Abstract
Sugarcane brown rust is a fungal disease with global distribution that is caused by Puccinia melanocephala, and is responsible for severe yield and sucrose losses in sugarcane production. The most effective strategy of controlling the disease is breeding and cultivating resistant varieties. Bru1 is a major gene that has been demonstrated to confer resistance to P. melanocephala strains from different parts of the globe. R12H16 and 9O20-F4 are two molecular markers that have been previously reported to be closely associated with Bru1, and are often used in brown rust resistance identification during sugarcane germplasm evaluation activities. In the present study, a novel type of molecular marker based on four primers is used to detect Bru1 among 200 sugarcane ancestral species and landraces from the China National Germplasm Repository of Sugarcane (NGRS). According to the results, among the 200 tested materials, 125 materials, such as 51NG63, Katha, and Uba, Bru1 could be detected based on bands corresponding to those in POJ2878, indicated that the 125 (62.5%) materials contained Bru1, and the detection rates of Bru1 in 69 sugarcane ancestral species and 131 landraces were 73.9% and 56.5%, respectively. These results indicated that the sugarcane ancestral species and landraces conserved in the China NGRS contain abundant brown rust-resistant materials, which could provide resources that could facilitate the breeding of brown rust-resistant sugarcane varieties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sugarcane brown rust is a globally distributed and destructive sugarcane disease caused by the fungus Puccinia melanocephala H. Sydow & P. Sydow (Purdy 1983). Sugarcane is a perennial vegetatively propagated crop, susceptible to a variety of pathogens. In addition, the adaptability of pathogenic fungi would influence the duration of disease resistance in sugarcane, making brown rust a persistent threat that degrades sugarcane germplasm resources. Yield losses attributable to sugarcane brown rust in susceptible cultivars can be as high as 50%, which considerably and adversely affects the stable development of the global sugar production industry (Comstock 1992; Hoy and Hollier 2009; Wang et al. 2017). Evaluation, screening of resistant parents and scientific selection, and subsequent breeding and planting of resistant varieties of sugarcane represent the most economical and effective strategy of controlling sugarcane brown rust (Li et al. 2016). Brown rust 1 (Bru1) is a major sugarcane rust resistance gene originally identified in sugarcane cultivar R570 (Daugrois et al. 1996; Asnaghi et al. 2004). Bru1 has been demonstrated to exhibit broad-spectrum resistance to brown rust isolates from different countries and regions (Asnaghi et al. 2001). R12H16 and 9O20-F4 are two molecular markers closely related to Bru1 that have been developed and used extensively in the identification and evaluation of resistance to brown rust in sugarcane germplasms (Costet et al. 2012).
The China National Germplasm Repository of Sugarcane (NGRS) is located in Kaiyuan city, Honghe Prefecture, Yunnan Province, covering an area of 3.33 ha. Numerous valuable sugarcane germplasm resources such as Saccharum ancestral species (Saccharum spontaneum L., Saccharum robustum Brandes & Jesw. ex Grassl., Saccharum officinarum L., Saccharum barberi Jesw., and Saccharum sinense Roxb.), genus in the family Poaceae related to the genus Saccharum, and sugarcane hybrids are conserved in the nursery. The Saccharum ancestral species are important genetic resources for sugarcane breeding activities (Wu et al. 2014). Among them, the contribution of S. robustum to sugarcane breeding is mainly in the form of enhanced lodging resistance, drought tolerance, and perennial ratoon ability (Wu et al. 2014), while Saccharum officinarum L. is the main source of yield-associated traits, such as high sugar content, thick stalks, and smut resistance gene in sugarcane breeding activities. In addition, Saccharum barberi and S. sinense were the earliest domesticated cultivars, and they exhibit high ratoon persistence and stress resistance traits (Wu et al. 2014). Sugarcane landraces are also valuable genetic resources since they can broaden the genetic base and provide locally adapted genes for the improvement of sugarcane crops (Gashaw et al. 2018).
