Abstract
Genome sequencing of Enterococcus faecium M3K31, a strain isolated from griffon vultures, previously revealed the presence of three sequences encoding for bacteriocins, namely enterocin P, enterocin HF, and a SRCAM 602-like bacteriocin. In this work, we describe the SRCAM 602-like bacteriocin that we named faerocin MK. Mature faerocin MK consists of 43 residues and contains an YGNGV-motif and two cysteine residues at positions 10 and 15, consistent with other class IIa bacteriocins. Faerocin MK and SRCAM 602 were chemically synthesized and their scope of activity was tested. Faerocin MK is active against a wide range of Gram-positive organisms while SRCAM 602 was not active against any tested organism. Although both peptides are more structured in trifluoroethanol, faerocin MK has an α-helical character nearly twice that of SRCAM 602. Nucleotide sequence analysis revealed that the faerocin MK precursor is produced with a 28-amino acid signal peptide and that an immunity gene follows the structural faerocin MK gene. Heterologous expression of the faerocin MK operon showed that faerocin MK and its immunity protein were successfully expressed in other organisms. This indicates that the bacteriocin is secreted through the sec pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Enterococcus species are widespread among plants, insects, animal products, and also within the human gastrointestinal tract. Their abundancy is an advantage that can be exploited, particularly as they possess beneficial attributes, such as the potential to produce a great variety of bacteriocins [1]. The diverse nature of these antimicrobial peptides allows enterococci to be active against a broad scope of organisms; of particular interest is the activity against food-borne pathogens and food spoilage organisms [1, 2]. In addition to their use in food products, enterococci have also been investigated as potential probiotics. However, the presence of virulence factors in many strains of enterococci raises the issue of safety when used in food products or as probiotics [2, 3]. Often cited as an opportunistic pathogen, enterococci are among the most prevalent nosocomial pathogens and are responsible for endocarditis, bacteremia, and urinary tract infections, among others [2, 4]. With the advent of whole genome sequencing, bacteriocinogenic enterococci can be discovered that lack virulence factors and might have potential as probiotics [5]. Analysis of the recently sequenced genome of Enterococcus faecium M3K31, isolated from griffon vultures, Gyps fulvus, indicates that this particular strain does not contain virulence factor genes [6]. Further genome mining revealed the presence of genes encoding enterocin P, enterocin HF, and a SRCAM 602-like bacteriocin. Enterocin P is a sec-dependent class IIa bacteriocin that contains an YGNGV-motif [7]. Enterocin HF also belongs to class IIa bacteriocins and is produced as a precursor with an N-terminal double-glycine leader peptide. The 3D structure of enterocin HF was recently elucidated [8]. The SRCAM 602-like peptide resembles multiple SRCAM peptides that were claimed to have a wide scope of antimicrobial activity against both Gram-positive and Gram-negative bacteria [9]. However, subsequent studies were unable to detect the production of several of these peptides and their genetic determinants were not found in the corresponding bacterial genomes [10, 11]. It was shown that the inhibitory activity towards Gram-negative bacteria by Paenibacillus polymyxa NRRL B-30509, that was claimed to produce SRCAM 602, could be attributed to the production of lipopeptides [12]. This prompted us to investigate the activity and structure of this SRCAM-like bacteriocin from E. faecium M3K31, which we named faerocin MK (FaeMK), and compare it to that of the reported SRCAM 602 peptide. In this study, we show that FaeMK is a 43-residue class IIa bacteriocin that is active against Gram-positive organisms and is likely to be structurally similar to enterocin HF. The FaeMK precursor contains a signal peptide that enables the bacteriocin to access the general secretion pathway. The FaeMK operon, containing the structural bacteriocin gene and the immunity gene, was cloned and successfully expressed in heterologous strains, which confirms the sec-dependent nature of this bacteriocin.
Materials and Methods
Bacterial Cultures
Gram-positive strains were grown using All Purpose Tween medium (Difco, Sparks, MD) at 25 °C, except for Staphylococcus aureus ATCC 25923 that was grown on tryptic soy broth medium (Difco). Gram-negative strains were grown using Luria Broth Base, Miller medium (Difco) at 37 °C. If required, chloramphenicol was added to the media at 5 μg/mL for Carnobacterium maltaromaticum UAL26 and 10 μg/mL for Enterococcus faecium BFE 900.
