Abstract
Lymphatic filariasis is one of the major diseases that belong to the category of neglected tropical illness. Filarial nematodes are the cause of the disease and are transmitted to humans via blood-feeding arthropod vectors. Drugs such as Albendazole, Ivermectin and diethylcarbamazine are administered either individually or in combination to overcome the progress of the lymphatic filariasis. These drugs have some minor side effects like temporary hair loss, dizziness, nausea etc. The filarial parasites have multifunctional proteins including the Glutathione-s-transferase (GST) enzyme. This study aims at the identification of a natural molecule that has the potential to bind with the GST enzyme, which plays a major role in detoxification of endogenous electrophilic compounds. Thus the binding interrupts the detoxification process within the filarial parasite, Brugia malayi. A medicinal plant Calotropis procera, owing to its anthelmintic properties was searched for the presence of potential phytocompounds. The phytocompounds were docked against the homology modeled GST enzyme using the MOE software. The results were screened and analyzed based on the Lipinski rule of 5. N-octanoate was the phytocompound obtained based on molecular docking, subjected to molecular dynamics. These results require further in vitro and in vivo validation to consider n-octanoate as a potential drug candidate for lymphatic filariasis treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lymphatic filariasis or Filariasis is one of the neglected tropical diseases caused by the nematode filarial parasites Wuchereria bancrofti, Brugia malayi and Brugia timori. Filariasis causes permanent limb disability and can even cause permanent malformations of several body parts (Ottesen et al. 2008). The filarial parasites are thread-like parasitic nematodes (roundworms) and are transmitted to the human host through blood-feeding arthropod vectors. The adult worms reside in specific tissues within the host where they release thread-like microfilariae into the blood. The circulating microfilariae enter the vector during blood-feeding and develop into an infective larva ready for human transmission. The intensity of the disease in the host depends on the locations of the tissue preferred by the microfilariae and matured worms. The matured worms inhabit lymph vessels, causing lymphatic inflammation and dysfunction due to blockage and host reactions and gradually lead to lymphedema and fibrosis (Stillwaggon et al. 2016). Prolonged and recurrent infection with these worms can lead to a buildup of excess tissue in the affected area, as in elephantiasis (Cross 1996).
Published articles on filariasis emphasize the importance of Glutathione-s-transferases (GST) in chronic infections. GSTs are multifunctional proteins that belong to an enzyme family which filarial parasitic worms depend for their survival in the host (Bhargavi et al. 2005). GSTs are major detoxification enzymes that neutralize cytotoxic compounds and protect cells and tissues from the reactive oxygen species attack. They catalyze the conjugation of Glutathione (GSH) to endogenous electrophilic compounds that are less harmful (Bhoj et al. 2020). These enzymes protect the tissue from damage caused by free radicals, superoxides and aids in the intracellular transport of non-catalytic carrier proteins (Sommer et al. 2001). GST's involvement in drug resistance and biosynthesis of arachidonic acid metabolites makes GST a more attractive target for therapeutics (Van Ommen et al. 1991). Studies prove that GST of humans are structurally different from worms and can be used as a potential target for developing anti-filarial drugs (Brophy et al. 2000).
The present drugs have poor inhibition on adult parasitic worms. Current treatment includes Mass Drug Administration of drugs such as Albendazole, Ivermectin, Doxycycline and Diethylcarbamazine etc. individually or in combination (Brophy and Pritchard 1994). Intake of these drugs shows some trivial side effects like dizziness, temporary hair loss, nausea, fever, headache, and joint pain etc. The mechanism of action and side effects of the drugs used in Filariasis treatment are presented in Table 1. This scenario makes us ponder over an alternative drug that possesses potentiality against the disease with no side-effects.
Traditional medicines from natural resources- phytocompounds have long been promising substitutes in the treatment of various diseases. They exhibit favorable potency, bearable toxicity and curable properties. Calotropis procera Linn. is a common shrub mostly found as a weed around India (Iyadurai et al. 2020), attributed to warm and dry areas, the sub- Himalayan regions and the southern regions of the country. Different parts of C. procera have been reported to possess different therapeutic activities such as proteolytic (Atal and Sethi 1962) antimicrobial, larvicidal (Al-Rowaily et al. 2020), nematocidal, anticancer, anti-inflammatory (Basu and Chaudhuri 1991; Kumar and Basu 1994). Its flowers possess digestive and tonic properties.
