Abstract
The past decade has brought a significant rise in antimicrobial resistance, and the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter species) have considerably aggravated a threat to public health, causing nosocomial infections worldwide. The objective of the current study was to isolate novel probiotic strain with antimicrobial activity against multidrug-resistant ESKAPE pathogens. For this purpose, eighteen breastfed infant faeces were collected and lactic acid bacteria (LAB) with antagonistic activity were isolated. Out of 102 anaerobic LAB isolated, only nine exhibited inhibitory activity against all ESKAPE pathogens. These selected nine isolates were further characterized for their probiotic attributes such as lysozyme tolerance, simulated gastrointestinal tolerance, cellular auto-aggregation and cell surface hydrophobicity. Bile salt deconjugation and cholesterol-lowering capacity was also determined. Among all nine, isolate LBM220 was found to possess superior probiotic potential. Confirmatory identification of isolate LBM220 was done by both 16S rRNA sequence analysis and mass spectrometric analysis using MALDI-TOF. Based on BLAST result, isolate LBM220 was identified as Lactobacillus gasseri. Phylogenetic analysis of Lactobacillus gasseri LBM220 [accession number MN097539] was performed. Also, detailed safety evaluation study of Lact. gasseri LBM220 showed the presence of intrinsic antibiotic resistance and the absence of hemolytic, DNase, gelatinase and toxic mucinolytic activity. Time kill assay was also performed to confirm the strong kill effect of Lact. gasseri LBM220 on all six multidrug resistant ESKAPE pathogens. Thus, Lact. gasseri LBM220 can be utilized and explored as potential probiotic with therapeutic intervention.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With advent of the antibiotic era, the exaggerated and imprudent utilization of antibiotics has led to the cumulative acquisition of resistant traits in many human pathogens resulting in the rapid emergence of multidrug-resistant (MDR) bacteria, which are practically beyond any treatment regimen [1]. Multidrug-resistant pathogens impart major burden on healthcare systems, such as elevated rates of mortality and morbidity, diagnostic uncertainties, exorbitant treatment costs and lack of trust in medicines available in the market [2]. For example, the total cost of extended hospitalization due to bloodstream infections caused by third-generation cephalosporin-resistant Enterobacteriaceae, methicillin-susceptible Staphylococcus aureus (MSSA) or methicillin-resistant Staphylococcus aureus (MRSA) was found to be EUR 970,000 in a retrospective cohort study on ten European hospitals, which participated in Infection Control Program, Switzerland [2]. The term ‘ESKAPE’ has been introduced by Infectious Diseases Society of America and hospital-based surveillance studies, for a group of six pathogens, including both Gram negative and Gram positive bacterial species. These are Enterococcus faecium, Staphylococcus aureus, Klebseilla pneumonia, Acinetobater baumannii, Pseudomonas aeruginosa and Enterobacter species [3, 4]. These nosocomial ‘ESKAPE bacteria’ embody paradigms of pathogenesis and disease transmission and have derived mechanisms to counterattack the repercussions of antibiotics as an adaptive trait to survive. Thus, multidrug-resistant ESKAPE pathogens are a major problem to public health systems worldwide and are likely to increase in the near future [5]. Recently, decline in new class of antibiotics coming to the market have further aggravated the antibiotic resistant problem. Therefore, in the last decade, the use of additives with antimicrobial potential has gained momentum as it does not induce antimicrobial resistance. Among those additives, probiotics are considered to be one among the most appropriate alternatives as they tend to impart several health benefits to the host, particularly by ameliorating intestinal microbial balance. Probiotics are described as ‘live microorganisms which when consumed in sufficient amount impart several health benefits to the host’ [6]. The majority of probiotics belong to Lactic acid bacteria (LAB), common among them are the species of genus Lactobacillus, which belong to ‘Generally regarded as safe’ (GRAS) status. LAB is a heterogeneous class of bacteria that possess common metabolic attributes, such as the production of lactic acid, which is their major end product of carbohydrate fermentation. They also produce numerous metabolites like bacteriocins, organic acids and hydrogen peroxide which contribute to their antimicrobial activity. Apart from its inhibitory property, probiotics provide several other potential health benefits, such as lowering of cholesterol [7], anti-obesity [8], cancer suppression [9], anti-allergic [9], anti-diabetic [10], improved digestion [11], alleviation in oxidative stress-related diseases [12], lactose intolerance and irritable bowel syndrome [13]. In order to qualify the strain to be prospective probiotic, it must fulfil certain conditions such as tolerance to simulated gastrointestinal stress, cellular adhesion ability and production of inhibitory substances [14]. Also, it must be safe for use based on parameters such as antibiotic susceptibility, toxin production and mucin degradation [15]. Thus, the objective of the present study was to characterize the LAB isolated from exclusively breast-fed infant faeces, select the LAB isolates that possessed antimicrobial activity against multidrug-resistant ESKAPE pathogens, followed by in vitro determination of their functional probiotic attributes. Finally, the most potential probiotic isolate was identified and evaluated for its safety aspect.
Materials and Methods
Collection of Samples
A total of eighteen healthy, exclusively breastfed infants (< 9 months), were included in the study. The faecal samples were obtained directly from the diaper in a sterile container and kept in cold till further processing. As per exclusion criteria, premature infants and infants on antibiotics or probiotics were not considered from enrolment. The present work was conducted according to stipulated guidelines. Written informed consent was obtained from both the parents after briefing the research objectives. The study was approved by the institutional ethical review board of Dr. Ram Manohar Lohia Institute of Medical Sciences, Lucknow (Ref. No. 2784/RMLIMS/2018).
Bacterial Strains
The clinical multidrug resistant (MDR) ESKAPE pathogens Enterococcus faecium, Staphylococcus aureus, Klebseilla pneumonia, Acinetobater baumannii, Pseudomonas aeruginosa and Enterobacter aerogenes were included in the study. All pathogenic MDR strains were procured from Department of Microbiology, Dr. Ram Manohar Lohia Institute of Medical Sciences, Lucknow.
Isolation of LAB with Inhibitory Activity Against MDR Pathogens
Faecal samples (10 g) were mixed with 90 ml peptone water and vortexed for 5 min. The homogenized samples were then serially diluted with 0.85% (w/v) normal saline and 100 μl dilutions were inoculated on De Man Rogosa Sharpe agar (MRS; Himedia, India), followed by anaerobic incubation at 37 °C. After 48 h, pure colonies were obtained and characterized using Gram stain, cell morphology and catalase reaction. In the second set of experiments, plates of Gram-positive and catalase-negative isolates, presumptive of LAB were individually overlaid with Brain heart infusion agar (BHI; Oxoid, Basingstoke, UK) seeded with clinical MDR ESKAPE pathogens as given above (approx. 106cfu/ml). Plates overlaid with indicator pathogens without LAB isolates served as control. Both test and control plates were incubated for 24 h at 37 °C. Bacterial colonies exhibiting inhibition were picked and maintained in MRS broth containing 30% (v/v) glycerol and stored at − 80 °C. For further study, isolates were subcultured twice in MRS broth anaerobically at 37 °C for 48 h.
