain biochemical reactions even if corresponding genes were not found in their genomes is not exceptional. For example, the recent comparison of fungal genomes and screening of enzyme activities resulted in this controversial observation in several cases (Eichlerová et al. 2015). Busk et al. (2014) have reported that the fungi Serpula lacrymans and Postia placenta have no cellobiohydrolase genes although known as cellulose-degraders. Another recent report shows that a Luteibacter isolate was able to degrade cellulose in the absence of any typical cellulase (López Mondéjar et al. 2016). It is very likely that the observed activity may be due to the activity of other cellulases, such endoglucanases or β-glucosidases acting on cellooligosaccharides. Wide substrate specificity is not exceptional among microbial cellulases (Baldrian and Valášková 2008). For example, the purified enzyme of Daedalea quercina described as a β-glucosidase also exhibited the activity of β-xylosidase and cellobiohydrolase (Valášková and Baldrian 2006). Our observation supports the idea that genome sequencing and generic classification of predicted proteins into GH families using the predictions based on Hidden Markov Models (Yin et al. 2012) is not sufficient to describe the biochemical potential of a microorganism and that functional screening is essential in this respect.
The fact that the genome of Bosea sp. FBZP-16 contains numerous genes for many other GH enzymes not yet tested for activity by enzyme assays, combined with these findings, support the role of Bosea sp. FBZP-16 in the degradation of lignocellulose plant material. Bosea sp. FBZP-16 demonstrated the ability to produce exoxylanase and β-xylosidase, with the presence of their corresponding genes, which are reported to be the main components responsible for effective conversion of xylan fraction of biomass to monomeric xylose (Bhalla et al. 2015). These data indicate that Bosea sp. FBZP-16 is a promising source of (hemi)cellulolytic enzymes. The availability of these complete genomic sequences of the genus Bosea enables also sequence comparisons, which can provide valuable information for the biotechnological application of these microbes. Here, we have determined the recoverable cellulases and hemicellulases released following microbiological, biochemical, and next-generation sequencing biochemical screening steps. The bacterial strains used in this research presented the ability to produce either cellulases or xylanases, or both enzymes. Furthermore, the enzyme profiling of the isolates indicate that they can have capacity of producing multiple extracellular enzymes. Because of the sheer number of enzymes that are required to hydrolyse plant biomass to fermentable oligosaccharide, organisms which produce (hemi)cellulases along with other polysaccharides degrading enzymes will be of great commercial importance. Our results demonstrate the advantage of using several complementary approaches for identifying the role of bacteria in decomposition, and that it is impossible to infer from genomic data alone what a microbe is decomposing cellulose or hemicellulose. The obtained results in the present study indicate that the assayed bacteria whether it be exoglucanases with activity on microcrystalline cellulose or endoglucanases with activity on soluble cellulose such as CMC can be potential candidates for their application in the production of fermentable sugars without leaving out other areas of environmental interest.
References
Adams AS, Jordan MS, Adams SM, Suen G, Goodwin LA, Davenport KW, Currie CR, Raffa KF (2011) Cellulose-degrading bacteria associated with the invasive woodwasp Sirex noctilio. ISME J 5:1323–1331
Amore A, Parameswaran B, Kumar R, Birolo L, Vinciguerra R, Marcolongo L, Ionata E, La Cara F, Pandey A, Faraco V (2015) Application of a new xylanase activity from Bacillus amyloliquefaciens XR44A in brewer’s spent grain saccharification. J Chem Technol Biotechnol 90:573–581
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 9:75
Baldrian P (2009) Microbial enzyme-catalyzed processes in soils and their analysis. Plant Soil Environ 55:370–378
Baldrian P, López-Mondéjar R (2014) Microbial genomics, transcriptomics and proteomics: new discoveries in decomposition research using complementary methods. Appl Microbiol Biotechnol 98:1531–1537
Baldrian P, Šnajdr J (2011) Lignocellulose-degrading enzymes in soils. In: Shukla G, Varma A (eds) Soil enzymology, soil biology, vol 22. Springer, Berlin, pp. 167–186
Baldrian P, Valášková V (2008) Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol Rev 32:501–521
Berlin A, Balakshin M, Gilkes N, Kadla J, Maximenko V, Kubo S, Saddler J (2006) Inhibition of cellulase, xylanase and β-glucosidase activities by softwood lignin preparations. J Biotechnol 125:198–209
Bhalla A, Bischoff KM, Sani RK (2015) Highly thermostable xylanase production from a thermophilic Geobacillus sp. strain WsUcF1 utilizing lignocellulosic biomass. Front Bioeng Biotechnol 3(84):1–8
Bremner JM (1960) Determination of nitrogen in soil by the Kjeldahl method. J Agric Sci 55(1):11–33
Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F (2015) RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365, 1–6
Busk PK, Lange M, Pilgaard B, Lange L (2014) Several genes encoding enzymes with the same activity are necessary for aerobic fungal degradation of cellulose in nature. PLoS ONE. doi:10.1371/journal.pone.0114138
Eichlerová I, Homolka L, Žifčáková L, Lisá L, Dobiášova P, Baldrian P (2015) Enzymatic systems involved in decomposition reflects the ecology and taxonomy of saprotrophic fungi. Fungal Ecol 13:10–22
