Abstract
Recently, bacterial laccases has drawn researchers’ interest due to their ability to overcome high pH and salt concentration conditions compared to fungal laccases. Here we report a novel micro-aerobic cultivation strategy for enhancing CotA laccase expression and study its application for dye decolorization. Micro-aerobic cultivation of Escherichia coli BL21 (DE3) strain carrying pT7-FLAG-MAT-TAG-1-CotA had significantly enhanced CotA laccase activity up to 13903 U/L. The most unique findings of this investigation are that micro-aerobic cultivation strategy enhanced the reactive oxygen species production which consequently led to the over expression of CotA laccase gene. Malachite green, Crystal violet, Congo red and Bromophenol blue were efficiently decolorized by using purified CotA laccase without presence of any mediators at pH 6 and 9. These results provide a great platform for the dynamic production and application of bacterial laccase in industry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacterial laccases have a great advantage over fungal laccases due to their activation and stabilization at alkaline conditions, elevated temperatures and mediums supplemented with high concentration of copper ions [1, 2]. A thermo stable CotA laccase in Bacillus subtilis, a 65-kDa protein, takes part in the production of the melanin like brown spore pigment in the endospore coat. It is also responsible for the defense provided by the spore coat against H2O2 and UV light [3]. CotA-laccase is 513 amino acids [4] and it is reported for industrial textile waste water detoxification and decolorization [5], and for catalyzing phenolic acids dimerization [6].
Low production yields of the enzyme by native strains are considered as the main obstacle of bacterial laccases applications in industry [7]. Easily handling and cultivable organisms can be used to overcome this problem and facilitate the higher production of these enzymes [8]. Escherichia coli is considered the most preferable recombinant protein expression system due to its rapid growth and easy genetic manipulation and considered as the most dynamic system for industrial biocatalysts [9–11]. Culture conditions also have a strong effect in controlling the formation of soluble proteins in vivo. The insufficient content of copper in the cytoplasm upon heterologous expression of CotA laccases in E. coli is disadvantageous due to production of an inactive enzyme. This problem can be resolved by adding copper to the medium and expressing CotA laccase under micro-aerobic conditions for obtaining fully copper-loaded enzyme [12]. E. coli has the ability to grow in oxygen availability and deficiency in which, it can take the advantage of oxygen deficiency which exists during micro-aerobic cultivation and preserve the redox balance [13–15].
The objective of the current study was to construct a dynamic recombinant E. coli strain integrated with optimization process for efficient bacterial laccase expression. Isolation, cloning and expression of CotA laccase encoding gene using pT7-FLAG-MAT-TAG-1 expression vector which contains T7 promoter was performed in order to obtain soluble and active CotA laccase. Once the high CotA laccase expression strain was constructed, fermentation process optimization was carried out. After that, we developed a novel micro-aerobic cultivation strategy in order to study its effect on ROS production and CotA laccase gene expression. The purified CotA laccase produced from micro-aerobic cultivation was tested for its potential application in dye decolorization.
Materials and Methods
Materials and Strains
Easy Pure Genomic DNA, Quick Plasmid mini Prep, Gel Extraction, PCR Purification kits, Taq DNA polymerase, chemically competent Trans 10 E. coli cells, E. coli strain BL21 (DE3) pLysS, primers, T4 DNA ligase and restriction enzymes were purchased from TransGen Biotech (Beijing, China). 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonicacid) (ABTS), isopropyl beta-d thiogalactoside (IPTG), PT7-FLAG-MAT-TAG-1 vector and ampicillin were bought from Sigma-Aldrich (St. Louis, MO, USA). B. subtilis 168 bacterial strain which obtained from China General Microbiological Culture Collection Center (Beijing, China), was cultivated in Luria–Bertani (LB) medium at 30 °C and 180 rpm for 12 h.