Confirmation and Characterization of the Inhibitory Substances in Culture Supernatant
As the major objective of the present study was to isolate Gram-positive probiotic bacteria for controlling the growth of MDR ESKAPE pathogens, the faecal isolates that showed the antagonistic activity at first screening were then selected and evaluated for their production of inhibitory substances using the agar well diffusion assay as described by Yu et al. [16]. Cell-free culture supernatant (CFCS) of selected LAB isolates was collected by centrifugation (12,000×g, 15 min,4 °C) from overnight cultures. After filter sterilization using a 0.22 μ filter, the supernatant of each isolate was divided into four aliquots. First aliquot was neutralized using 1 M NaOH, followed by heating at 80 °C for 15 min. In second aliquot, 0.5 mg/mL catalase (Himedia, India) enzyme was added to determine inhibitory activity due to hydrogen peroxide, while 1 mg/ml pronase (Himedia, India) was added in third aliquot to determine bacteriocin production. The last aliquot of CFCS without any adjustment served as control. Sterile culture plates containing 20 ml Mueller Hinton broth (MHB; Oxoid, Basingstoke, UK) in 1.2% (w/v) agar were then seeded with each indicator ESKAPE pathogen. A hole of 6 mm diameter was punctured aseptically into the agar layer and filled with CFCS (80 μl). The plates were incubated aerobically at 37 °C for 24 h. Thereafter, inhibition zone sizes were measured, recorded and expressed as weak (7–9 mm), intermediate (10–13 mm), strong (14–16 mm), and very strong (> 17 mm) according to Sirichokchatchawan et al. [17]. Each assay was performed in triplicates and mean + SD is presented.
In Vitro Determination of Functional Probiotic Properties
Resistance to Lysozyme
In order to assess the ability to survive through the oral cavity, isolates were tested for lysozyme tolerance as described by Turchi et al. [18] with slight modifications. Overnight grown LAB were harvested by centrifugation (8000×g, 15 min, 4 °C), washed twice with PBS (pH 6.5) and resuspended in 2 ml PBS supplemented with 100 mg/L lysozyme (Himedia, India). Cell suspensions without lysozyme served as control. After incubating anaerobically at 37 °C for 90 min, viable cell counts were determined by plating 50 μl diluted cultures onto MRS agar. Assays were done in three replicates and results expressed as mean of log cfu/ml ± SD. Percent viability was calculated using the formula:
Tolerance to Simulated Gastric and Intestinal Juices
Resistance of LAB isolates to simulated gastrointestinal environment was determined according to modified protocol of de Moraes et al. [19]. LAB cells were collected by centrifugation (8000×g, 15 min, 4 °C), washed twice and resuspended in freshly prepared simulated gastric fluid containing 2.0 g/L pepsin in PBS adjusted to pH 2.0. Viable counts were determined after 120 min anaerobic incubation at 37 °C. Similarly, LAB cells were suspended in simulated intestinal fluid, containing 250 mg/L pancreatin (Himedia, India) and 0.3% w/v oxgall (Sigma-Aldrich) in PBS at pH 8.0 and incubated for 120 min at 37 °C. Viability was determined before and after exposure to test conditions by surface plating on MRS agar. Assay was performed in triplicates and expressed as a mean of log cfu /ml ± SD. Percent viability was calculated using the formula:
Cell Surface Hydrophobicity
Adhesion of LAB isolates to hydrocarbons was carried out using both xylene and n-hexadecane. Overnight grown LAB were harvested by centrifugation (12,000×g, 15 min, 4 °C), washed twice and resuspended in PBS to obtain absorbance in the range of 0.8–1.0 at 600 nm (Ao). To 3 ml of cellular suspension, 1 ml of each hydrocarbon was mixed followed by thorough vortexing for 5 min and 1 h incubation for phase separation. Then, A600 value (A) of the aqueous layer was determined using Nanodrop spectrophotometer (DS-11, Denovix, USA), and results were expressed as a percentage of hydrophobicity (% H) = (A0 − A) / A0 × 100, where A0 and A were OD values before and after extraction with organic solvent, respectively. The assay was carried out in three replicates.
Auto-aggregation Ability
Overnight grown LAB were harvested by centrifugation (12,000×g, 15 min, 4 °C), washed twice and resuspended in PBS to obtain absorbance in the range of 0.8–1.0 at 600 nm (Ao). After thorough vortexing, cellular suspensions were incubated for time t (4 h and 24 h). Thereafter, A600 value (At) was determined using Nanodrop spectrophotometer, and results were expressed as a percentage of auto-aggregation (%A) = (A0 − At) / A0 × 100, where At is OD value at time t = 4 h or 24 h and Ao is the OD at t = 0 h. The assay was carried out in three replicates.
Bile Salt Deconjugation Ability
Qualitative bile salt deconjugation ability of selected LAB isolates was determined using agar plate assay as described by Shehata et al. [20] with some modifications. The 24-h grown cultures of LAB isolates (10 μl) were spotted on MRS agar plates which were supplemented with 0.5% (w/v) sodium taurodeoxycholate (Himedia, India) and 0.04% calcium chloride (Himedia, India). All plates were then incubated anaerobically at 37 °C for 72 h. The positive result was confirmed by opaque halo around colonies formed due to precipitation of bile salt. MRS agar plate without bile salt served as control.
Cholesterol-Lowering Property
Ability of LAB to reduce cholesterol in spent broth was determined as per the protocol of Rastogi et al. [21]. Overnight cultures were inoculated in MRS broth suspended with 0.8% (w/v) oxgall (Sigma-Aldrich) and 0.1 g/L water-soluble cholesterol (Sigma-Aldrich) and incubated at 37 °C for 72 h under anaerobic condition. Sterile MRS broth without test organism served as control. After incubation, the supernatant was collected and used for quantifying cholesterol reduction. Briefly, 2 ml KOH (45% w/v) and 3 ml ethanol were added to 1 ml of clear supernatant, mixed and heated at 60 °C for 10 min. After cooling, 5 ml hexane was added and allowed to stand for phase separation. The hexane layer was transferred to a clean glass tube and evaporated. The residue obtained was dissolved in O-phthalaldehyde and vortexed. After mixing, 2 ml conc. sulphuric acid was added and absorbance (A) was read at 552 nm.