Bacteriocin Assays
FaeMK and SRCAM 602 were chemically synthesized and acquired in 98% purity from GenScript USA Inc. (Piscataway, NJ). Prior to use, the sequence and purity of the peptides were verified with mass spectrometry in-house. The minimum inhibitory concentrations (MICs) were tested using a spot-on-lawn assay. The peptides were tested against the indicator strains listed in Table 1. Using the growth conditions discussed above, the strains were grown overnight and 100 μL of each indicator strain was used to inoculate soft agar (0.75% w/v) and overlaid on solid agar plates using the appropriate media. A 2-fold series dilution of the peptides was prepared from a 64 μM stock solution and 10 μL of each concentration was spotted onto the agar plate. After an overnight incubation, the plates were examined for zones of inhibition and MICs were determined.
Circular Dichroism Spectroscopy
An OLIS DSM 17 CD spectrophotometer with a thermally controlled quartz cell with a 0.2-mm path length was used. Data were collected during five scans from 185 to 260 nm in 1 nm increments at 20 °C. FaeMK and SRCAM 602 solutions were prepared in 0, 25, and 50% 2,2,2-trifluoroethanol (TFE) in water at concentrations of 0.83 and 1.52 mg/mL, respectively. Results are reported in molar ellipticity units (deg × cm2 / dmol) and the percent α-helicity of the two peptides was calculated using: \( \frac{3000-{\theta}_{222}}{39000}\times 100\% \) [17].
Sequence Alignment and Homology Modeling
Clustal Omega was used to align the sequences of FaeMK with SRCAM 602, durancin GL, and hiracin JM79 [18]. A 3D model was created using SWISS-MODEL and enterocin HF as the model template [19].
Construction of Plasmids
A 552-bp DNA fragment containing the FaeMK operon was obtained from BioBasic Inc. (Markham, ON, Canada). The DNA fragment was designed with two different restriction sites, SacI and XbaI, to enable the cloning of this DNA fragment into the multiple cloning site of the broad host range shuttle vector pMG36c [20]. The ligation mixture was electroporated into C. maltaromaticum UAL26 according to the method described previously [21]. The resulting plasmid, pMG36c-faeMKI, was isolated from UAL26 transformants using the GeneJET Plasmid Miniprep Kit (Fermentas Canada Inc., Burlington, ON, Canada). To facilitate the isolation of the DNA, UAL26 cells were first treated with 5 mg/mL lysozyme for 30 min at 37 °C before cell lysis. The isolated plasmid DNA was sequenced to confirm the accurate sequence of pMG36c-faeMKI. Plasmid pMG36c-faeMKI was then electroporated into E. faecium BFE 900 as described above.
Results and Discussion
DNA Sequence of Faerocin MK Operon
A gene encoding a bacteriocin that is highly similar (85% identity) to that of the putative peptide SRCAM 602 was detected on the genome of E. faecium M3K31 [6]. Analysis of the nucleotide sequence reveals that this bacteriocin, which we termed faerocin MK (FaeMK), consists of 43 amino acids and is produced as a precursor with a 28-amino acid leader peptide (Fig. 1). FaeMK contains the YGNGV-motif that is characteristic of class IIa bacteriocins [23]. The leader peptide of FaeMK precursor resembles a sec-dependent signal peptide with a positively charged N-terminus, a stretch of hydrophobic amino acids, and cleavage site following an alanine residue. Bacteriocins produced with this type of leader peptide can access the general secretion pathway of the cell and do not require a dedicated transport machinery for secretion [14]. Antimicrobial peptides that belong to this class kill sensitive cells by membrane permeabilization [24]. Immediately downstream of the structural gene for FaeMK, faeMK, a putative immunity gene, faeI, is located that could encode a peptide of 94 amino acids. Both faeMK and faeI are preceded by a putative ribosomal binding site (RBS). A putative promoter, with TTGTCA as the -35 region and TTTTAT as the -10 region, was identified upstream of the FaeMK structural gene. In addition, a 13-bp inverted repeat is located between the -10 region and the RBS of faeMK. The function of this repeat is unclear but it might suggest that the expression of the FaeMK operon is regulated. A BLAST search for the FaeMK peptide showed high homology with durancin GL (98% identity), and hiracin JM79 (86% identity) (Fig. 2). Durancin GL and hiracin JM79 are sec-dependent bacteriocins produced by Enterococcus durans 41D and Enterococcus hirae DCH5, respectively [22, 25]. Comparison between the DNA sequence of the FaeMK operon and that of durancin GL showed 99% identity. The structural gene faeMK showed two mutations compared to the structural gene for durancin GL: one is a silent mutation and the other is a mutation that results in a substitution at position 14 of the FaeMK peptide (Figs. 1 and 2). Remarkably, the DNA sequence immediately upstream of the ATG start codon of faeMK is different and shows no sequence homology with the DNA sequence upstream of the structural gene for durancin GL (Fig. 1). In contrast to the DNA sequence upstream of faeMK, no inverted repeat was reported between the promoter and RBS of the durancin GL structural gene [22]. Another major difference is the fact that the durancin GL operon is situated on a plasmid whereas the FaeMK operon is located on the chromosome.