This study focuses on proposing a potent lead molecule against filarial elephantiasis from traditional medicinal plant C. procera. Homology modeling, Molecular docking and molecular dynamics studies are performed on a potential drug target of filarial parasite—Glutathione-s-transferase with phytocompounds retrieved from C. procera.
Materials and methods
Target selection
The sequence of Glutathione s transferase of B. malayi was retrieved from UniProt (UniProt ID: A0A0H5S7P0) (2021) having 208 residues and was considered for homology modeling. Homology modeling is a molecular modeling technique, which builds a 3D structure from a template homologous protein structure. In this study, the protein was modeled using an online tool—swiss model (Peitsch 1996). The sequence was uploaded in the swiss model to search for homologous protein templates. Template 5d73.1.A (structure of pi-class glutathione S-transferase of W. bancrofti, which was identified as the causative organism of > 90% filariasis cases) (Farrar et al. 2013) is selected from the results with the following values: GMQE: 0.95, QMEAN: − 0.11, QSQE: 1.00. The modeled structure of Glutathione S transferase of B. malayi has single A chain of 208 residues starting from MET to final residue GLN.
In this study, the binding sites used for docking were retrieved from literature and includes Tyr7, Phe8, Gly12, Leu13, Trp 38, Lys42, Gln49, Leu50, Ser 63, His98, Thr99, Tyr 101, Thr102, Tyr106 and Val202 (Domadia et al. 2008, Kalani et al. 2014 and Mathew et al. 2011).
Target validation
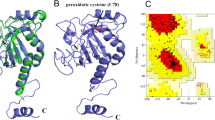
The homology modeled structure was validated using multiple tools like ERRAT, Verify3D and Procheck. (Colovos and Yeates 1993; Laskowski et al. 1993). The modeled protein was uploaded in the SAVESv6.0 (ERRAT—DOE-MBI Structure Lab UCLA) to estimate the ERRAT, Verify 3D and Procheck plots. ERRAT identifies and calculates the statistics of non-bonded interactions between different atom types and generates “the error function versus position of a 9-residue sliding window” plot based on the data from highly refined structures. Verify 3D determines the compatibility of an atomic model (3D) by using its own amino acid sequence (1D). A structural class based on the atomic model’s location and environment (alpha, beta, loop, polar, nonpolar, etc.) is assigned and compared with the results of the best structures. Procheck analyzes residue-by-residue geometry and overall structure geometry (Ramachandran plot, chi1-chi2 plot, main-chain parameters, side chain parameters, residue properties, main-chain bond lengths, main chain bond angles and planar groups) of modelled structure to provide the stereochemical quality of the protein.
Ligand structure collection
Phytocompound composition of C. procera were listed out from literature surveys and 47 ligand structures were retrieved from PubChem database in.sdf file format (Kim et al. 2021).
ADME prediction
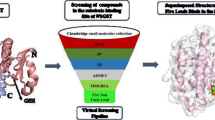
ADME (Absorption, Digestion, Metabolism and Elimination) properties of the phytocompounds were calculated using Swiss ADME. Swiss ADME calculated the ALogp, MLogP, WLogP, Molecular weight, Solubility, GI (Gastrointestinal) absorption, and blood–brain barrier permeability, PAINS (Pan Assay Interfering Compounds) of the selected phytocompounds. The canonical smiles of the phytocompounds retrieved from PubChem were given as an input for ADME prediction. Physicochemical properties of ligands were calculated using DataWarrior (Sander and Freyss 2015). The ligands were filtered under Lipinski rule of 5, mutagenicity and PAINS.