Cloning of CotA Laccase Gene, Production and Purification
CotA laccase gene of B. subtilis 168 was isolated by following the instructions of “Easy Pure Genomic DNA Kit”. B. subtilis 168 CotA gene (GenBank Accession No. 936018) was amplified by PCR using forward (HKCotAN1) and reverse (HKCotAN3) primers indicated in Table 1. PCR was conducted by using an automated thermo cycler (T100 Thermo cycler, Bio-Rad), and the program was adjusted as: 4 min at 95 °C, 30 s at 95 °C, 30 s at 55 °C, 2 min at 72 °C, step 2 repeated for 32×, and final extension at 72 °C for 10 min. Ethidium bromide stained DNA bands were visualized by Image Lab Software, Bio-Rad. Pure B. subtilis 168 CotA DNA was isolated by following the instructions of “Easy Pure Quick Gel Extraction Kit”. PT7-FLAG-MAT-TAG-1 plasmid was extracted by following the instructions of “Easy Pure Plasmid mini Prep Kit”. The purified CotA gene fragments and pT7-FLAG-MAT-TAG-1 plasmid were digested by HindIII and KpnI restriction enzymes. All fragments were incubated with restriction enzymes for 3 h at 37 °C. Purification of the DNA fragments was conducted by using PCR purification Kit. CotA DNA fragment was ligated with pT7-FLAG-MAT-TAG-1 plasmid using T4 DNA ligase. The recombinant plasmid vector was transformed into chemically competent Trans 10 E. coli cells which were used for cloning procedures, propagation, and amplification of the plasmid constructs. Extraction of the recombinant plasmids from chemically competent Trans 10 E. coli cells was performed, and then digested with HindIII and KpnI. Transformation of the recombinant plasmids into chemically competent E. coli BL21 (DE3) was done to functionally express CotA laccase. Successful transformation of produced strains was confirmed by sequencing.
250 mL flasks containing 50 mL LB medium, 100 μg/mL ampicillin and 0.5 mM CuCl2 were inoculated with 0.5 mL of E. coli BL21 (DE3) cells carrying pT7-FLAG-MAT-TAG-1-CotA vector and grown at 37 °C and 200 rpm. After the culture optical density reached 0.7 at OD600, 1 mM IPTG was added for expression induction and the temperature was lowered to 26 °C. After overnight incubation at 130 rpm, cells were separated from the culture medium by 5000×g centrifugation for 10 min. Cell sediment suspension was performed by 20 mM Tris–HCl buffer (pH 7.6). After that, cells were disrupted for 2 min by using BRANSON digital sonifier (pulse on 5 s, pulse off 3 s). Finally, the cell lysate was heated for 20 min at 70 °C followed by cooling for another 20 min in ice, and then centrifugation (14,000×g, 20 min, 4 °C) to remove cell debris and denatured proteins.
The purification of recombinant CotA laccase proteins from the soluble fraction was performed by using Ni-NTA agarose column. PT7-FLAG-MAT-TAG-1-CotA contains MAT-TAG which facilitates the purification through the immobilized metal affinity chromatography. The column was previously equilibrated with balance buffer which composed of 20 mM Tris–HCl buffer (pH 7.5) and 140 mM NaCl. A blue fluorescence protein fraction was bound to the sorbent, when the supernatants of the bacterial extract were filtered through the nickel column. Fractions containing laccase were collected by 20 mM potassium phosphate buffer (PPB) (pH 6) adjusted with 0.2 M imidazole and 0.15 M NaCl. Purified laccase fractions were dialyzed against distilled water over night to remove NaCl, and then concentrated by ultra-filtration (3 kDa molecular weight cut-off membrane). Bradford assay was used to determine CotA laccase concentration using bovine serum albumin (BSA) [16]. Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was utilized for determining the purity and molecular mass of CotA enzyme by using Mini-PROTEAN Tetra electrophoresis system from Bio-Rad. 12% acrylamide gel was used according to Laemmli method [17]. Coomassie brilliant blue stain G-250 was used for gel staining [18] and the purified enzyme molecular weight was estimated by the standard protein marker.
Optimization Process for CotA Laccase Expression
CotA laccase were expressed as described above using the following different growth conditions; induction times (3, 6, 9, 12, 18, 20, 22, 24, 26, 28 h), induction temperatures (18, 20, 25, 30, 35 °C), IPTG concentrations (0.5, 1, 1.5, 2, 2.5 and 3 mM), and addition time of IPTG (0.5, 1, 1.5, 2 and 2.5 h), respectively.
To scrutinize the effect of micro-aerobic cultivation, the optimum cultural conditions were used to design the experiment. Cells were grown under shaking incubation at 130 rpm (25 °C) for 26 h, and then converted into micro-aerobic conditions by simply switching off the shaking function for 4, 6, 8, 10, 12 h, respectively. Optimization experiments were conducted in triplicates.