Nucleotide sequence of the faerocin MK operon. The deduced amino acid sequences for faeMK and faeI are shown below the nucleotide sequence. The putative -35 and -10 promoter regions, as well as putative RBSs, are underlined. An inverted repeat identified upstream of faeMK is double underlined. Nucleotides that are different from those reported for the durancin GL operon [22] are highlighted in gray. The vertical arrow indicates the cleavage site of the faerocin MK precursor
Characterization of Faerocin MK
The peptide sequence for FaeMK was compared to the reported SRCAM 602 sequence, hiracin MJ79, and durancin GL (Fig. 2). This homologous sequence alignment highlights the conserved YGNGV-motif, with a single conserved mutation in the SRCAM 602 and hiracin MJ79 peptides at position 8 yielding an YGNGL-motif instead. A consequential inconsistency in the SRCAM 602 sequence is the presence of a single cysteine residue at position 10. In contrast, the other three peptides contain two cysteine residues at positions 10 and 15. This allows for the formation of a disulfide bridge, a feature found on all other class IIa bacteriocins that is attributed to providing structural stability at the N-terminus [23].
The FaeMK and the reported SRCAM 602 peptides were chemically synthesized for this study. Activity assays showed that FaeMK inhibits many of the Gram-positive bacteria tested and is particularly potent against Listeria and Carnobacterium spp., showing a MIC for strains of these genera at the low nanomolar range (Table 1). FaeMK was not active against the Gram-negative strains tested. The inhibition of Gram-positive bacteria and lack of activity against Gram-negative bacteria are consistent with other class IIa bacteriocins [23]. However, the observed activity of SRCAM 602 was not consistent with that reported in literature [9]. SRCAM 602 showed no activity against Gram-positive or Gram-negative organisms (Table 1).
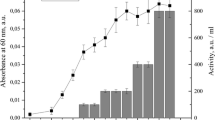
The relative structures of the two peptides were analyzed using circular dichroism spectroscopy (Fig. 3). The α-helicity of both peptides was determined as a percentage using a structure inducing solvent, trifluoroethanol. The averaged results indicate a 28 and 16% α-helical character for FaeMK and SRCAM 602, respectively, in a 1:1 solution of water:trifluoroethanol. This result prompted us to create a model of FaeMK based on the published 3D structure of enterocin HF as a template, which is the most closely matched class IIa bacteriocin with a known structure, having a 46% identity with FaeMK [8]. The resulting 3D model showcases the expected hairpin at the N-terminus, with the cysteine residues responsible for the disulfide bridge formation highlighted in gray, and a single α-helix structure observed in typical class IIa bacteriocins (Fig. 4) [23].
Heterologous Expression of Faerocin MK
To investigate whether we could achieve FaeMK production in heterologous hosts, a 552-bp fragment containing the faeMKI operon that includes a 16-bp nucleotide sequence immediately upstream of the ATG start codon for faeMK was cloned behind the constitutive P32 promoter of the shuttle vector pMG36c. The DNA ligation mixture was transformed into C. maltaromaticum UAL26 giving pMG36c-faeMKI in the resulting UAL26 transformant. UAL26 containing pMG36c-faeMKI was used in a deferred inhibition test with UAL26 as the indicator strain and UAL26 containing pMG36c without an insert was used as a negative control. A small, but clear inhibition zone produced by UAL26 containing pMG36c-faeMKI could be detected whereas the negative control showed no inhibition (Fig. 5a). To test heterologous FaeMK production in enterococci, plasmid pMG36c-faeMKI was isolated from the UAL26 transformant and introduced into E. faecium BFE 900. BFE 900 containing pMG36c without an insert was again used as a negative control. Deferred inhibition test using BFE 900 as the indicator strain showed that FaeMK production could also be achieved in E. faecium BFE 900 (Fig. 5b). To determine whether UAL26 and BFE 900 containing pMG36c-faeMKI showed a decrease in sensitivity to FaeMK, a spot-on-lawn test was performed using a 2-fold serial dilution of FaeMK. UAL26 and BFE 900 containing pMG36c-faeMKI were 8-fold and 32-fold less sensitive to FaeMK, respectively, compared to the control strains that contained pMG36c. These results indicate that the faeMKI operon is also responsible for immunity and that faeI is the immunity gene. This is in agreement with the identification of the immunity gene for durancin GL, which is identical to that for FaeMK [22]. The finding that heterologous production of FaeMK can be achieved without the additional cloning of a dedicated bacteriocin transport machinery confirms the proposal that FaeMK is a sec-dependent bacteriocin.
