Abstract
Auxin response factor (ARF) acts as a vital component of auxin signaling and participates in growth, development, and stress responses in plants. In the present study, we comprehensively analyzed kiwifruit’s (Actinidia chinensis) ARF genes (AcARFs) and their involvement in abiotic stress response. We identified a total of 41 AcARFs encoding ARFs in the A. chinensis genome. AcARF genes were characterized by the classic ARF_resp and a B3 domain and primarily localized on the cytoplasm and nucleus. AcARFs were categorized into eight subgroups as per the phylogenetic analysis. Synteny analysis showed that 35 gene pairs in AcARF family underwent segmental and whole genome duplication events. Promoter cis-element prediction revealed that AcARFs might be involved in abiotic factors related to stress response, which was later assessed and validated by qRT-PCR based expression analysis. Additionally, AcARFs showed tissue-specific expression. These findings extend our understanding of the functional roles of AcARFs in stress responses. Taken together, the systematic annotation of the AcARF family genes provides a platform for the functional and evolutionary study, which might help in elucidating the precise roles of the AcARFs in stress responses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Auxin is a plant hormone of utmost significance as it plays crucial roles in the course of plant growth and development, such as embryogenesis, flower and fruit development, apical dominance, tropic response, root architecture, and vascular development (Aloni et al. 2006; Esmon et al. 2006; Kazan 2013; Mattsson et al. 2003; Woodward and Bartel 2005). Auxin exerts its effects via the modulation of gene expression of a myriad of gene families, such as small auxin up RNA (SAUR), gretchen hagen 3 (GH3), indole-3-acetic acid (Aux/IAA) and auxin response factor (ARF) (Wang et al. 2018; Yuan et al. 2019; Zhou et al. 2019). ARFs function as transcription factors and are crucial components of the auxin signalling pathway. They modulate auxin-regulated genes (ARGs) by binding explicitly to the auxin response elements (AuxRE, 5’ TGTCTC 3’) in the promoter of ARGs (Berendzen et al. 2012). ARFs contain a conserved N-terminal DNA-binding domain (DBD) and a C-terminal dimerization domain (CTD) (Tiwari et al. 2003). The DBD of ARFs binds to AuxRE in ARG promoter regions. The C-terminal dimerization domain is identical to that of Aux/IAA proteins apportioned to domains III and IV; besides, it facilitates homodimerization or heterodimerization of the ARFs or numerous other Aux/IAA proteins (Ulmasov et al. 1999). ARFs also encompass a non-conserved middle region (MR), present between DBD and CTD. The ARFs mediated transcriptional activation or repression is based on the amino acid composition. The activator function is imparted by the QSL-rich (glutamine (Q), serine (S), leucine (L) amino acids) and repressor activity by the S-rich (serine), SPL-rich (serine, proline, leucine), and SL / G-rich (serine, leucine and/or glycine) MR in ARFs (Ulmasov et al. 1999).