Measurement of ROS Production
ROS effect was determined for pT7-FLAG-MAT-TAG-1-CotA laccase under aerobic cultivation (26 h) and different micro-aerobic cultivation (2, 4, 6, 8, 10 h). Samples were collected and followed by 5000 rpm centrifugation for 10 min. Cells were suspended after that in 1 mL 100 mM PPB (pH 7.4) then followed by 2 min sonication and 14,000×g centrifugation for 20 min. Supernatants were diluted ×10,000 using phosphate buffered saline (PBS) then, treated with 2′,7′-di-chlorofluorescin diacetate (DCFH-DA) according to Reactive Oxygen Species Assay Kit from Beyotime Institute of Biotechnology (Haimen, China). Finally, the micro-plate containing the mixture was added at 37 °C (30 min) for heating. Spectra Max M5 Multi-Mode Micro-plate Reader (Molecular Devices, California, USA) was used for collecting the data at 488 nm.
Analytical Procedures
TRIzol Reagent (Invitrogen, USA) was used for total RNA isolation. E. coli BL21 (DE3) cells carrying pT7-FLAG-MAT-TAG-1-CotA obtained at 26 h aerobic cultivation and 2, 4, 6, 8, and 10 h micro-aerobic cultivation were collected by centrifugation and frozen in liquid nitrogen. Bacterial cells collected from 0.5 mL liquid broth was suspended by mixing in 1 mL TRIzol reagent and followed by 5 min incubation at room temperature. After that, Chloroform (0.2 mL) was added and mixed vigorously followed by 3 min incubation at room temperature then, the mixture was centrifuged for separation of the aqueous phase which contained RNA from the organic phase. 0.5 mL isopropyl alcohol was added to recover the RNA from the aqueous phase by precipitation. The pellet of RNA was rinsed with 75% pre-cooled ethanol, air-dried and dissolved in 30 µL de-ionized water treated with diethylpyrocarbonate, DEPC (Sigma) to prevent RNA degradation by RNases. The quality and concentration of the total RNA was evaluated at 260 and 280 nm by Q5000 UV–Vis Spectrophotometer (Qua well, USA). RNA was stored at −80 °C until required. About 1547 nanograms of total RNA were processed with RNase-Free DNase (Promega, USA). The cDNA preparation from the total RNA was performed with GoScript Reverse Transcriptase (Promega) using Oligo(dT)15 primer. The RNA and reverse transcription primer was mixed with GoScript reaction mix (total volume 20 µL). The mixture was introduced to PCR cycle [5 min annealing (25 °C), 1 h extension (42 °C) then 15 min inactivation (70 °C)]. After that, cDNA was quantified with GoTaq qPCR Master Mix (Promega) using forward primer MCotA-F and reverse primer MCotA-F where 16S rRNA was used as reference gene (Table 1). 7500 Fast Real-Time PCR system from Applied Biosystems, California USA was used for PCR performance under the following conditions: GoTaq Hot start polymerase activation at 95 °C for 2 min (1 cycle) then 40 cycles of amplification (95 °C, 60 °C, for 15 s and 1 min at each temperature, respectively) and dissociation for 1 cycle at 68 °C. \({{\text{2}}^{-\Delta \Delta {{\text{C}}_{\text{t}}}}}\) method described by Livak and Schmittgen [19] was applied for qRT-PCR data analysis. Samples data normalization was compared to the results of the parallel qRT-PCR assay of the prepared cDNA from the 16S rRNA gene.
Dry cell weight (DCW) was deliberated for cells produced from the experiment of optimizing the induction times and micro-aerobic conditions. Cell collection was performed in pre-weighed tubes by 10,000×g centrifugation for 10 min followed by washing the pellets three times with de-ionized water. Cells were dried at 80 °C.
Spectrophotometric measurement of laccase activity was performed by using ABTS at 25 °C. Extracted CotA laccase was diluted 10× by adding 50 µL from the supernatant to 450 µL 20 mM Tris–HCl buffer, pH 7.6. Diluted samples were kept on ice during measurement. Oxidation of ABTS (Ɛ = 36,000 M−1 cm−1) in 0.1 M PPB (pH 6) was measured at 420 nm. The enzyme quantity needed for oxidation of 1 µmol of ABTS per minute indicates one unit of the enzyme activity. All measurements were done in triplicates.