Molecular docking
Molecular docking studies were performed in Molecular Operating Environment (MOE) (ULC 2020). The modeled structure of GST was considered as target protein and ligand molecules (13 ligands) that satisfy Lipinski rule of 5, PAINS (0 alerts) and are non-toxic in nature were considered for docking. Pre-docking procedures such as protein preparation, binding site identification, and ligand database creation were done prior to docking. The purified target structure was loaded, and the structure was prepared to resolve the issues such as H-count, charge, library, etc. (Kute and Thorat 2014). The Protonate 3D tool was used to protonate the target structure after structure preparation and default parameters were used. The binding sites were defined using the sequence editor by selecting the residues and its corresponding atoms. The selected residues were highlighted in the graphic window. A database (.mdb file format) was created for importing ligand structures into a single file in MOE. Docking was performed using the Dock application from the compute tool panel and force field OPLS AA was used for this study. The parameters set for the docking were as follows; Receptor: MOE file, Site: Selected residues, Ligand: MDB file, Method:—Placement: Triangle Matcher; score: London dG; Poses:5, Refinement: rigid receptor; score: Affinity dG; Poses: 5. A file was created explicitly for the output storage and specified in the dock parameter setting panel. Post docking evaluations were performed by analyzing each docking conformation individually. The receptor-ligand interaction analysis approach was used to analyze Hydrogen- bond interactions in favorable sites and S-score. Hydrogen bond interactions between residues of defined binding sites and phytocompounds were considered as favorable H-bond interactions.
Binding affinity calculation was performed using PyRx software by vina wizard (Dallakyan and Olson 2015), the binding affinity of ligands were estimated and analyzed for better interactions. Binding affinity is the strength of the binding interaction of a biomolecule to a ligand/ binding partner.
Molecular dynamics
For molecular dynamics, GROMACS (version 2020.1), an open-source molecular dynamics software, was used (Berendsen et al. 1995). There was a separation of the docked complex into protein and ligand. The force field used was the CHARMM36 all-atom force field, and the CGenFF server supported the ligand parameters and topologies compatible with the above force field. The separated protein and ligand were developed into a complex and ligand topology and parameter files were added to the protein topology file, resulting in the topology file of the complex. In the solvation process the complex was solvated using water. In order to prepare the complex for energy minimization, required ions were added into the solvated complex. Energy minimization was carried out in order to bring the complex into its minimum potential energy. Restraining the ligand and treatment of temperature coupling groups was accompanied by energy minimization. The complex was regulated by equilibration of volume- NVT ensemble (constant Number of particles, Volume, and Temperature) followed by equilibration of pressure- NPT ensemble (Number of particles, Pressure, and Temperature). An MD simulation run for 40 ns was initiated. The trajectories of docked complexes were analyzed through 40 ns of molecular dynamics simulation. The simulated complex was analyzed for Root Mean Square Deviation, Root Mean Square Fluctuation, Radius of gyration, number of hydrogen bonds and interaction energy.
Result and discussion
Glutathione-s-transferase (GST) is the multifunctional protein that comprises major detoxification enzymes. It mainly helps the organism's macromolecules to stay intact from the attack of electrophiles and they catalyze the conjugation of Glutathione to the electrophiles. They are mostly found in the cytosol and in addition to catalyzing the conjugation to electrophiles they perform some peroxidase and isomerase activities. Since it has a significant role in the parasite worm’s life cycle, it can be considered as a drug target.
Sequence from UniProt was used to search for homologous protein templates, and template 5d73.1.A was selected as it had the following parameter values; GMQE (Global Model Quality Estimation): 0.95, QMEAN: − 0.11, QSQE (Quaternary Structure Quality Estimate): 1.00 showed that template 5d73.1.A was a suitable model.
ERRAT plots show a quality factor of 93.9547 which shows that the model has a good quality (Messaoudi et al. 2013). 96.14% of the residues had passed the average 3D- 1D score > = 0.2 when validated by Verify3D (Ullah et al. 2012). In the Ramachandran plot, 0% residues were found in the disallowed region inferring that the model has a good secondary structure (Waghmare et al. 2016).
Glutathione-s-transferase was taken as the target and phytocompounds structures from the plant C. procera were taken as ligands for docking studies. The molecular properties and ADMET descriptors of 47 ligands were calculated and were screened under Lipinski's rule of 5, PAINS, and toxicity. Out of 47, only 13 ligands satisfied Lipinski's rule of 5, PAINS (0 alerts), and non-mutagenic property (Table 2).
The Molecular Operating Environment software was used for molecular interaction study. The protein preparation was done for eliminating the water molecules and adding explicit hydrogens. Binding sites were selected from the literature survey, defined as follows Tyr7, Phe8, Gly12, Leu13, Trp 38, Lys 42, Gln49, Leu50, Ser63, Arg95, His98, Thr99, Tyr101, Thr102, Tyr106 and Val202. Ligands were written into a single database file in MOE and the ligands and protein were subjected to docking.