Cholesterol reduction was calculated as follows:
Identification of Selected LAB
Identification by Mass Spectrometric Analysis
To identify, the 24-h grown pure culture of most potential LAB isolate was used, from which pinch of colony was taken and placed on specialized disposable slide using a toothpick. Immediately afterward, cells were lysed with 0.5 μL of formic acid (25% v/v) and allowed to dry at room temperature. Thereafter, 1 μL of matrix solution (3.1% (w/v) α-cyano-4-hydroxycinnamic acid) was added and allowed to dry. The prepared slide was analysed using Vitek® MS-Plus mass spectrometer (bioMériux, Marcyl’Etoile, France) in linear positive-ion mode, across the mass to-charge ratio range of 2000 to 20,000 Da. The sample was irradiated with 50 laser shots per second at 50 Hz. The equipment performed the calibration using Escherichia coli ATCC 8739, prior to the analysis of the sample. The results were obtained and displayed by the Myla v2.4 middleware software.
Molecular Identification
Genomic DNA of the selected LAB isolate was extracted using DNA extraction kit (Himedia, India) according to manufacturer’s protocol and stored at − 20 °C. 16S ribosomal RNA (rRNA) gene amplification was performed in a thermocycler using universal primers 27F and 1492R. PCR products obtained were separated by electrophoresis in 0.8% (w/v) agarose gels in 0.5 × TAE buffer at 100 V for 1 h. Gels were stained in 0.5 × TAE buffer containing 0.5 μg/ml ethidium bromide (Sigma Diagnostics, USA). The resulting amplicons obtained were sequenced with primers 785F and 907R using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). The sequences obtained were aligned and compared to known sequences in GenBank using the National Centre for Biotechnology Information (NCBI) software tool BLAST. Based on the highest hit scores, the strain was identified as Lactobacillus gasseri. Sequences of lactobacilli strains nearest to the identified isolate LBM220 were retrieved from GenBank database and aligned using Clustal Omega. Phylogenetic analysis involved 16S rRNA nucleotide sequences of 17 strains using MEGA X program. Phylogenetic relationship was inferred using neighbour-joining method while evolutionary distances were computed using the Maximum Composite Likelihood method.
Time-Kill Assay with CFCS of Lact. gasseri LBM220 on Multidrug-Resistant ESKAPE Pathogens
Time-kill assay was determined by treating multidrug resistant ESKAPE pathogens with CFCS of Lact. gasseri LBM220 as described by Zhang et al. [22] with slight modification. To perform, 500 μl of pathogen suspension (108 cfu ml−1) was added into 20 ml of either CFCS or CFCS at pH 6.5. For control, MRS broth (pH 6.5) was taken. Test and control tubes were incubated aerobically at 37 °C. Aliquots were removed at regular intervals of 2 h (t = 0, 2, 4, 8 h), serially diluted and plated on BHI agar to assess the viability of pathogens after co-incubation with culture supernatant. The assay was performed in triplicates and results expressed as mean of log cfu ml−1 ± SD.
Safety Evaluation of Lact. gasseri LBM220
Antibiotic Susceptibility
Antibiotic susceptibility of Lact. gasseri LBM220 was determined using modified Kirby Bauer disc diffusion method against 14 clinically relevant antibiotics (Oxoid, Basingstoke, UK), namely ampicillin (10 μg), cefoxitin (40 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamicin (120 μg), tetracycline (30 μg), penicillin (10 μg), erythromycin (10 μg), cefotaxime (30 μg), vancomycin (30 μg), fosfomycin (200 μg), tobramycin (10 μg), linezolid (30 μg) and doxycycline (30 μg). Each antibiotic disc was dispensed on MRS agar inoculated with 0.5 McFarland turbid cultures. After anaerobic incubation at 37 °C for 48 h, inhibition zones were recorded, and results from the three independent experiments were interpreted as sensitive or resistant based on CSLI 2018 guidelines [23].
Blood Hemolysis Test
Fresh overnight culture of Lact. gasseri LBM220 was streaked on 5% (w/v) Sheep Blood Agar (BD Scientific, India) and anaerobically incubated at 37 °C for 48 h. Positive hemolytic colonies were examined for β-hemolysis (clear zones around colonies), α-hemolysis (greenish zones around colonies) or γ-hemolysis (no clear zones around colonies). Staph. aureus ATCC 25923 was used as positive control.
DNase and Gelatinase Test
Twenty-four-hour grown culture was inoculated on DNase agar (Oxoid, Basingstoke, UK) and anaerobically incubated at 37 °C for 72 h. After incubation, plates were flooded with 3% (v/v) HCl, kept for 8 min and examined for clear halo in case of positive colonies. Similarly, toxic gelatinase producing ability was confirmed by spotting 10 μl Lact. gasseri LBM220 onto surface of MRS agar supplemented with 3% (w/v) gelatin (Himedia, India). Gelatin hydrolysis was observed as opaque halo around colonies after anaerobic incubation for 72 h. Staph. aureus ATCC 25923 was used as a positive control for both the experiments.
Mucin Degradation Ability
Toxicity of Lact. gasseri LBM220 in degrading gastric mucin in vitro was carried out according to the modified protocol of Martín et al. (2006). Partially purified 0.5% (w/v) hog gastric mucin (HGM; Sigma-Aldrich, USA) and 1.5% (w/v) agarose (Himedia, India) were added to the anaerobic culture medium without glucose for the experiment. The modified media was then seeded with overnight culture of test organism, followed by anaerobic incubation at 37 °C for 72 h. After incubation, plates were stained with 0.1% (w/v) amido black in acetic acid, kept for 30 min and washed with 1.2 M acetic acid. Positive colonies were observed for mucin lysis zone. Salmonella typhimurium and Shigella flexneri served as positive control.
Accession Number
The nucleotide sequence of 16S rRNA of strain was deposited at the GenBank database under the following accession number: Lact. gasseri LBM220 (MN097539).
Statistical Analysis
All the experiments were carried out in triplicates and mean ± standard deviation (SD) of experimental data was calculated using Microsoft Excel 2010, Microsoft Corporation (USA).
Results and Discussion
Isolation of LAB with Antimicrobial Activity Against MDR Pathogens
All eighteen faecal samples were cultured anaerobically for isolation and a total of 102 anaerobic Gram-positive, catalase-negative LAB isolates were obtained. All these pure cultures were evaluated for their antagonistic potential against multidrug-resistant ESKAPE pathogens by employing a double-agar-layer assay as described previously. After comparison with controls, only nine isolates among all tested LAB isolates showed significant antagonistic effect against all indicator MDR pathogens, with variable degree of antagonism. Thus, for further determination of inhibitory substances and characterization of probiotic potential, only these nine isolates (LBV12, LBN16, LBX18, LBS310, LBP218, LBL19, LBQ12, LBM220, LBM108) were selected.