In recent years, sugarcane researchers have identified and evaluated the resources in the NGRS continuously; however, the activities are limited by manpower and material resources. Many evaluations are limited to random germplasms and are not yet systematically comprehensive, resulting in blind parental selection. We adopted a four-primer molecular marker for rapid identification of Bru1 (Guo et al. 2016) from 200 sugarcane ancestral species and landraces conserved at the NGRS. The aim of the present study is to identify and evaluate the brown rust resistance in sugarcane germplasm resources of NGRS, which will facilitate not only the breeding of brown rust-resistant sugarcane varieties but also research on the pathogenic mechanisms of sugarcane brown rust.
Materials and Methods
Plant Materials
The experimental materials were 69 ancestral species, and 131 landraces including sugarcane varieties POJ2878 (+CK), which harbors Bru1 and Co281 (−CK), which does not harbor Bru1 as control plant materials (Racedo et al. 2013) (Table 1). All the materials were obtained from the NGRS and provided by the National Crop Germplasm Resources Center.
Genomic DNA Extraction and Molecular Detection of Bru1
Fresh young leaves of the selected plant materials were sampled, quick-frozen in liquid nitrogen, and the genomic DNA isolated using the EasyPure Plant Genomic DNA Kit (TransGen Biotech, Beijing), according to the manufacturer’s instructions. The genomic DNA was detected by electrophoresis on a 1.2% (m/v) agarose gel and ultra-low volume visible spectrophotometer (BioDrop, USA) and then diluted to a final concentration of 20 ng/μL for immediate use or stored at − 20 °C for later use.
The PCR amplification primers used are introduced in the patent of Guo et al. (2016), including P1: AGGTTTGTCTGGTGGGAACT, P2: CTTACCCTGTGTGCAATGGG, P3: TTTATAAATTACCATTAAGGACCAT, and P4: CAGCATTTAAGATGGCATAGGCGTC. The above primers were entrusted to Biotech Engineering (Shanghai) Co., Ltd. for DNA synthesis. With the prepared genomic DNA as a template, and the P1, P2, P3 and P4 as primers, 2× EasyTaq PCR SuperMix (+ dye) was used to perform PCR amplification. The PCR instrument used was a Bio-rad C 1000 Touch thermal cycler (Bio-Rad, Hercules, CA, USA). The amplification reaction system consisted of a total volume of 12.5 μL, including 1.0 μL of DNA template, 0.3 μL (10 μmol/L) for each of the four primers, 6.25 μL of 2× EasyTaq PCR SuperMix and 4.05 μL of ddH2O. The amplification reaction thermocycling conditions were pre-denaturation for 5 min at 94 °C; denaturation for 30 s at 94 °C, annealing for 30 s at 56 °C, extension for 40 s for 72 °C, 35 cycles in total; extension for 7 min at 72 °C, and storage at 12 °C. After the amplification, 1 μL of HaeIII and 1.5 μL of 10× buffer were added to each PCR system, and incubated at 37 °C for 10 min, and then incubated at 65 °C for 10 min to inactivate the enzyme. Afterward 8.0 μL and 2.0% agarose gel was used to perform electrophoresis. The bands obtained were compared with the bands from POJ2878. Materials with expected nucleotide lengths of 389 bp and 606 bp bands, as in POJ2878, were considered to harbor Bru1. Additionally, the experiment was repeated with 2× EasyPfu PCR SuperMix (-dye), which was 18-fold fidelity compared to 2× EasyTaq PCR SuperMix, and the PCR results of the two enzymes were consistent, thus confirming the reliability of the results.
Results
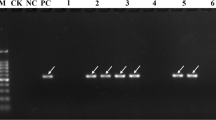
According to the PCR amplification results, the PCR products of the four primers in the positive control material POJ2878 were digested with HaeIII and showed specific 606-bp and 389-bp bands, while there were no corresponding bands in the negative control material, Co281 and water (Fig. 1). Among the 200 tested materials, 51NG63 (lane 6), Katha (lane 10), Uba (lane 12), and 122 other materials, two bands corresponding to the bands in POJ2878 was detected, and the results of the repeated tests were consistent, indicating that the 125 materials (62.5%) harbored the brown rust resistance gene, Bru1. Some materials, such as Daye (lane 2), Kaiyuanlvpi (lane 20), and Hongbanguozhe (lane 31), displayed only one between the 389-bp and 606-bp bands, which may indicate that the materials are brown rust-resistant gene locus exchange strains, which could be the resources that could be exploited for the cloning of the brown rust resistance gene (Table 1).