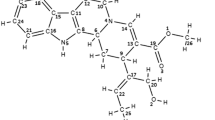
The docking result had 374 poses and top-scored conformations (13 poses) per ligand were selected based on H-bond interactions and S score and also binding affinity of the ligands towards the receptor was calculated (Table 3). The compound with desired hydrogen bond interactions (hydrogen bond in the defined binding sites) and least binding affinity were selected from Table 3. Phytocompound n-Octanoate showed desired hydrogen bond interaction with the given binding sites such as Trp 38, Lys 42, Gln49, have higher binding affinity and exhibit druggable characteristics (Table 4). The 2D plot of interactions and hydrophobic interactions of n-Octanoate with Glutathione-s-transferase was studied (Fig. 1a and b). This ligand has the least binding affinity; hence it has the highest binding stability with the target protein (Glutathione-s-transferase). The ligand n-Octanoate resulted in better docking scores and satisfied Lipinski's rule of 5.
a Hydrophobic surface of interaction n-Octanoate with Glutathione-s-transferase, b 2D interactions of n-Octanoate with Glutathione-s-transferase, c Hydrophobic surface of interaction n-Octanoate with Glutathione-s-transferase after simulation, d 2D interactions of n-Octanoate with Glutathione-s-transferase after simulation
The docked complex was subjected to molecular dynamics using GROMACS. The temperature was equilibrated at 300 K and the pressure was equilibrated at 1 bar for a time for MD run, 40 ns. Figure 1c and d depicts the hydrogen bond interactions of the ligand with the protein and residues Lys 42 and Trp 38 maintained hydrogen bonds with the ligand n- Octanoate after the simulation. i.e., two out of three hydrogen bonds were still intact through the simulation until the end of simulation which points that the compound n- Octanoate is a successful binder with the receptor protein. The RMSD (Root Mean Square Deviation) (Fig. 2a) plot and RMSF (Root mean square fluctuation) plot (Fig. 2b) of the complex after simulation were obtained. The RMSD deviates in between 0.1 and 0.2 nm, stabilizes at 0.15 nm from 20 ns up to the end of simulation, showing less deviation in RMSD (Cob-Calan et al. 2019). The part of the protein that fluctuates mostly during the period of simulation can be understood from RMSF, infers in the flexibility of protein in a binding state. Residue 208 shows the greater fluctuation among the total protein residues. The Radius of Gyration defines the rigidity of the system, greater degree of fluctuation results in the inconsistency throughout the simulation shows the compactness of the system. The RoG of the complex fluctuated in between 1.7 and 1.75 m, with an average of 1.725 nm and found more fluctuating at 25 ns inferring in the higher compactness/ rigidity of the system (Fig. 2c) (Turner et al. 2019). The total number of hydrogen bonds in the complex throughout the simulation was also estimated and a maximum of 182 hydrogen bonds, a minimum of 140 hydrogen bonds, an average of 165 bonds were found to be in the complex (Fig. 2d). Interaction energies of the complex including Lennard- Jones short-range and Coulombic short-range potential energies were calculated. The Lennard–Jones short-range potential energy of the complex was found to be − 31.5848 kJ/mol with a total drift of 53.4165 kJ/mol (Fig. 3a) and Coulombic shot-range potential energy, − 6.63557 kJ/mol with a total drift of 9.61891 kJ/mol (Fig. 3b).
In the future, several factors will be playing a prominent role in the prevalence of lymphatic filariasis. A study suggests that changing patterns of climate will be interfering in the spread of parasitic diseases. A re-emergence of parasitic diseases is possible due to global warming and its associated changes in the environment (Rodó et al. 2013; Wu et al. 2016). The migration to metropolitan cities and international trade/Global market can be a potential channel to spread transmittable diseases (Irvine et al. 2015). Mass Drug Administration (MDA) is controlling the disease to an extent (Kalyanasundaram et al. 2020). Mutation of parasitic nematodes can decrease the efficiency of MDA (Kwarteng et al. 2016). Additional measures like morbidity management, vector control and surveillance must be employed more efficiently to minimize the spread of the disease (Famakinde 2018) and (Irvine et al. 2015) Vector control, reducing human—vector contact and improvising the standard of living can be crucial in eliminating filariasis (Rebollo and Bockarie 2017).