Concluding Remarks
We demonstrated that faerocin MK is a class IIa bacteriocin, matching both its activity and structure to previously reported bacteriocins in this class. Our heterologous expression studies indicate that this operon, containing only faeMK and faeI, is sufficient for the expression and secretion of FaeMK as well as providing immunity for the host organism. In addition, we conclude that the previously reported SRCAM 602 peptide sequence would yield an inactive and loosely structured product [9]. This confirms our previous conclusion that SRCAM 602 is not a bacteriocin produced by P. polymyxa NRRL B-30509 [12]. It is interesting to note that E. faecium M3K31 encodes three bacteriocins with an YGNGV-motif, two of which are sec-dependent bacteriocins. Future studies could investigate the methods of production of these bacteriocins in E. faecium M3K31, a strain that has great potential to be used as a probiotic.
References
Franz CMAP, van Belkum MJ, Holzapfel WH, Abriouel H, Gálvez A (2007) Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol Rev 31:293–310
Giraffa G, Carminati D, Neviani E (1997) Enterococci isolated from dairy products: a review of risks and potential technological use. J Food Prot 60(6):732–738. https://doi.org/10.4315/0362-028X-60.6.732
Franz CMAP, Huch M, Abriouel H, Holzapfel W, Gálvez A (2011) Enterococci as probiotics and their implications in food safety. Int J Food Microbiol 151(2):125–140. https://doi.org/10.1016/j.ijfoodmicro.2011.08.014
Aguirre M, Collins MD (1993) Lactic acid bacteria and human clinical infection. J Appl Bacteriol 75(2):95–107. https://doi.org/10.1111/j.1365-2672.1993.tb02753.x
Acedo JZ, Ibarra Romero C, Miyata ST, Blaine AH, McMullen LM, Vederas JC, van Belkum MJ (2017) Draft genome sequence of Enterococcus canintestini 49, a potential probiotic that produces multiple bacteriocins. Genome Announc 5(40):e01131–e01117
Arbulu S, Frantzen C, Lohans CT, Cintas LM, Herranz C, Holo H, Diep DB, Vederas JC, Hernández PE (2016) Draft genome sequence of the bacteriocin-producing strain Enterococcus faecium M3K31, isolated from griffon vultures (Gyps fulvus subsp. fulvus). Genome Announc 4(2):e00055–e00016
Cintas LM, Casaus P, Håvarstein LS, Hernández PE, Nes IF (1997) Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with board antimicrobial spectrum. Appl Environ Micobiol 63:4321–4330
Arbulu S, Lohans CT, van Belkum MJ, Cintas LM, Herranz C, Vederas JC, Hernandez PE (2015) Solution structure of enterocin HF, an antilisterial bacteriocin produced by Enterococcus faecium M3K31. J Agric Food Chem 63(49):10689–10695. https://doi.org/10.1021/acs.jafc.5b03882
Svetoch EA, Stern NJ, Eruslanov BV, Kovalev YN, Volodina LI, Perelygin VV, Mitsevich EN, Mitsevich IP, Pokhilenko VD, Borzenkov VN, Levchuk VP, Svetoch OE, Kudriavtseva TY (2005) Isolation of Bacillus circulans and Paenibacillus polymyxa strains inhibitory to Campylobacter jejuni and characterization of associate bacteriocins. J Food Prot 68(1):11–17. https://doi.org/10.4315/0362-028X-68.1.11
Lohans CT, van Belkum MJ, Li J, Vederas JC (2015) Characterization of bacterial antimicrobial peptides active against Campylobacter jejuni. Can J Chem 93(4):381–388. https://doi.org/10.1139/cjc-2014-0411
van Belkum MJ, Lohans CT, Vederas JC (2015) Draft genome sequences of Paenibacillus polymyxa NRRL B-30509 and Paenibacillus terrae NRRL B-30644, strains from a poultry environment that produce tridecaptin A and paenicidins. Genome Announc 3(2):e00372–e00315