PCR amplification and enzyme digestion results for resistant brown rust gene, Bru1, in some test materials. Note: M1: Trans 2K Plus DNA Marker, M2: 100-bp Plus DNA Ladder. +CK: Positive control material POJ2878, −CK: Negative control material Co281, 0CK: ddH2O, 1: Sichuanbaizhe, 2: Daye, 3: Guilinzhuzhe, 4: Jiangxizhuzhe, 5: Tonganguozhe, 6: 51NG63, 7: Mungo, 8: Badila, 9: Pansahi, 10: Katha, 11: HATUNI, 12: Uba, 13: 57NG208, 14: Qianweiluze, 15: Nagori, 16: 48Mouna, 17: Zopilota, 18: Songxizhuzhe, 19: Wenshanzhuzhe, 20: Kaiyuanlvpi, 21: Zhibaozipi, 22: Niuzhe, 23: 51NG3, 24: Fujiandaye, 25: Taoshanzhe, 26: Hongluohan, 27: 14NG124, 28: 96NG14, 29: 28NG251, 30: Guguoguozhe, 31: Hongbanguozhe, 32: Nagans, 33: Tanzhouzhuzhe, 34: Guangzezhuzhe; 35: Gengmazhe, 36: Sichuanluzhe, 37: Deyangdayezi, 38: Heqingcaoganzhe, 39: Loether, 40: Black Cheribon, 41: 27MQ1124, 42: Chunwuni
Discussion
Bru1 is a major sugarcane brown rust resistance gene originally discovered in the sugarcane cultivar R570, and has been confirmed to exhibit broad-spectrum resistance to brown rust isolates from different countries and regions (Li et al. 2016; Daugrois et al. 1996; Asnaghi et al. 2004). Two molecular markers closely related to Bru1 (R12H16 and 9O20-F4) were confirmed to be able to stably and effectively detect sugarcane brown rust resistance gene, Bru1. In recent years, Li et al. have used the two molecular markers to detect Bru1 in some key sugarcane breeding parents (Li et al. 2016), cultivated original species (Li et al. 2015a), wild core germplasms (Li et al. 2015b), and novel elite varieties/lines (Li et al. 2017) in the NGRS. Their studies have revealed high numbers of potentially excellent brown rust resistance resources and indicates that there could be other brown rust resistance genes.
A recent study demonstrated that R12H16 and 9O20-F4-RsaI are effective for the detection of Bru1 among modern sugarcane varieties; however, it is relatively ineffective for evaluating wild resources, and 9O20-F4-RsaI may yield false positives (Wang et al. 2017; Li et al. 2018). Consequently, Wang et al. (2017) developed an improved Bru1 diagnostic marker, 9O20-F4-HaeIII, which produces a band of 389 bp and eliminates the false positive bands detected by the 9O20-F4-RsaI marker in S. spontaneum. The study also reported another novel dominant Bru1 diagnostic marker R12E03-2. However, detection using two primer pairs, R12H16 and 9O20-F4-HaeIII, would require two PCR systems and two electrophoresis reactions. The new primers designed by Guo et al. (2016) were used in the present study to identify the sugarcane brown rust resistance gene rapidly. In addition, the combination of four primers in the present method can reduce the two PCR and electrophoresis requirements to single reaction, which improves detection efficiency and reduces the associated costs considerably. The identification results of the present study are consistent with some results reported for cultivated species by Li et al. (2015a), excluding 27MQ1124, which was reported to not harbor Bru1. Notably, the results of our repeated tests also indicated that 27MQ1124 harbored Bru1.