El-Naggar NEl-A, Abdelwahed NAM, Saber WIA, Mohamed AA (2014) Bioprocessing of some agro-industrial residues for endoglucanase production by the new subsp.; Streptomyces albogriseolus subsp. cellulolyticus strain NEAE-J. Braz J Microbiol, 743–751
Florencio C, Couri S, Farinas CS (2012) Correlation between agar plate screening and solid-state fermentation for the prediction of cellulase production by Trichoderma strains. Enzy Res
Gao D, Haarmeyer C, Balan V, Whitehead TA, Dale BE, Chundawat SPS (2014) Lignin triggers irreversible cellulase loss during pretreated lignocellulosic biomass saccharification. Biotechnol Biofuels, 7(175):1–13
Girard M-C, Schvartz C, Jabiol B (2011) Etude des sols: description, cartographie, utilisation. Les constituants organiques des sols. Dunod, Paris
Grossman RB, Reinsch TG (2002) Bulk density and linear extensibility. In: Dane JH, Topp GC. (eds) Methods of soil analysis, part 4. physical methods. SSSA Book Series vol 5, pp 201–228. Soil Science Society of America, Inc., Madison
Hayakawa M, Nonomura H (1987) Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J Ferment Technol 65:501–509
Jung S, Song Y, Kim HM, Bae H-J (2015) Enhanced lignocellulosic biomass hydrolysis by oxidative lytic polysaccharide monooxygenases (LPMOs) GH61 from Gloeophyllum trabeum. Enzyme Microb Technol 77:38–45
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kitouni M, Boudemagh A, Oulmi L, Reghioua S, Boughachiche F, Zerizer H, Hamdiken H, Couble A, Mouniee D, Boulahrouf A, Boiron P (2005) Isolation of actinomycetes producing bioactive substances from water, soil and tree bark samples of the north–east of Algeria. Journal de Mycologie Médicale 15:45–51
Kuster E, Williams ST (1964) Selective media for the isolation of Streptomycetes. Nature 202:928–929
López-Mondéjar R, Zühlke D, Becher D, Riedel K, Baldrian P (2016). Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci Rep. doi: 10.1038/srep25279
Maki M, Leung KT, Qin W (2009) The prospects of cellulase-producing bacteria for the bioconversion of lignocellulosic biomass. Int J Biol Sci 5:500–516
Meier-Kolthoff JP, Klenk HP, Göker M (2014) Taxonomic use of DNA G + C content and DNA–DNA hybridization in the genomic age. Int J Syst Evol Microbiol 64:352–356
Miller GL (1959) Use of dinitrosaiicyiic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Nandimath AP, Kharat KR, Gupta SG, Kharat AS (2016) Optimization of cellulase production for Bacillus sp. and Pseudomonas sp. soil isolates. Afr J Microbiol Res 10:410–419
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214
Pochon J, Tardieux P (1962) Techniques d’analyse en microbiologie du sol. Editions de la Tourelle, Saint Mandé: 22
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Salvachúa D, Martínez AT, Tien M, López-Lucendo MF, García F, de los Ríos V, Martínez MJ, Prieto A (2013) Differential proteomic analysis of the secretome of Irpex lacteus and other white-rot fungi during wheat straw pretreatment. Biotechnol Biofuels 6(115):1–14
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Song HY, Lim HK, Kim DR, Lee KI, Hwang IT (2014) A new bi-modular endo-β-1,4-xylanase KRICT PX-3 from whole genome sequence of Paenibacillus terrae HPL-003. Enzyme Microb Technol 54:1–7
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci (USA) 101:11030–11035
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
U.S.S.L.S. (United State Salinity Laboratory Staff) (1954) Diagnosis and improvement of saline and alkali soils. US Department of Agriculture, Handbook No 60, U. S. Gov. Print. Office, Washington DC
Valášková V, Baldrian P (2006) Degradation of cellulose and hemicelluloses by the brown rot fungus Piptoporus betulinus - production of extracellular enzymes and characterization of the major cellulases. Microbiology 152:3613–3622
Větrovský T, Baldrian P (2013) Analysis of soil fungal communities by amplicon pyrosequencing: current approaches to data analysis and the introduction of the pipeline SEED. Biol Fertil Soils 49:1027–1037
Větrovský T, Steffen KT, Baldrian P (2014) Potential of cometabolic transformation of polysaccharides and lignin in lignocellulose by soil actinobacteria. PLoS ONE, 9, e89108
Williams ST, Davies FL (1965) Use of antibiotics for selective isolation and enumeration of actinomycetes in soil. J Gen Microbiol 38:251–261
Woo HL, Hazen TC, Simmons BA, DeAngelis KM (2014) Enzyme activities of aerobic lignocellulolytic bacteria isolated from wet tropical forest soils. Syst Appl Microbiol 37:60–67
Yin Y, Mao X, Yang JC, Chen X, Mao F, Xu Y (2012) dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451
Acknowledgements
The authors gratefully acknowledge financial support by the Algerian Ministry of Higher Education and Scientific Research, Laboratory of Applied Microbiology, Faculty of Natural and Life Sciences, University of Bejaia, 06000 Bejaia, Algeria. This work was also supported by the Ministry of Education, Youth and Sports of the Czech Republic (LM2015055, LD15086) and by the institutional research concept of the Institute of Microbiology of the Czech Academy of Sciences, Praha (RVO61388971).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Houfani, A.A., Větrovský, T., Baldrian, P. et al. Efficient screening of potential cellulases and hemicellulases produced by Bosea sp. FBZP-16 using the combination of enzyme assays and genome analysis. World J Microbiol Biotechnol 33, 29 (2017). https://doi.org/10.1007/s11274-016-2198-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-016-2198-x