Dye Decolorization Test for CotA Laccase
The experiment was conducted by using three different textile dyes, Malachite green (λmax = 618), Brilliant yellow (λmax = 490), Crystal violet (λmax = 594), two azo dyes, Congo red (λmax = 495), Methyl red (λmax = 410), and Bromophenol blue (λmax = 592). The reaction mixture contained 5 mM from the dye dissolved in 0.1 M PPB (pH 6) or Tris–HCl buffer (pH 9) and 100 U/L from purified CotA laccase was used to start the reaction. After that, the reaction was incubated under gentle shaking at 55 °C for 12 h. The same reaction mixture without addition of CotA laccase considered as control sample. Thereafter, every 3 h samples were taken and the relative decrease in absorbance was calculated according to the following equation:
All measurements were conducted in triplicates and the means of decolorization percent was calculated.
Results and Discussion
CotA Laccase Gene Cloning and Expression in Recombinant E. coli
pT7-FLAG-MAT-TAG-1 is an E. coli expression vector (4832 bp) and has Met-N-terminal FLAG peptide sequence. This peptide sequence is considered to be a folding enhancer because of the extensions of hydrophilic peptides which exist through refolding and can efficiently promote the folding yield of the recombinant protein [20]. The plasmid vector also contains C-terminal MAT-Tag for metal binding proteins which help in purification and production of soluble proteins especially for those which are usually produced in the form of inclusion bodies such as the CotA laccase. Sequence analysis confirmed the success of the cloning procedures.
CotA laccase was successfully expressed with high volumetric activity 1474 ± 196 U/L (specific activity 5.2 ± 0.2 U/mg). Heating the supernatant at 70 °C is a very useful treatment as it enriches most of the heat resistant proteins and it is also useful for the denaturation of most E. coli proteins [6]. The purified enzyme can be shown from the SDS-PAGE as indicated at Fig. 1. ~65 kDa bands related to the prognosticated size of CotA laccase were observed. The concentration of soluble protein with about 90% purity was approximately 285 ± 4.1 µg/mL as assessed by Bradford assay.
Process Optimization Required for Enhancing CotA Laccase Activity
Optimization process showed great influence on enhancing CotA laccase expression. The highest laccase activity for pT7-FLAG-MAT-TAG-1-CotA among the different induction times was found to be 3010 ± 70 U/L at 26 h induction. After 26 h of incubation time, laccase activity started to decrease rapidly as the cultivation time was extended as shown in Fig. 2a. The low recombinant protein production after 26 h induction time, during stationary or late exponential growth phase, was assigned to the depletion of the medium nutrients concentration [21, 22]. These factors caused the reduction of the cell metabolic activity during the later growth phases and for that reason, gene expression decreased. Optimizing induction temperatures indicated that increasing the temperature over 25 °C did not enhance laccase activity in which the activity decreased rapidly at 35 °C. Figure 2b indicates that optimum induction temperature was found at 25 °C. Decreasing induction temperatures to 20 and 18 °C led to the complete repression of CotA laccase activity. Different induction temperatures have a significant influence on CotA laccase activity. It seems that CotA laccase expression prefer 25 °C to be produced in the soluble form where it is easy to accumulate as inclusion bodies under other tested temperatures [1, 10]. As indicated in Fig. 2c, the maximum CotA laccase activity was accomplished at 2 mM IPTG concentration. Most of literatures studied recombinant CotA laccase expression were using 1 mM IPTG concentration [23, 24]. This difference comes from the fact that these literatures were using different expression systems and the pT7-FLAG-MAT-TAG-1 expression vector used in this study is the first time to be used for CotA laccase expression. As shown in Fig. 2c, the enzyme activity decreased after 2 mM IPTG concentration as result of the drop of metabolic burden in E. coli. The competition for the expression machinery and the side reactions due to the activity of the recombinant proteins are main reasons for originating this burden which can lead to the decline of the biomass yield, cell productivity and viability [25, 26]. Figure 2c elucidates this fact; the growth of the control (E. coli BL21 (DE3) cells without the recombinant plasmid) is more viable when compared to E. coli BL21 (DE3) cells carrying pT7-FLAG-MAT-TAG-1-CotA. Increasing addition time of IPTG to 2 h led to the optimum activity of the enzyme where it reached to 3910 ± 101 U/L. CotA laccase activity started to decrease after increasing IPTG addition time more than 2 h as indicated in Fig. 2d. Optimization of bacterial expression conditions resulted to the enhancement of the of CotA laccase specific activities by 3.2 fold (16.8 U/mg ± 0.7) when compared to non optimized one (5.2 ± 0.2 U/mg).