Characterization of Antimicrobial Activity of Inhibitory Metabolites
The antimicrobial activity of LAB is primarily caused due to production of antimicrobial peptides (AMPs) such as bacteriocins, hydrogen peroxide and organic acids, such as lactic acid, acetic acid and propionic acid, and these metabolites may have diverse mechanisms of action. Thus, in order to elucidate the presence of inhibitory metabolites in CFCS of nine selected LAB isolates, agar well diffusion assay against all multidrug-resistant ESKAPE pathogens was employed and interpretations were done on the basis of inhibition zone. The results showed that the CFCS of all nine cultures treated with 1 mg/mL pronase did not affect their inhibitory activity against the indicator pathogens, demonstrating their inability to produce inhibitory peptides such as bacteriocin. Similarly, results of CFCS treated with 0.1 mg/mL catalase also did not affect their antimicrobial activity, indicating no hydrogen peroxide production. However, neutralized culture-free supernatant (pH 6.5) of all isolates did not inhibit test pathogens, as no significant zone of inhibition was observed. This indicates that their antimicrobial activity is mainly attributed due to organic acid production. Studies have shown that LAB strains can produce organic acids through heterofermentative pathway. These acids may tend to interact with plasma membrane of bacteria, inducing intracellular acidification and protein denaturation. Bacteriocidal effect linked to the most common organic acid, lactic acid is probably due to the induction of morphological and physiological changes in cellular membrane leading to leakage of cellular contents [24]. In the present study, cell-free culture supernatant of selected isolates, without any treatment, was found to inhibit all multidrug-resistant ESKAPE pathogens to varying levels (moderate to very strong), ranging from 9 to 21 mm zone size as represented in Table 1. Of all isolates, LBL19 displayed maximum inhibition halo against Ent. faecium (MDR) of 21 mm diameter; while LBM220 was found to be the most effective against Enterobacter aerogenes (MDR) and Ac. baumannii (MDR) with an inhibition zone of 18 mm. LBQ12 and LBM16 also showed very strong inhibition (> 17 mm) against Staph. aureus (MDR) and Kl. pneumonia (MDR) respectively. Similar to our results, Abdelhamid et al. [25] reported high antagonistic activity of six probiotics against multidrug-resistant E.coli with inhibition zone (13–14 mm). In another study, Kumar et al. [26] reported the antimicrobial activity of Lact. plantarum and Lact. acidophilus against multidrug-resistant enteroaggregative E.coli. Similarly, the inhibitory ability of milk fermented with Lact. casei strain shirota against common multidrug-resistant bacteria, including Ps. aeruginosa, Ac. baumannii, Methicillin-resistant Staph. aureus, ESBL-producing E.coli and Kl. pneumonia, had been studied [26].
Lysozyme Tolerance
In order to retain viability in the oral cavity, LAB must resist the antibacterial activity of lysozyme present in oral secretion. All nine isolates were tested in the presence of 0.1% (w/v) lysozyme for 90 min and the results as presented in Table 2 have revealed high lysozyme resistance, ranging from a minimum mean survival value of 78.94 ± 1.89% to a maximum value of 95.149 ± 1.339%. Five out of nine selected strains showed > 90% mean survival percentage, with strain LBM220 giving a value of 91.5%. Our results demonstrating high tolerance among isolates can be correlated with earlier reports [27]. Resistance of Gram-positive bacteria to lysozyme may be due to variation in peptidoglycan structure in the cell wall and the physiological state of the cell. Sirichokchatawan et al. [17] also reported > 80% survival of five LAB (isolated from pig sample) in presence of 0.1% lysozyme for 30 mins. Similarly, two isolates Lact. fermentum KJ03 and Lact. plantarum KJ03 have shown more than 90% mean survival in the presence of (100 mg/L) lysozyme after 20 min incubation [28].
Simulated Gastrointestinal Tolerance
Bacteria once ingested reach the human gastrointestinal tract and encounters the hostile environment of the stomach and duodenum. Only those microorganisms, surviving this exposure, will be able to subsist and colonize the gut, thus making it an important attribute required in a potential probiotic LAB. Considering this, all nine LAB isolates from infant faeces were exposed to the combined effect of gastric and intestinal fluids in simulated GIT transit tolerance assay and the results of their percent mean survivability are presented in Table 2. It was observed that after 120 min exposure to simulated gastric fluid at pH 2.0 containing 2.0 g/L pepsin, isolates LBQ12 and LBM220 exhibited the highest retention with cell viability of 93.61 ± 0.93% and 89.19 ± 0.76% respectively. However, the isolates LBX18 and LBP218 showed a drastic reduction in cell counts with the mean value of 3.35 ± 0.76 and 3.78 ± 0.18 log cfu mL−1 respectively. Rest all other isolates exhibited moderate survival in range of 84.07 ± 0.55 to 62.46 ± 1.58%. Studies have shown that LAB employs various mechanisms to overcome the damage caused due to acidic stress in the gut, which includes maintaining the intracellular pH either by proton-translocating ATPase mediated proton removal from cell or by producing negatively charged molecules [29]. As soon as bacteria passed through the stomach, they enter the small intestine where they mix with bile. Since bile tends to disorganize the membrane structure of living cells [30], it imparts another stress to combat for probiotics. To assess this stress, the nine LAB isolates in our study were subjected to simulated intestinal fluid containing 250 mg/L pancreatin and 0.3% w/v oxgall at pH 8.0. The results showed high variation in mean viable population after 3 h in which isolates LBM220 and LBM108 displayed the highest mean survivability of 95.30 ± 0.37 and 91.06 ± 0.86% respectively, whereas isolates LBP218, LBV12 and LBX18 did not show significant viability (less than 45%). Other isolates LBN16, LBS310, LBL19 and LBQ12 exhibited mean survival rates in range of 86.20 ± 0.81% to 63.11 ± 0.98%. Similarly, Rodrigues da Cunha et al. [31] also reported tolerance of 30 Lact. gasseri isolates from infant faeces to 0.25% oxgall to varying extent. High variation in viability among isolates can be correlated with earlier studies [32, 33].