Lohans CT, Huang Z, van Belkum MJ, Giroud M, Sit CS, Steels EM, Zheng J, Whittal RM, McMullen LM, Vederas JC (2012) Structural characterization of the highly cyclized lantibiotic paenicidin A via a partial desulfurization/reduction strategy. J Am Chem Soc 134(48):19540–19543. https://doi.org/10.1021/ja3089229
Gursky LJ, Martin NI, Derksen DJ, van Belkum MJ, Kaur K, Vederas JC, Stiles ME, McMullen LM (2006) Production of piscicolin 126 by Carnobacterium maltaromaticum UAL26 is controlled by temperature and induction peptide concentration. Arch Microbiol 186(4):317–325. https://doi.org/10.1007/s00203-006-0147-z
Worobo RW, van Belkum MJ, Sailer M, Roy KL, Vederas JC, Stiles ME (1995) A signal peptide secretion-dependent bacteriocin from Carnobacterium divergens. J Bacteriol 177(11):3143–3149. https://doi.org/10.1128/jb.177.11.3143-3149.1995
Schillinger U, Kaya M, Lücke FK (1991) Behavior of Listeria monocytogenes in meat and its control by a bacteriocin-producing strain of Lactobacillus sake. J Appl Bacteriol 70(6):473–478. https://doi.org/10.1111/j.1365-2672.1991.tb02743.x
Franz CMAP, Worobo RW, Quadri LE, Schillinger U, Holzapfel WH, Vederas JC, Stiles ME (1999) Atypical genetic locus associated with constitutive production of enterocin B by Enterococcus faecium BFE 900. Appl Environ Microbiol 65(5):2170–2178
Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, Weisgraber KH (2000) Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry 39(38):11657–11666. https://doi.org/10.1021/bi000099m
Sievers F, Higgins DG (2014) Clustal omega. Curr Protoc Bioinformatics 48:3.13.1–3.13.16
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22(2):195–201. https://doi.org/10.1093/bioinformatics/bti770
van de Guchte M, van der Vossen JM, Kok J, Venema G (1989) Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl Environ Microbiol 55(1):224–228
van Belkum MJ, Stiles ME (2006) Characterization of the theta-type plasmid pCD3.4 from Carnobacterium divergens, and modulation of its host range by RepA mutation. Microbiology 152(1):171–178. https://doi.org/10.1099/mic.0.28294-0
Du L, Somkuti GA, Renye JA Jr (2012) Molecular analysis of the bacteriocin-encoding plasmid pDGL1 from Enterococcus durans and genetic characterization of the durancin GL locus. Microbiology 158(Pt_6):1523–1532. https://doi.org/10.1099/mic.0.055624-0
Drider D, Fimland G, Héchard Y, McMullen LM, Prévost H (2006) The continuing story of class IIa bacteriocins. Microbiol Mol Biol Rev 70(2):564–582. https://doi.org/10.1128/MMBR.00016-05
Chikindas ML, García-Garcerá MJ, Driessen AJ, Ledeboer AM, Nissen-Meyer J, Nes IF, Abee T, Konings WN, Venema G (1993) Pediocin PA-1, a bacteriocin from Pediococcus acidilactici PAC1.0, forms hydrophilic pores in the cytoplasmic membrane of target cells. Appl Environ Microbiol 59(11):3577–3584
Sánchez J, Diep DB, Herranz C, Nes IF, Cintas LM, Hernández PE (2007) Amino acid and nucleotide sequence, adjacent genes, and heterologous expression of hiracin JM79, a sec-dependent bacteriocin produced by Enterococcus hirae DCH5, isolated from Mallard ducks (Anas platyrhynchos). FEMS Microbiol Lett 270(2):227–236. https://doi.org/10.1111/j.1574-6968.2007.00673.x
Acknowledgements
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Alberta Government (QEII Scholarship Program).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Chiorean, S., Vederas, J.C. & van Belkum, M.J. Identification and Heterologous Expression of the sec-Dependent Bacteriocin Faerocin MK from Enterococcus faecium M3K31. Probiotics & Antimicro. Prot. 10, 142–147 (2018). https://doi.org/10.1007/s12602-017-9374-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-017-9374-7