Germplasm resources are the foundations of breeding activities. The China NGRS has conserved more than 3800 sugarcane germplasm resources, and it is one of the invaluable gene banks hosting resources that could be used for the genetic improvement of sugarcane varieties in China and in other countries globally (Li et al. 2016). Investigating the origins of sugarcane resources, systematically evaluating the resources, and selecting varieties with desirable traits, such as brown rust resistance, are essential for the breeding of sugarcane varieties that are resistant to pathogens and environmental stress. Sugarcane brown rust is an economically important disease globally, and one of the most common and most destructive diseases in the sugarcane industry in China (Gao et al. 2019). Although the application of pesticides can alleviate the disease, it increases input costs and has potentially adverse effects on ecosystems. Therefore, breeding and cultivation of disease-resistant varieties are the most cost-effective strategies for preventing and managing sugarcane brown rust (Li et al. 2016; Racedo et al. 2016).
Sugarcane ancestors are the major progenitors of sugarcane hybrids. Local breeds also exert their unique advantages in breeding. Screening of brown rust-resistant materials from the two groups of materials could provide suitable parental resources for the breeding of brown rust-resistant varieties. Considering systematic brown rust identification and evaluation studies have not been carried out using the NGRS resources, we plan to evaluate all the resources in the NGRS in to enhance our understanding of the resources through comprehensive and systematic studies.
In the present study, we conducted a molecular study to detect brown rust resistance gene, Bru1, in sugarcane ancestors and native species conserved in the NGRS. The detection rate of Bru1 in 200 sugarcane ancestors and local species was 62.5%, suggesting that the resources harbored brown rust-resistant materials. Among the 69 sugarcane ancestors and 131 local species tested, 73.9% and 56.5%, respectively, harbored the gene, indicating that the two groups of resources had high brown rust resistance materials. However, the levels of resistance in the test materials require further evaluation by manual inoculation with P. melanocephala spore suspensions. Although considerable progress has been made with regard to research of sugarcane brown rust, the underlying mechanism of Bru1 in brown rust resistance, in addition to the mechanisms of other brown rust resistance genes, remains unclear. Therefore, screening excellent resistance parents, identifying and isolating brown rust disease-causing genes, and clarifying the underlying pathogenic mechanisms require attention in future research.
References
Asnaghi, C., A. D’hont, J. Glaszmann, and P. Rott. 2001. Resistance of sugarcane cultivar R570 to Puccinia melanocephala isolates from different geographic locations. Plant Disease 85(3): 282–286.
Asnaghi, C., D. Roques, S. Ruffel, C. Kaye, J.Y. Hoarau, H. Télismart, J.C. Girard, L.M. Roboin, A.M. Risterucci, L. Grivet, and A. D’Hont. 2004. Targeted mapping of a sugarcane rust resistance gene (Bru1) using bulked segregant analysis and AFLP markers. Theoretical and Applied Genetics 108(4): 759–764.
Comstock, J.C. 1992. Effect of rust on sugarcane growth and biomass. Plant Disease 76: 175–177.
Costet, L., L. Le Cunff, S. Royaert, L.M. Raboin, C. Hervouet, L. Toubi, O. Garsmeur, Y. Rousselle, J. Pauquet, S. Nibouche, J.C. Glaszmann, J.Y. Hoarau, and A. D’ Hont. 2012. Haplotype structure around Bru1 reveals a narrow genetic basis for brown rust resistance in modern sugarcane cultivars. Theoretical and Applied Genetics 125(5): 825–836.
Daugrois, J.H., L. Grivet, D. Roques, J.Y. Hoarau, H. Lombard, J.C. Glaszmann, and A. D’Hont. 1996. A putative major gene for rust resistance linked with a RFLP marker in sugarcane cultivar ‘R570.’ Theoretical and Applied Genetics 92(8): 1059–1064.
Gao, X.N, R. Liu, Y.W. Qi. 2019. Research progress on sugarcane brown rust. China Plant Protection 39(11): 26–30.
Gashaw, E.T., F. Mekbib, and A. Ayana. 2018. Sugarcane landraces of Ethiopia: Germplasm collection and analysis of regional diversity and distribution. Advances in Agriculture 2018(6): 1–18.
Guo, J.L., Y.H. Lu, H.B. Wang, L.P. Xu, Y.X. Que, D. Liu, N.Y. Xiao, P.H. Chen. 2016. Molecular marker primers for rapid identification of brown rust resistance gene loci in sugarcane and their application: China. ZL201610271677.8.