Cellular Adhesion Ability
For effective colonization of probiotics in the human gut, they must adhere firmly to intestinal lining restraining their removal with the intestinal flow. Bacterial cell surface characteristics determine their adhesion ability, such as hydrophobicity. The higher the hydrophobicity, the higher will be their adhesion capacity. Cell-surface hydrophobicity (CSH) of all nine LAB isolates was determined using cellular partition to apolar solvents—xylene and n-hexadecane. Results demonstrated wide variations among different isolates with percent cell surface hydrophobicity, ranging from 9.53 to 73.79% (Fig. 1). The highest hydrophobicity of 73.79 ± 1.33 and 69.23 ± 1.40% was recorded against xylene and n-hexadecane respectively with LBM220, followed by LBM108 showing 63.47 ± 1.74 and 54.36 ± 1.45% adhesion. As presented in Fig. 1, other LAB isolates have moderate to low affinity with both hydrocarbons. The variations observed in hydrophobicity among isolates can be correlated with earlier reports [34, 35]. This variable affinity may be attributed due to specific functional groups and surface charges present at the cellular membrane. All test isolates were also examined for their auto-aggregation capacity, which is another prerequisite factor needed for colonization of gastrointestinal tract. The aggregation of isolates was observed at two different time intervals, viz. 4 h and 24 h and as represented in Fig. 2, the mean values ranged between 7.59 and 75.45%. After a period of 24 h, culture LBQ12 had highest auto-aggregation activity (75.45 ± 0.60%), followed by LBM220 (67.01 ± 0.68%) and LBP218 (59.44 ± 0.97%), while cultures LBL19 and LBV12 exhibited the lowest auto-aggregation of 19.91 ± 1.71% and 18.67 ± 1.38% respectively. The values increased at longer incubation time (up to 24 h). The auto-aggregation capacity of cells also plays an important role in the alleviation of pathogen colonization [36]. Our results are in line with that of Kassaa et al. [37] and Puniya et al. [35]. Among all isolates, LBM220 displayed good hydrophobicity and auto-aggregation, thereby confirming its ability to adhere, persist and propagate in GIT, qualifying it as potential probiotic.
Bile Salt Deconjugation and Cholesterol-Lowering Property
Bile salt deconjugation ability in bacteria is due to the production of bile salt hydrolase (BSH) enzyme. This is considered a desirable attribute of candidate probiotic microorganisms as it provides an auxiliary health benefit to the host in reducing serum cholesterol and providing a shield from bile toxicity through bile salt detoxification. This in turn augments their survival and persistence in the duodenum and ileal tract [38]. The current study on selected LAB demonstrated variability in deconjugation of bile salt in vitro (Table 3). Isolates LBN16, LBQ12 and LBM108 formed visibly opaque white colonies with precipitation halo on sodium taurodeoxycholate supplemented MRS plates, while LBM220, LBS310 and LBX18 formed translucent colonies. The isolate LBL19 exhibited growth but no precipitation, whereas LBV12 and LBP218 did not grow at all. Precipitation is primarily caused due to BSH-mediated formation of free bile acids that are insoluble in water in low pH and LAB produce both BSH and acids that lower the culture pH [30]. Several authors reported the role of BSH activity in lowering serum cholesterol in vivo as bile salts produced by hepatocytes are released into duodenum, where BSH enzyme deconjugates the steroidal moiety from these bile salts making them poorly soluble in water, thereby reducing reabsorption in the intestinal tract and increasing its excretion in faeces [39, 40]. With this understanding, all nine LAB cultures were tested for cholesterol-lowering ability in spent broth in the presence of 0.8% oxgall. Strain LBM220 exhibited the highest cholesterol reduction (75.20 ± 0.35%), followed by LBS310 (68.10 ± 1.21%) and LBN16 (66.93 ± 1.56%). The lowest value was observed for LBV12, LBP218, LBL19 and LBM108 (< 20%). Results significantly demonstrate that the highest cholesterol reducing ability was possessed by only BSH-positive isolates.
Identification of Most Potential LAB Isolate
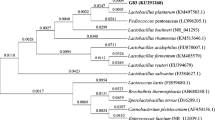
After probiotic evaluation of all nine LAB isolates which were chosen for their significant inhibition against multidrug-resistant ESKAPE pathogens, only isolate LBM220 showed the best oro-gastrointestinal stress tolerance, cellular-adhesion, BSH and cholesterol reducing ability. Thus, morphological, physiological and biochemical tests were done to identify isolate LBM220, while confirmatory identification was done by both, 16S rRNA sequence analysis and mass spectrometric analysis using MALDI-TOF. Strain LBM220 was found to be Gram-positive, catalase-negative non-spore forming bacilli that formed off-white pinpoint colonies, while its 16S rRNA gene sequence was identified as Lactobacillus gasseri (99% similarity), when compared with other bacterial strains in the GenBank database. The nucleotide sequence of Lactobacillus gasseri LBM220 was deposited in the GenBank database under accession number MN097539. Also, identification by mass spectrometry MALDI-TOF reveals genus and species of isolate LBM220 as Lactobacillus gasseri, with a high percentage of confidence (99%). The phylogenetic tree was based on 16S rRNA gene sequence analysis, depicting the phylogenetic relationships among Lactobacillus gasseri LBM220 strain and 16 Lactobacillus type strains obtained from the GenBank (given as supplementary file). Strain LBM220 formed a monophyletic clade with Lact. gasseri ATCC 33323. Figure 3 shows the 16S rRNA gene amplification product of LMB220 along with positive and negative controls.
Time-Kill Assay with CFCS of Lact. gasseri LBM220 on Multidrug-Resistant ESKAPE Pathogens
To further confirm the results obtained by agar-well diffusion assay for Lact. gasseri LBM220, the time-kill assay was also performed. The viability of all ESKAPE pathogens was assessed by incubating them up to 8 h with either CFCS or CFCS maintained at pH 6.5. The pathogens inoculated in MRS broth (at pH 6.5) were used as control. The aliquots were taken at four consecutive time intervals (t = 0, 2, 4 and 8 h) and assessed as described in methodology. The results, as displayed in Fig. 4, demonstrate that the survivability of each indicator pathogen decreased significantly by approx. 5–6 log cfu ml−1 after 2 h in the presence of Lact. gasseri CFCS, indicating its potential kill effect. The viability of four out of six test pathogens disappeared completely after contact for 4 h, except for Kl. pneumonia (MDR) and Ps. aeruginosa (MDR) that displayed minimal survival of 1 log cfu ml−1. After 8 h of incubation, all the six pathogens were non-viable in presence of CFCS. Whereas when the CFCS was neutralized to pH 6.5, no significant killing effect was observed in all test pathogens, further confirming the antimicrobial activity of isolate to be due to organic acids. Similarly, there was no loss in viability of test organisms grown in MRS broth maintained at pH 6.5.