Conclusion
The docking studies using Glutathione-S- transferase of B. malayi as the target intend to find a potent drug candidate for Lymphatic Filariasis. Phytocompounds from the plant C. procera were used as the ligand molecules. The phytocompound n-Octanoate have shown promising results and can be considered for the purpose. This ligand has H-bond interaction with the binding sites of GST. N-octanoate showed the least binding energy − 4.9 kcal/mol from the screened phytocompound database. Further in vitro and in vivo studies can be used to confirm these candidates as a potential drug that can be used against Filariasis.
References
Al-Rowaily SL, Abd-ElGawad AM, Assaeed AM, Elgamal AM, Gendy AE-NGE, Mohamed TA, Dar BA, Mohamed TK, Elshamy AI (2020) Essential oil of Calotropis procera: comparative chemical profiles, antimicrobial activity, and allelopathic potential on weeds. Molecules 25(21):5203
Atal C, Sethi P (1962) Proteolytic activity of some indian plants. Planta Med 10:77–90
Basu A, Chaudhuri AKN (1991) Preliminary studies on the antiinflammatory and analgesic activities of Calotropis procera root extract. J Ethnopharmacol 31:319–324
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91(1–3):43–56
Bhargavi R, Vishwakarma S, Murty US (2005) Modeling analysis of GST (glutathione-S-transferases) from Wuchereria bancrofti and Brugia malayi. Bioinformation 1:25
Bhoj PS, Bahekar S, Khatri V, Singh N, Togre NS, Goswami K, Chandak HS, Dash D (2020) Role of glutathione in chalcone derivative induced apoptosis of Brugia malayi and its possible therapeutic implication. Acta Parasitol 66:406–415
Brophy PM, Pritchard DI (1994) Parasitic helminth glutathione S-transferases: an update on their potential as targets for immuno-and chemotherapy. Exp Parasitol 79:89–96
Brophy PM, Campbell AM, van Eldik AJ et al (2000) β-Carbonyl substituted glutathione conjugates as inhibitors of O. volvulus GST2. Bioorg Med Chem Lett 10:979–981
Cob-Calan NN, Chi-Uluac LA, Ortiz-Chi F, Cerqueda-García D, Navarrete-Vázquez G, Ruiz-Sánchez E, Hernández-Núñez E (2019) Molecular docking and dynamics simulation of protein β-tubulin and antifungal cyclic lipopeptides. Molecules 24(18):3387
Cross JH (1996) Filarial nematodes. Med Microbiol, 4th edn
Dallakyan S, Olson AJ (2015) Chemical biology. Chem Biol Methods Protoc 1263:220–243
Domadia PN, Bhunia A, Sivaraman J et al (2008) Berberine targets assembly of Escherichia coli cell division protein FtsZ. Biochemistry 47:3225–3234
Famakinde DO (2018) Mosquitoes and the lymphatic filarial parasites: research trends and budding roadmaps to future disease eradication. Trop Med Infect Dis 3:4
Farrar J, Hotez PJ, Junghanss T et al (2013) Manson’s tropical diseases E-Book. Elsevier health sciences, Amsterdam
Irvine MA, Reimer LJ, Njenga SM et al (2015) Modelling strategies to break transmission of lymphatic filariasis-aggregation, adherence and vector competence greatly alter elimination. Parasit Vectors 8:1–19
Iyadurai R, Gunasekaran K, Jose A, Pitchaimuthu K (2020) Calotropis poisoning with severe cardiac toxicity A case report. J Fam Med Prim Care 9(8):4444
Kalani K, Kushwaha V, Sharma P et al (2014) In vitro, in silico and in vivo studies of ursolic acid as an anti-filarial agent. PLoS ONE 9:e111244
Kalyanasundaram R, Khatri V, Chauhan N (2020) Advances in vaccine development for human lymphatic filariasis. Trends Parasitol 36(2):195–205
Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE (2021) PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Research, [online] 49(D1), pp.D1388–D1395. Available at: https://academic.oup.com/nar/article/49/D1/D1388/5957164
Kumar VL, Basu N (1994) Anti-inflammatory activity of the latex of Calotropis procera. J Ethnopharmacol 44:123–125
Kute SS, Thorat SD (2014) A review on various software development life cycle (SDLC) models. Int J Res Comput Commun Technol 3:778–779