Hoy, J.W., and C.A. Hollier. 2009. Effect of brown bust on yield of sugarcane in Louisiana. Plant Disease 93(11): 1171–1174.
Li, W.F., H.L. Shan, R.Y. Zhang, C.H. Pu, X.Y. Wang, X.Y. Cang, J. Yin, Z.M. Luo, and Y.K. Huang. 2017. Resistance evaluation to brown rust and molecular detection of Bru1 gene in new elite sugarcane varieties /lines. Acta Agronomica Sinica 47(5): 667–674.
Li, W.F., X.Y. Wang, Y.K. Huang, H.L. Shan, R.Y. Zhang, J. Yin, and Z.M. Luo. 2015a. Identification of resistance to brown rust and molecular detection of Bru1 gene in 34 sugarcane cultivated original species. Molecular Plant Breeding 13(8): 1814–1821.
Li, W.F., X.Y. Wang, Y.K. Huang, R.Y. Zhang, H.L. Shan, J. Yin, and Z.M. Luo. 2015b. Identification of resistance to brown rust and molecular detection of Bru1 Gene in 31 Wild Core Sugarcane Germplasms. Acta Agronomica Sinica 41(5): 806–812.
Li, W.F., X.Y. Wang, Y.K. Huang, R.Y. Zhang, H.L. Shan, Z.M. Luo, and J. Yin. 2016. Identification of resistance to brown rust and molecular detection of Bru1 gene in 101 main sugarcane breeding parents in China. Acta Agronomica Sinica 42(9): 1411–1416.
Li, Z., Y.C. Su, Y. Qing, Y. Chen, S.W. Gao, Y.G. Zhang, Y.X. Que, and L.P. Xu. 2018. Molecular insights into brown rust resistance and potential epidemic based on the Bru1 gene in sugarcane varieties and new elite clones. Euphytica 214(10): 189.
Purdy, L.H. 1983. Sugarcane rust, a newly important disease. Plant Disease 67(11): 1292–1296.
Racedo, J., M.F. Perera, R. Bertani, C. Funes, V. González, M.I. Cuenya, A. D’Hont, B. Welin, and A.P. Castagnaro. 2013. Bru1 gene and potential alternative sources of resistance to sugarcane brown rust disease. Euphytica 191(3): 429–436.
Racedo, J., M.F. Perera, R. Bertani, C. Funes, V. Gonzalez, M.I. Cuenya, A. D’Hont, B. Welin, and A.P. Castagnaro. 2016. Molecular diagnostic of both brown and orange sugarcane rust and evaluation of sugarcane brown rust resistance in Tucuman, Argentina, using molecular markers associated with Bru1 a broad-range resistance allele. Sugar Tech 18(4): 414–419.
Wang, H.B., P.H. Chen, Y.Q. Yang, A. D’Hont, and Y.H. Lu. 2017. Molecular insights into the origin of the brown rust resistance gene Bru1 among Saccharum species. Theoretical and Applied Genetics 130: 2431–2443.
Wu, C.W., P.F. Zhao, and H.M. Xia. 2014. Modern cross breeding and selection techniques in sugarcane. Bei Jing: Science Press.
Acknowledgements
Thanks are due to Elixigen company for reviewing the manuscript for English accuracy.
Funding
This work was supported by the National Natural Science Foundation of China (31601362), National Crop Germplasm Resources Center (NCGRC-2020-42), and the Funding for the Protection of Species and Cultivar Resources from the Ministry of Agriculture and Rural Affairs of China (19200375).
Author information
Authors and Affiliations
Contributions
XJL carried out the experiment and wrote the manuscript. CHX, JM, HBL, CJL, XLL, XQL, and CYK participated in experimental work. XL conceived and designed the study.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, X., Xu, C., Mao, J. et al. Detection of Key Brown Rust Resistance Gene, Bru1, in 200 Sugarcane (Saccharum L.) Ancestral Species and Landraces Using a Four-Primer Molecular Marker. Sugar Tech 23, 838–842 (2021). https://doi.org/10.1007/s12355-021-00954-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12355-021-00954-y