Effect of cell-free culture supernatant (CFCS) of Lactobacillus gasseri LBM220 on cell viability of multidrug resistant (MDR) ESKAPE pathogens. Each MDR ESKAPE pathogen was incubated with either CFCS or CFCS maintained at pH 6.5. Aliquots were taken every 2 h (0–8 h), serially diluted and plated on brain heart infusion agar to determine colony counts. Pathogens incubated in MRS broth (at pH 6.5) served as controls. Results are expressed in log cfu ml−1 for each ESKAPE pathogen. *Each value is expressed as mean ± SD (n = 3)
Safety Evaluation of Lacto. gasseri LBM220
Safety evaluation is one of the most important parameters to be assessed before considering strain to be a probiotic [6].
Antibiotic Susceptibility
Resistance to different classes of antibiotics is a pertinent feature to investigate, as it being an acquired or inherent trait, varies within the Lactobacillus genus. When the resistance is acquired from exogenous DNA, the risk for horizontal transfer of resistance genes increases substantially. However, in most cases, antibiotic resistance of lactobacilli is natural and not of a transmissible type [41]. In the present study also, Lact. gasseri LBM220 was evaluated against fourteen antibiotics as per 2018 CLSI guidelines and was found to be sensitive to eleven of them, namely ampicillin, chloramphenicol, tetracycline, cefoxitin, penicillin, erythromycin, cefotaxime, doxycycline, fosfomycin, linezolid and vancomycin, while showing resistance towards gentamycin, tobramycin and ciprofloxacin. The findings of our study i.e. resistance to aminoglycoside (gentamycin and tobramycin) were similar to the findings of Rodrigues da Cunha et al. [31], who have also reported resistance of 30 strains of Lact. gasseri from infant faeces to aminoglycosides (gentamycin and amikacin). Several authors [42, 43] consider resistance to aminoglycoside as an intrinsic property of heterofermentative lactobacilli. This is probably due to the lack of cytochrome-mediated electron-transport, which mediates drug uptake in bacteria. Also, Jiang et al. [44] reported the presence of aminoglycoside resistance genes in lactobacilli. Thus, it can be stated that there is no risk of the spread of this resistance gene to pathogenic/opportunistic bacteria in GIT. Lact. gasseri LBM220 was also resistant to ciprofloxacin which belongs to class quinolone that acts by disrupting DNA replication owing to inhibition of enzymes DNA gyrase and topoisomerase IV. Our results are in concurrence with findings of Kõll et al. [45] who also reported ciprofloxacin resistance in four Lact. gasseri isolates from infants faeces. Reports from various other authors corroborate our results regarding high ciprofloxacin and aminoglycoside resistance among lactobacilli from human origin [31, 46].
Hemolysis, DNase and Gelatinase Production
Hemolysis is another factor that contributes to virulence among pathogenic strains, assisting in their iron availability and thereby causing anaemia and oedema to the host. Iron is a micronutrient that promotes growth of bacteria because it acts as a cofactor for numerous enzymes. According to Gaucher et al. [47], iron requirement of genus Lactobacillus is minimal and this provides additional ecological benefit to them to sustain in their natural environment, where they may be competing with pathogens. The present study reports the non-hemolytic activity of Lact. gasseri LBM220 in blood agar culture. Our study is in accordance with previous findings by Kõll et al. [45], who observed no hemolysis in 93 lactobacilli isolates from infant stool. Rodrigues da Cunha et al. [31] also reported no erythrocyte lysis among 30 Lact. gasseri strains from infant faeces. Similarly, in this study, Lact. gasseri LBM220 when tested for other virulence factors such as DNase and Gelatinase producing ability was found to be negative compared to control organism, Staph. aureus ATCC 25923, which showed β-hemolysis on blood agar and opaque halo around colonies for the other two evaluation tests.
Mucin Degradation Assay
Another important safety parameter to investigate is the ability of probiotic microorganisms to degrade mucin. A highly glycosylated protein on the surface of the intestinal wall, mucin provides the first line of defence and prevents the translocation of bacteria via their invasion through the intestinal lining. Bacterial translocation induces several serious infections such as bacteraemia, endocarditis or sepsis [48]. In view of this, Lact. gasseri LBM220 was examined for in vitro mucin degradation ability and results had displayed no zone of mucin degradation around the colonies. Positive controls used in our study, Sal. typhimurium and Sh. flexneri, had displayed significant mucin lysis zone around colonies. This confirms negligible activity of identified LAB to decompose the mucus layer, thus stating its safe use as probiotics. Our results are in concurrence with reports from other authors [19, 46], who also indicated the non-mucin degradation ability of several lactobacilli species.
Conclusion
The antimicrobial potential of probiotic strains against multidrug-resistant ESKAPE pathogens is an important area of study, and this work was an attempt to explore the antimicrobial potential of probiotic strains against drug resistant organisms. In the present study, nine isolates from the pool of 102 anaerobic LAB isolated from breast-fed infant faeces were found to demonstrate significant inhibition against all pathogens. When these isolates were evaluated for probiotic potential, only LBM220 strain possessed excellent oro-gastrointestinal tolerance, cell-adhesion ability, BSH and cholesterol-lowering ability. The strain identified as Lact. gasseri was also found to be safe for human consumption. Thus, our study stands significant in reporting successfully the potent role of autochthonous probiotic bacteria isolated from infant faeces in antagonizing all six multidrug-resistant ESKAPE pathogens, but for possible health claims in humans, in vivo animal model experiments must be done in the future.