Optimization of the best conditions for enhancing CotA laccase activity. a Induction time effect on CotA laccase activity and cell growth of E. coli cells with and without CotA gene. b Induction temperature effect on CotA laccase activity and cell growth of E. coli cells with CotA gene. c Effect of different IPTG concentration on CotA laccase activity and cell growth of E. coli cells with and without CotA gene. d Different addition time of IPTG. DCW dry cell weight
Mechanism of Micro-aerobic Cultivation Strategy for Enhancing Recombinant CotA Laccase Production
Micro-aerobic Cultivation Enhance CotA Laccase Activity
As shown in Table 2, micro-aerobic cultivation had a significant effect on enhancing CotA laccase activity. The optimum static incubation time required after 26 h shaking; for obtaining highest volumetric activity was 8 h at which, volumetric activity raised 3.58 fold (13,803 ± 275 U/L) compared to that of produced under aerobic cultivation (3857 ± 101 U/L). To our knowledge, it is the best values for both a native and a recombinant CotA laccase. Furthermore, instantly after the promotion of micro-aerobic conditions, active bacterial growth was decreased and the dry cell weight reached 1.3 ± 0.01 g/L while, under aerobic conditions, the growth reached to 1.8 ± 0.02 g/L as indicated in Fig. 2a. Micro-aerobic cultivation results are related to the alterations in the central metabolism of facultative aerobe E. coli; furthermore, aerobic cultivation is more dynamic for cell growth than alternative metabolic modes [15].
Micro-aerobic Cultivation Increases ROS Production
ROS level was increased rapidly from the 2nd h of micro-aerobic cultivation until reached the maximum at 8 h (52 ± 6.8 µmol/g-cells) as indicated in Fig. 3. Thereafter, ROS decreased rapidly and ended with concentration similar to that observed during aerobic cultivation. In oxic conditions E. coli produce ROS by taking the advantage of the autoxidation of its redox enzymes so that, E. coli would be able to perform respiration [27, 28]. The insufficient reduction of oxygen to water which occurs simultaneously during respiration and exposure to metals causes ROS generation. The oxidative damage produced due to ROS production in prokaryotic cells often lead to activation of antioxidant defense enzymes and regulatory transcription factors genes in order to protect itself against that oxidative damage [29]. Thereafter, the post-translational modifications are induced and give rise to structural and conformational changes within sensory proteins. These structural rearrangements change the protein activity and promote appropriate and regulated response on the transcriptional level [30]. It has been reported that copper chlorides are mostly act like catalyst to convert H2O2 to OH− and can be involved in oxidative stress formation inside the cell [31].The decrease in cell dry weight at 8 h micro-aerobic cultivation in this study indicates the massive production of ROS under these cultivation conditions. This enormous production of ROS decreases the rate of bacterial cell growth and the over expression of any enzyme synthesis can attenuate the cells and inhibit their growth [30, 32].
Micro-aerobic Cultivation Enhances CotA Laccase Gene Expression
The cDNA copy numbers indicated that the level of CotA gene expression during aerobic cultivation was very low. The amount of cDNA started to increase from 2 h micro-aerobic cultivation until reached to the maximum at 8 h. In this hour, the copy number of CotA gene transcript had increased significantly by 62 fold compared to that of aerobic cultivation (Fig. 3). The cultivation transition of E. coli from aerobic to micro-aerobic conditions leads to changes in gene expression, the physiology of metal ions and central metabolism [15]. Durão et al. [12] was the first paper to explore and describe the link between micro-aerobic growth conditions and copper incorporation with CotA laccase to enhance its activity but no data has been published for achieving a better understanding of the relation between micro-aerobic cultivation and enhancing CotA laccase gene expression. According to the results of this study, the induction of CotA laccase under micro-aerobic condition correlated well with the up-regulation related to mRNA expressions and begets subsequent excessive production of ROS.