Kwarteng A, Ahuno ST, Akoto FO (2016) Killing filarial nematode parasites: role of treatment options and host immune response. Infect Dis Poverty 5:1–6
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Mathew N, Srinivasan L, Karunan T et al (2011) Studies on filarial GST as a target for antifilarial drug development—in silico and in vitro inhibition of filarial GST by substituted 1, 4-naphthoquinones. J Mol Model 17:2651–2657
Messaoudi A, Belguith H, Ben HJ (2013) Homology modeling and virtual screening approaches to identify potent inhibitors of VEB-1 β-lactamase. Theor Biol Med Model 10:1–10
Ottesen EA, Hooper PJ, Bradley M, Biswas G (2008) The Global programme to eliminate lymphatic filariasis: health impact after 8 Years. PLoS Negl Trop Dis 2:e317
Peitsch MC (1996) ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem Soc Trans 24(1):274–279
Rebollo MP, Bockarie MJ (2017) Can lymphatic filariasis be eliminated by 2020? Trends Parasitol 33:83–92
Rodó X, Pascual M, Doblas-Reyes FJ et al (2013) Climate change and infectious diseases: can we meet the needs for better prediction? Clim Change 118:625–640
Sander T, Freyss J (2015) Mütevazı von Korff, christian rufener. datawarrior: kimya için veri görselleştirme ve analizini destekleyen açik kaynakli bir Program. J Chem Inf Model 55:460–473
Sommer A, Nimtz M, Conradt HS et al (2001) Structural Analysis and Antibody Response to the Extracellular Glutathione S-Transferases from Onchocerca volvulus. Infect Immun 69:7718–7728
Stillwaggon E, Sawers L, Rout J et al (2016) Economic costs and benefits of a community-based lymphedema management program for lymphatic filariasis in Odisha State, India. Am J Trop Med Hyg 95:877–884
Turner M, Mutter ST, Kennedy-Britten OD, Platts JA (2019) Molecular dynamics simulation of aluminium binding to amyloid-β and its effect on peptide structure. PLoS ONE 14(6):e0217992
ULC CCG (2020) Molecular operating environment (MOE), 2019.01
Ullah M, Ghosh T, Ishaque N, Absar N, Hira J (2012) A Bioinformatics Approach for Homology Modeling Binding Site Identification of Triosephosphate Isomerase from Plasmodium falciparum 3D7. J Young Pharm 4(4):261–266
(2021) UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. https://doi.org/10.1093/nar/gkaa1100
Van Ommen B, Ploemen J, Bogaards JJP et al (1991) Irreversible inhibition of rat glutathione S-transferase 1–1 by quinones and their glutathione conjugates. Struct Act Relatsh Mech Biochem J 276:661–666
Waghmare S, Buxi A, Nandurkar Y et al (2016) In silico sequence analysis, homology modeling and function annotation of leishmanolysin from Leishmania donovani. J Parasit Dis 40:1266–1269
Wu X, Lu Y, Zhou S et al (2016) Impact of climate change on human infectious diseases: Empirical evidence and human adaptation. Environ Int 86:14–23
Acknowledgements
I am grateful to Dr. Achuthsankar S. Nair, Head of the Department, Department of Computational Biology and Bioinformatics, University of Kerala for rendering all the facilities for the completion of the work. I am particularly thankful to Mr. Vinod M. P., Ms. Chinchu E. R., Mr. Ashish Sudharshan, Ms. Roshny Prasad, Dept. of Computational Biology & Bioinformatics, University of Kerala for the support and encouragement
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Optional: please review the submission guidelines from the journal whether statements are mandatory.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflicting financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mohan, A., Shaji, S., Padmanabhan, S. et al. The potentials of Calotropis procera against filarial elephantiasis: an in-silico approach. J Parasit Dis 46, 384–394 (2022). https://doi.org/10.1007/s12639-021-01456-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12639-021-01456-0