References
Waclaw B (2016) Evolution of drug resistance in bacteria. Adv Exp Med Biol 915:49–67. https://doi.org/10.1007/978-3-319-32189-9_5
Stewardson AJ, Allignol A, Beyersmann J, Graves N, Schumacher M, Meyer R, Tacconelli E, de Angelis G, Farina C, Pezzoli F, Bertrand X, Gbaguidi-Haore H, Edgeworth J, Tosas O, Martinez JA, Ayala-Blanco MP, Pan A, Zoncada A, Marwick CA, Nathwani D, Seifert H, Hos N, Hagel S, Pletz M, Harbarth S, the TIMBER Study Group (2016) The health and economic burden of bloodstream infections caused by antimicrobial-susceptible and non-susceptible Enterobacteriaceae and Staphylococcus aureus in European hospitals, 2010 and 2011: a multicentre retrospective cohort study. Euro Surveill https://doi.org/10.2807/1560-7917.ES.2016.21.33.30319
Zhen X, Lundborg CS, Sun X, Hu X, Dong H (2019) Economic burden of antibiotic resistance in ESKAPE organisms: a systematic review. Antimicrob Resist Infect Control 8:137. https://doi.org/10.1186/s13756-019-0590-7
Pendleton JN, Gorman SP, Gilmore BF (2013) Clinical relevance of the ESKAPE pathogens. Expert Rev Anti-Infect Ther 11(3):297–308. https://doi.org/10.1586/eri.13.12
Santajit S, Indrawattana N (2016) Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int 2016:2475067–2475068. https://doi.org/10.1155/2016/2475067
FAO/WHO (2006) Probiotics in food, health and nutritional properties and guidelines for evaluation. Rome: FAO Food and Nutrition Paper. http://www.fao.org/3/a-a0512e.pdf
Wang L, Guo MJ, Gao Q, Yang JF, Yang L, Pang XL, Jiang XJ (2018) The effects of probiotics on total cholesterol: a meta-analysis of randomized controlled trials. Medicine. 10. 1097/MD.0000000000009679
Wang ZB, Xin SS, Ding LN, Ding WY, Hou YL, Liu CQ, Zhang XD (2019) The potential role of probiotics in controlling overweight/obesity and associated metabolic parameters in adults: a systematic review and meta-analysis. Evid Based Complement Alternat Med 2019:1–14. https://doi.org/10.1155/2019/3862971
Nazir Y, Hussain SA, Abdul HA, Song Y (2018) Probiotics and their potential preventive and therapeutic role for cancer, high serum cholesterol, and allergic and HIV diseases. Biomed Res Int. 10. 1155/2018/3428437
Rittiphairoj T, Pongpirul K, Mueller NT, Li T (2019) Probiotics for glycemic control in patients with type 2 diabetes mellitus: protocol for a systematic review. Syst Rev 8:227. https://doi.org/10.1186/s13643-019-1145-y
Domingo SJJ (2017) Review of the role of probiotics in gastrointestinal diseases in adults. Gastroenterol Hepato 40(6):417–429. https://doi.org/10.1016/j.gastre.2016.12.001
Kobatake E, Nakagawa H, Seki T, Miyazaki T (2017) Protective effects and functional mechanisms of Lactobacillus gasseri SBT2055 against oxidative stress. PLoS One. https://doi.org/10.1371/journal.,pone.0177106
Staudacher H (2015) Probiotics for lactose intolerance and irritable bowel syndrome. Br J Community Nurs Suppl Nutrition: S12–S14. https://doi.org/10.12968/bjcn.2015.20.Sup6a.S12
Thakur N, Rokana N, Panwar H (2016) Probiotics: selection criteria, safety and role in health and diseases. J Innov Biol 3:259–270 https://www.semanticscholar.org/paper/Probiotics-%3A-Selection-criteria-%2C-safety-and-role-Thakur-Rokana/10c2b5d9f103e4f87bfe01146916c053ec9e2835
Bejar W, Farhat-Khemakhen A, Smaoui S, Mohamed M, Ben M, Badis F, Lofti A, Emmanuelle M, Samir M, Hichem B, Hichem C (2011) Selection of Lactobacillus plantarum TN627 as a new probiotic candidate based on in vitro functional properties. Biotechnol Bioproc E 16:1115–1123. https://doi.org/10.1007/s12257-011-0198-0
Yu HJ, Chen YF, Yang HJ, Yang J, Li CK, Kwok LY, Zhang HP, Sun TG (2014) Screening for Lactobacillus plantarum with potential inhibitory activity against enteric pathogens. Ann Micro 65(3):1257–1265. https://doi.org/10.1007/s13213-014-0963-3
Sirichokchatchawan W, Pupa P, Praechansri P, Am-In N, Tanasupawat S, Sonthayanon P, Prapasarakul N (2018) Autochthonous lactic acid bacteria isolated from pig faeces in Thailand show probiotic properties and antibacterial activity against enteric pathogenic bacteria. Microb Pathog 119:208–215. https://doi.org/10.1016/j.micpath.2018.04.031
Turchi B, Mancini S, Fratini F, Pedonese F, Nuvoloni R, Bertelloni F, Ebani VV, Cerri D (2013) Preliminary evaluation of probiotic potential of Lactobacillus plantarum strains isolated from Italian food products. World J Microbiol Biotechnol 29(10):1912–1922. https://doi.org/10.1007/s11274-013-1356-7
de Moraes GMD, de Abreu LR, do Egito AS, Salles HO, da Silva LMF, Nero LA, Todorov SD, dos Santos KMO (2017) Functional properties of Lactobacillus mucosae strains isolated from brazilian goat milk. Probiotics & Antimicro Prot 9(3):235–245. https://doi.org/10.1007/s12602-016-9244-8
Shehata MG, Sohaimy SA, El-Malak A, El-Sahn YMM (2016) Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Ann Agric Sci 61(1):65–75. https://doi.org/10.1016/j.aoas.2016.03.001
Rastogi S, Mittal V, Singh A (2019) In vitro evaluation of probiotic potential and safety assessment of Lactobacillus mucosae strains isolated from Donkey’s lactation. Probiotics and Antimicrob Proteins 12:1045–1056. https://doi.org/10.1007/s12602-019-09610-0
Zhang Y, Zhang L, Du M, Yi H, Guo C, Tuo Y, Han X, Li J, Zhang L, Yang L (2011) Antimicrobial activity against Shigella sonnei and probiotic properties of wild lactobacilli from fermented food. Microbiol Res 167(1):27–31. https://doi.org/10.1016/j.micres.2011.02.006
Clinical and Laboratory Standards Institute (2018) Performance standards for antimicrobial disk susceptibility tests; approved standard - 12th ed. M02-A13. Clinical and Laboratory Standards Institute, Wayne, PA https://clsi.org/standards/products/microbiology/documents/m02/
Wang C, Chang T, Yang H, Cui M (2015) Antibacterial mechanism of lactic acid on physiological and morphological properties of Salmonella enteritidis, Escherichia coli and Listeria monocytogenes. Food Control 47:231–236. https://doi.org/10.1016/j.foodcont.2014.06.034
Abdelhamid AG, Esaam A, Hazaa MM (2018) Cell free preparations of probiotics exerted antibacterial and antibiofilm activities against multidrug resistant E. coli. Saudi Pharm J 26(5):603–607. https://doi.org/10.1016/j.jsps.2018.03.004