Effect of CotA Laccase for Dye Decolorization
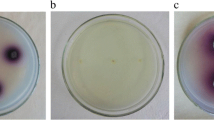
Three textile dyes, two azo dyes and bromophenol blue were used for studying the decolorization ability of the purified CotA laccase. The results indicated that the purified enzyme had the ability to decolorize most of tested dyes efficiently without the presence of any mediator at pH 6. As shown in Fig. 4a, crystal violet was decolorized efficiently (95%) after 12 h incubation while, bromophenol blue, malachite green and congo red were decolorized by about 81, 62 and 59%, respectively. On the other hand, methyl red and brilliant yellow dyes were only decolorized by about 32 and 19%, respectively. These results without the presence of any mediator in the decolorization reaction makes CotA laccase promising candidate in waste water treatment from dyes. Most of mediators are very expensive and generates highly unstable radical intermediates which are environmentally toxic [33]. In order to investigate the ability of CotA laccase for dye decolorization under alkaline conditions, the decolorization reaction was carried out at pH 9. Figure 4b indicated that CotA laccase decolorized the textile dyes malachite green and crystal violet more efficiently (67 and 97%, respectively) under alkaline conditions. The degradation of each dye using CotA laccase was not in the same level due to the difference of the structure, size, charge density and steric hindrances, which play important role for extending the degradation rate. The simple molecule which has a low number of aromatic rings is degraded at a great rate when compared to the complex molecule [34]. The reason of the high decolorization of Crystal violet and bromophenol than malachite green is that Malachite green is classified as triarylmethane dye. Triarylmethane dyes need longer time for decolorization or it could be decolorized to a certain extent [35]. Azo dyes are very difficult to be degradable compared to the other studied dyes as it contains aromatic poly phenolic components. Most dye industries prefer to use azo dyes due to its unique and quality properties. Azo dyes having –N=N– groups which are widely used in industry because of their simple synthesis method. The azo groups are generally connected to benzene and naphthalene rings. These azo groups could be cleaved by oxidative enzymes especially which have high activity [36]. The reason of the high decolorization of Congo red than methyl red is that the azo compounds with hydroxyl or amino groups were efficiently degraded than those with methyl or methoxy groups [36]. Rodrigues et al. [37] reported that textile effluent was characterized by alkaline pH and high temperature in which, most of fungal laccases cannot maintain their activities under such drastic conditions but bacterial laccases are much more stable under these conditions [11]. These disadvantages of fungal laccases are because of their low thermostability, acidic pH optimum and susceptibility to high concentrations of chloride; thus, fungal laccases will lose their activities when they will be used for textile dye decolorization. The other reason is that fungal laccases usually need a redox mediator to degrade the synthetic dyes efficiently with high-redox potential. On the other hand, bacterial laccases have high pH stability at alkaline conditions even at a rather extreme pH of 11 [10] and are resistant to high temperature and chloride concentrations [38]. Subsequently, CotA laccase from B. subtilis have a potential application in the field of waste water treatment from textile and azo dyes.
Conclusions
pT7-FLAG-MAT-TAG-1 expression vector is favorable for cloning and expression of soluble CotA laccase. An efficient process optimization was successfully increased CotA laccase specific activities by 3.2 fold. Micro-aerobic cultivation had significant influence on CotA laccase as it enhanced its volumetric activity by 3.58 fold. The micro-aerobic cultivation strategy we developed enhanced the synthesis of active holo-enzyme, the intracellular ROS production and CotA gene expression. High efficiency of purified CotA laccase in dye decolorization under ph 6 and 9 without the presence on any mediator was successfully achieved. These properties make CotA laccase promising candidate in industry.