Multiple members of the ARF family have been identified and well-characterized in numerous plant species subsequent to the cloning of the Arabidopsis thaliana’ first ARF gene, AtARF1. As per the genomic analyses, a total of 23 ARF members in A.thaliana, 39 in Populus trichocarpa, 25 in rice (Oryza sativa), 17 in Eucalyptus grandis, 22 in tomato (Solanum lycopersicon), 19 in sweet orange (Citrus sinensis), and 31 in maize and Brassica rapa have been identified so far (Bouzroud et al. 2018; Kalluri et al. 2007; Li et al. 2015a; Mun et al. 2012; Overvoorde et al. 2005; Wang et al. 2007; Xing et al. 2011; Yu et al. 2014). Besides, the functional significance of ARFs in various plant growth and development processes has been well elucidated. For instance, the functional role of AtARF3 and AtARF4 in leaf and floral organ patterning coincides (Pekker et al. 2005; Sessions et al. 1997), AtARF5 / MONOPTEROS (MP) participates in embryonic development and vascular tissue formation (Hardtke and Berleth 1998), AtARF7 and AtARF19 both control lateral root formation (Okushima et al. 2007; Wilmoth et al. 2005), and AtARF6 and AtARF8 both regulate flowering process (coordinating stamen development, petal expansion, anther dehiscence, and gynoecium maturation) (Nagpal et al. 2005). As per the recent studies, tomatoes have multiple ARF genes, such as SlARF2, SlARF7, SlARF9, which are involved in leaf morphogenesis, development of flowers and fruits and fruit ripening (de Jong et al. 2009; Hao et al. 2015; Wilmoth et al. 2005). As per the previous reports, as transcriptional regulators, ARFs directly contribute to auxin signal transposition (Guilfoyle and Hagen 2007; Ljung 2013) and are involved in other many stress responses and hormonal cross-talks (Guilfoyle and Hagen 2007; McAtee et al. 2013) in Arabidopsis (Kalve et al. 2020; Okushima et al. 2005), rice (Jain and Khurana 2009), citrus (Li et al. 2015a), strawberry (Wang et al. 2019), banana (Hu et al. 2015b), tomato (Bouzroud et al. 2018), and chickpea (Singh et al. 2017). In Arabidopsis, multiple ARF genes participate in abiotic stress responses (Ji and Jiang 2015; Kalve et al. 2020; Okushima et al. 2005). In tomatoes, the SlARF gene mediates salt, drought, and flooding stress response, and the SlARF7 gene mediates cross-talks between auxin and GA signaling during the fruit and plant development in tomatoes (Bouzroud et al. 2018; de Jong et al. 2011; Kumar et al. 2011). Similarly, multiple ARF genes mediate abiotic stress responses against desiccation, salt, and cold stresses (Jain and Khurana 2009; Wang et al. 2007). In chickpea, CaARF genes significantly regulate abiotic stress (Singh et al. 2017).
Kiwifruit is a widely consumed and health-promoting fruit, and one of the most important fruit crops globally. The cultivated kiwifruit, Hongyang, is a heteroploid hybrid variety (2n = 2x = 58) derived from wild A. chinensis (Huang et al. 2013). Kiwifruits are frequently exposed to different abiotic stresses, such as drought, cold, salinity, and hormonal stress. In this study, we used the data from open-access databases for genome-wide identification and characterization of the ARF family of A. chinensis. We performed comparative phylogenetic analysis, gene structure and conservative motifs prediction, gene duplication events analysis, promoter cis-elements, and subcellular location prediction. Besides, we employed quantitative real-time PCR (qRT-PCR) to assess the expression profiles of the 12 AcARF genes in different stress conditions such as stress signalling induced by different phytohormones (IAA; abscisic acid, ABA; gibberellins, GA; salicylic acid, SA; jasmonic acid, JA), along with drought and salt stresses. The outcomes of our current study provide a platform for future research and highlight the importance of targeting ARF genes to obtain stress-resistant kiwifruit varieties.
Materials and methods
Identification and annotation of ARF genes in kiwifruit
The kiwifruit genome and proteome datasets were downloaded from Ensembl Plants (http://plants.ensembl.org/index.html). The A. thaliana ARF protein data set were downloaded from TAIR (https://www.arabidopsis.org/) (Swarbreck et al. 2008). The Hidden Markov Model (HMM) profiles of the ARF family were employed with default parameters and a cutoff value of 0.01 to identify the ARF genes from the A. chinensis genome. All candidate ARF genes containing ARF conserved domains were further examined via the Pfam (http://pfam.wustl.edu) and SMART (http://smart.embl-heidelberg.de) databases (Schultz et al. 1998). All the AcARF sequences were validated as a member of the ARF family by using the NCBI conserved domain database (http://www.omicsclass.com/article/310). Physical and chemical (molecular weight, MW, and the isoelectric point, pI) characterization of AcARF proteins was done using an online tool, ProtParam (http://web.expasy.org/protparam/) (Wilkins et al. 1999). The subcellular loci were predicted with WoLF PSORT (Horton et al. 2007).
Phylogenic analysis of AcARFs and AtARFs
The full-length ARF proteins were aligned using Clustal X (Chenna et al. 2003) and visually adjusted using BioEdit V7.2 (Tippmann 2004). The Neighbor-Joining (NJ) phylogenetic trees (Silva et al. 2005) were constructed using Mega V7.0 with 1000 as the bootstrap replicates (Kumar et al. 2016).