Tiengrim S, Thamlikitkul V (2012) Inhibitory activity of fermented milk with Lactobacillus casei strain Shirota against common multidrug-resistant bacteria causing hospital-acquired infections. J Med Assoc Thail 95(Suppl 2):S1–S5
Kumar M, Dhaka P, Vijay D, Vergis J, Mohan V, Kumar A, Kurkure NV, Barbuddhe SB, Malik SV, Rawool DB (2016) Antimicrobial effects of Lactobacillus plantarum and Lactobacillus acidophilus against multidrug-resistant enteroaggregative escherichia coli. Int J of Antimicrob Agents 48(3):265–270. https://doi.org/10.1016/j.ijantimicag.2016.05.014
Jampaphaeng K, Cocolin L, Maneerat S (2017) Selection and evaluation of functional characteristics of autochthonous lactic acid bacteria isolated from traditional fermented stinky bean (Sataw-Dong). Ann Microbiol 67(1):25–36. https://doi.org/10.1007/s13213-016-1233-3
Kudo H, Sasaki Y (2019) Intracellular pH determination for the study of acid tolerance of lactic acid bacteria: methods and protocols. Methods Mol Biol. https://doi.org/10.1007/978-1-4939-8907-2_4
Begley M, Hill C, Gahan CG (2006) Bile salt hydrolase activity in probiotics. Appl Environ Microbiol 72(3):1729–1738. https://doi.org/10.1128/AEM.72.3.1729-1738.2006
Rodrigues da Cunha L, Fortes Ferreira CL, Durmaz E, Goh YJ, Sanozky-Dawes R, Klaenhammer T (2012) Characterization of Lactobacillus gasseri isolates from a breast-fed infant. Gut Microbes 3:15–24. https://doi.org/10.4161/gmic.19489
Shokryazdan P, Sieo CC, Kalavathy R, Liang JB, Alitheen NB, Jahromi MF, Ho YW (2014) Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. Biomed Res Int 2014:1–16. https://doi.org/10.1155/2014/927268
Kumari A, Angmo K, Monika BTC (2016) Probiotic attributes of indigenous Lactobacillus spp. isolated from traditional fermented foods and beverages of north-western Himalayas using in vitro screening and principal component analysis. J Food Sci Technol 53(5):2463–2475. https://doi.org/10.1007/s13197-016-2231-y
Collado MC, Meriluoto J, Salminen S (2008) Adhesion and aggregation properties of probiotic and pathogen strains. Eur Food Res Technol 226:1065–1073. https://doi.org/10.1007/s00217-007-0632
Puniya M, Kumar RM, Panwar H, Kumar N, Ramneek P, AK (2016) Screening of lactic acid bacteria of different origin for their probiotic potential. J Food Process Technol https://doi.org/10.4172/2157-7110.1000545, 07
Boris S, Suárez JE, Vázquez F, Barbés C (1998) Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect Immun 66:1985–1989. https://doi.org/10.1128/IAI.66.5.1985-1989.1998
Kassaa IA, Hamze M, Hober D, Chihib NE, Drider D (2014) Identification of vaginal lactobacilli with potential probiotic properties isolated from women in north Lebanon. Microb Ecol 67:722–734
Šrka H, Milada P, Kateřina D (2017) Importance of microbial defence systems to bile salts and mechanisms of serum cholesterol reduction. Biotechnol Adv 36(3):682–690. https://doi.org/10.1016/j.biotechadv.2017.12.005
Ishimwe N, Daliri EB, Lee BH, Fang F, Du G (2015) The perspective on cholesterol-lowering mechanisms of probiotics. Mol Nutr Food Res 59(1):94–105. https://doi.org/10.1002/mnfr.201400548
Costabile A, Buttarazzi I, Kolida S, Quercia S, Baldini J, Swann JR, Brigidi P, Gibson GR (2017) An in vivo assessment of the cholesterol-lowering efficacy of Lactobacillus plantarum ECGC 13110402 in normal to mildly hypercholesterolaemic adults. Plos One. https://doi.org/10.1371/journal.,pone.0187964
Sharma C, Gulati S, Thakur N, Singh BP, Gupta S, Kaur S, Mishra SK, Puniya AK, Gill JPS, Panwar H (2017) Antibiotic sensitivity pattern of indigenous lactobacilli isolated from curd and human milk samples. 3 Biotech. https://doi.org/10.1007/s13205-017-0682-0
Nawaz M, Wang J, Zhou A, Ma C, Wu X, Moore JE, Millar BC, Xu J (2011) Characterization and transfer of antibiotic resistance in lactic acid bacteria from fermented food products. Curr Microbiol 62(3):1081–1089. https://doi.org/10.1007/s00284-010-9856-2
Dec M, Urban-Chmiel R, Stępień-Pyśniak D, Wernicki A (2017) Assessment of antibiotic susceptibility in Lactobacillus isolates from chickens. Gut Pathog 9:54. https://doi.org/10.1186/s13099-017-0203-z
Jiang M, Zhang F, Wan C, Xiong Y, Shah NP, Wei H, Tao X (2016) Evaluation of probiotic properties of Lactobacillus plantarum WLPL04 isolated from human breast milk. J Dairy Sci 99(3):1736–1746. https://doi.org/10.3168/jds.2015-10434
Kõll P, Mändar R, Smidt I, Hütt P, Truusalu K, Mikelsaar RH, Shchepetova J, Krogh-Andersen K, Marcotte H, Hammarström L, Mikelsaar L (2010) Screening and evaluation of human intestinal lactobacilli for the development of novel gastrointestinal probiotics. Curr Microbiol 61(6):560–566. https://doi.org/10.1007/s00284-010-9653-y
Martín R, Jiménez E, Olivares M, Marín ML, Fernández L, Xaus J, Rodríguez JM (2006) Lactobacillus salivarius CECT 5713, a potential probiotic strain isolated from infant feces and breast milk of a mother-child pair. Int J Food Microbiol 112(1):35–43. https://doi.org/10.1016/j.ijfoodmicro.2006.06.011
Gaucher F, Bonnassie S, Rabah H, Marchand P, Blanc P, Jeantet B, Jan G (2019) Review: adaptation of beneficial propionibacteria, lactobacilli, and bifidobacteria improves tolerance toward technological and digestive stresses. Front Microbiol https://doi.org/10.3389/fmicb.2019.00841, 10
Abe F, Muto M, Yaeshima T, Iwatsuki K, Aihara H, Ohashi Y, Fujisawa T (2010) Safety evaluation of probiotic bifidobacteria by analysis of mucin degradation activity and translocation ability. Anaerobe 16(2):131–136. https://doi.org/10.1016/j.anaerobe.2009.07.006
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(PDF 240 kb)
Rights and permissions
About this article
Cite this article
Rastogi, S., Mittal, V. & Singh, A. Selection of Potential Probiotic Bacteria from Exclusively Breastfed Infant Faeces with Antagonistic Activity Against Multidrug-Resistant ESKAPE Pathogens. Probiotics & Antimicro. Prot. 13, 739–750 (2021). https://doi.org/10.1007/s12602-020-09724-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-020-09724-w