References
Brander, S., Mikkelsen, J.D., Kepp, K.P.: Characterization of an akali- and halide-resistant laccase expressed in E. coli: CotA from Bacillus clausii. PLoS ONE 9, e99402 (2014)
Sharma, P., Goel, R., Capalash, N.: Bacterial laccases. World J. Microbiol. Biotechnol. 23, 823–832 (2007)
Dwivedi, U.N., Singh, P., Pandey, V.P., Kumar, A.: Structure–function relationship among bacterial, fungal and plant laccases: review. J. Mol. Catal. B 68, 117–128 (2011)
Lončar, N., Božić, N., Lopez-Santin, J., Vujčić, Z.: Bacillus amyloliquefaciens laccase—from soil bacteria to recombinant enzyme for wastewater decolorization. Bioresour. Technol. 147, 177–183 (2013)
Guan, Z.B., Shui, Y., Song, C.M., Zhang, N., Cai, Y.J., Liao, X.R.: Efficient secretory production of CotA-laccase and its application in the decolorization and detoxification of industrial textile wastewater. Environ. Sci. Pollut. Res. 22, 9515–9523 (2015)
Koschorreck, K., Richter, S.M., Ene, A.B., Roduner, E., Schmid, R.D., Urlacher, V.B.: Cloning and characterization of a new laccase from Bacillus licheniformis catalyzing dimerization of phenolic acids. Appl. Microbiol. Biotechnol. 79, 217–224 (2008)
Dubé, E., Shareck, F., Hurtubise, Y., Daneault, C., Beauregard, M.: Homologous cloning, expression, and characterization of a laccase from Streptomyces coelicolor and enzymatic decolourisation of an indigo dye. Appl. Microbiol. Biotechnol. 79, 597–603 (2008)
Lu, L., Wang, T.N., Xu, T.F., Wang, J.Y., Wang, C.L., Zhao, M.: Cloning and expression of thermo-alkali-stable laccase of Bacillus licheniformis in Pichia pastoris and its characterization. Bioresour. Technol. 134, 81–86 (2013)
Mohorčič, M., Benčina, M., Friedrich, J., Jerala, R.: Expression of soluble versatile peroxidase of Bjerkandera adusta in Escherichia coli. Bioresour. Technol. 100, 851–858 (2009)
Ihssen, J., Reiss, R., Luchsinger, R., Thöny-Meyer, L., Richter, M.: Biochemical properties and yields of diverse bacterial laccase-like multicopper oxidases expressed in Escherichia coli. Sci. Rep. (2015). doi:10.1038/srep10465
Santhanam, N., Vivanco, J.M., Decker, S.R., Reardon, K.F.: Expression of industrially relevant laccases: prokaryotic style. Trends Biotechnol. 29, 480–489 (2011)
Durão, P., Chen, Z., Fernandes, A.T., Hildebrandt, P., Murgida, D.H., Todorovic, S., Pereira, M.M., Melo, E.P., Martins, L.O.: Copper incorporation into recombinant CotA laccase from Bacillus subtilis: characterization of fully copper loaded enzymes. J. Biol. Inorg. Chem. 13, 183–193 (2008)
Salmon, K.A., Hung, S.P., Steffen, N.R., Krupp, R., Baldi, P., Hatfield, G.W., Gunsalus, R.P.: Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J. Biol. Chem. 280(15), 15084–15096 (2005)
Constantinidou, C., Hobman, J.L., Griffiths, L., Patel, M.D., Penn, C.W., Cole, J.A., Overton, T.W.: A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281(8), 4802–4815 (2006)
Partridge, J.D., Sanguinetti, G., Dibden, D.P., Roberts, R.E., Poole, R.K., Green, J.: Transition of Escherichia coli from aerobic to micro-aerobic conditions involves fast and slow reacting regulatory components. J. Biol. Chem. 282, 11230–11237 (2007)
Bradford, M.M.: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976)
Laemmli, U.K.: Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 (1970)
Wilson, C.M.: Staining of proteins on gels: comparison of dyes and procedures. Methods Enzymol. 91, 236–247 (1983)
Livak, K.J., Schmittgen, T.D.: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) method. Methods 25, 402–408 (2001)
Nygren, P.A., Stefan, S., Uhlén, M.: Engineering proteins to facilitate bioprocessing. Trends Biotechnol. 12, 184–188 (1994)
Bentley, W.E., Davis, R.H., Kompala, D.S.: Dynamics of induced cat expression in Escherichia coli. Biotechnol. Bioeng. 38, 749–760 (1991)
Duan, K.J., Lin, M.T., Hung, Y.C., Lin, C.T., Chen, C.W., Sheu, D.C.: Production of GST-SOD fusion protein by recombinant E coli XL1 Blue. J. Chem. Technol. Biotechnol. 75, 722–728 (2000)
Brissos, V., Pereira, L., Munteanu, F.D., Paulo, A.C., Lígia O., Martins, L.O.: Expression system of CotA-laccase for directed evolution and high-throughput screenings for the oxidation of high-redox potential dyes. Biotechnol. J. 4, 558–563 (2009)
Wang, C., Cui, D., Lei Lu, L., Zhang, N., Yang, H., Zhao, M., Dai, S.: Cloning and characterization of CotA laccase from Bacillus subtilis WD23 decoloring dyes. Ann. Microbiol. 66, 461–467 (2016)
Hu, J.H., Wang, F., Liu, C.Z.: Development of an efficient process intensification strategy for enhancing Pfu DNA olymerase production in recombinant Escherichia coli. Bioprocess Biosyst. Eng. 38, 651–659 (2015)
Lecina, M., Sarró, E., Casablancas, A., Godia, F., Cairo, J.J.: IPTG limitation avoids metabolic burden and acetic acid accumulation in induced fed-batch cultures of Escherichia coli M15 under glucose limiting conditions. Biochem. Eng. J. 70, 78–83 (2013)
Imlay, J.A.: The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat. Rev. Microbiol. 11(7), 443–454 (2013)
Imlay, J.A.: Diagnosing oxidative stress in bacteria: not as easy as you might think. Curr. Opin. Microbiol. 24,124–131(2015)
Lucana, D.O.O., Wedderhoff, I., Groves, M.R.: ROS-mediated signalling in bacteria: zinc-containing Cys-X-X-Cys redox centres and iron-based oxidative stress. J. Signal Transduct. (2012). doi:10.1155/2012/605905
Dalton, T.P., Shertzer, H.G., Alvaro Puga, A.: Regulation of gene expression by reactive oxygen. Annu. Rev. Pharmacol. Toxicol. 39, 67–101 (1999)
Kimura, T., Nishioka, H.: Intracellular generation of superoxide by copper sulphate in Escherichia coli. Mutat. Res. 389, 237–242 (1997)
Artsatbanov, V., Vostroknutova, G., Shleeva, M., Goncharenko, A., Zinin, A., Ostrovsky, D., Kapreliants, A.: Influence of oxidative and nitrosative stress on accumulation of diphosphate intermediates of the non-mevalonate pathway of isoprenoid biosynthesis in corynebacteria and mycobacteria. Biochemistry 77, 362–371 (2012)
Moldes, M.D., Sanromán, A.: Amelioration of the ability to decolorize dyes by laccase: relationship between redox mediators and laccase isoenzymes in Trametes versicolor. World J. Microbiol. Biotechnol. 22, 1197–1204 (2006)
Chairin, T., Nitheranont, T., Watanabe, A., Asada, Y., Khanongnuch, C., Lumyong, S.: Biodegradation of bisphenol a and decolorization of synthetic dyes by laccase from white-rot fungus, Trametes polyzona. Appl. Biochem. Biotechnol. 169, 539–545 (2013)
Xu, F.: Oxidation of phenols, anilines and benzenethiols by fungal laccases: correlation between activity and redox potentials as well as halide inhibition. Biochemistry 35, 7608–7614 (1996)
Kanagaraj, J., Senthilvelan, T., Panda, R.C.: Degradation of azo dyes by laccase: biological method to reduce pollution load in dye wastewater. Clean Technol. Environ. Policy 17, 1443–1456 (2015)
Rodrigues, C.S.D., Madeira, L.M., Boaventura, R.A.R: Treatment of textile effluent by chemical (Fenton’s Reagent) and biological (sequencing batch reactor) oxidation. J. Hazard Mater. 172, 1551–1559 (2009)
Liu, H., Cheng, Y., Du, B., Tong, C., Liang, S., Han, S., Zheng, S., Lin, Y.: Overexpression of a novel thermostable and chloride-tolerant laccase from Thermus thermophilus SG0.5JP17-16 in Pichia pastoris and its application in synthetic dye decolorization. PLoS ONE (2015). doi:10.1371/journal.pone.0119833
Acknowledgements
The funding of this study was supported by the National Key Technology Research and Development Program of China (No. 2015BAK45B01), the National Natural Science Foundation of China (No. 21476242), and the National Basic Research Program (973 Program) of China (No. 2013CB733600). We would like to acknowledge the Chinese Government for the financial support of the Ph. D scholarship awarded to Nadia A. Samak.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Samak, N.A., Hu, J., Wang, K. et al. Development of a Novel Micro-Aerobic Cultivation Strategy for High Potential CotA Laccase Production. Waste Biomass Valor 9, 369–377 (2018). https://doi.org/10.1007/s12649-016-9824-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12649-016-9824-6