In silico characterization of AcARFs
The chromosomal location of genes was retrieved from the A. chinensis genome browser by Scipio (Keller et al. 2008). The chromosomal location of the AcARF genes was visualized by MapChart V2.1 (Voorrips 2002). MCScan X with default parameters was employed to examine the gene duplication events in the AcARF genes (Wang et al. 2012). To study the relationship between the orthologous ARF genes obtained from kiwifruit and A. thaliana, a syntenic analysis map was constructed using DSP software. The AcARF gene structure was extracted from Ensembl Plants and visualized using GSDS V2.0 (http://gsds.cbi.pku.edu.cn) (Hu et al. 2015a). The Pfam (Finn et al. 2016) and MEME suite (Bailey et al. 2009) were used to reveal and visualize conserved motifs. The promoters (1500 bp before ATG) of the AcARF genes were obtained from the Ensembl Plants, and the cis-elements were predicted by PlantCARE (Lescot et al. 2002).
Plant materials
Kiwifruit ‘Hongyang’ (A. chinensis) plants were cultivated in Bairui Kiwifruit Research Institute located at Shaanxi, China (33° 42’N, 107° 39’E) from 2017 to 2019. Different tissues, including roots, stems, leaves (the fourth young leaves from the shoot), flowers, small green fruit (approximately 30 days after flower), and ripe fruit (approximately 120 days after flower) were collected from “Hongyang” trees (aged 5 years) during the 2020 growing season. The 2-year-old ‘Hongyang’ kiwifruit seedlings were used for hormonal treatments. All these seedlings were planted in 25 cm diameter pots in the greenhouse set to the temperature of 28 °C. The branch containing three to six pieces of leaves were treated with phytohormones. Leaves were sprayed with 100 μM·L−1 of phytohormones. The leaves sprayed with plain double distilled water (ddH2O) were treated as the controls, and the leaf samples were collected at 0, 1, 6, and 12 h post-treatment (hpt). The salty stress was induced by irrigating each seedling with 2 L of 200 mM·L−1 NaCl solution, and seedlings irrigated with ddH2O were treated as controls. The leaves were collected at 1, 6, 12, 24, and 48 hpt. Drought stress was induced by halting water supply to seedlings, and the leaves were collected at 48, 96, 144, and 168 hpt; these seedlings were re-watered after leaves collection, i.e., 48 hpt, in accordance with the previous study (Jing and Liu 2018). All the leaf samples were flash-frozen in liquid nitrogen and stored at − 80 °C for RNA extraction. For each treatment, six different leaves were sampled from six different seedlings.
RNA extraction and qRT-PCR
Total RNAs were extracted from around 100–200 mg of frozen leaf samples using a TRIZOL Reagent (Invitrogen) according to the manufacturer’s instructions. Genomic DBA contamination was removed by using the Turbo DNA-free™ kit (Ambion). The total RNA was reverse transcribed into cDNA by the Invitrogen reverse transcription kit (SuperScript III Reverse Transcriptase). The primers were designed using NCBI’s primer design tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) (Supplementary Table 5). qRT-PCR was performed using ABI Quant Studio tm 6 Flex Real-Time PCR System with SYBR-Green PCR Master Mix. The reaction mixture (20 μL) contained 10 μL SYBR Green Master Mix (ROX), 1 μL forward specific primer (10 μM), 1 μL reverse specific primer (10 μM), 1 μL cDNA (30 ng · μL−1) template, and 7 μL ddH2O. DNA amplification was performed by using the following thermocycling program: 95 °C for 3 min, 40 cycles of 95 °C for 20 s, 60 °C for 30 s, and 72 °C for 30 s, followed by 71 cycles with an increasing temperature gradient of 0.5 °C per cycle from 60 to 95 °C for 30 s. ACT2 gene (GenBank: EF063571) was used as an internal standard to calculate the relative fold change as per the comparative cycle threshold (2−ΔΔCt) method (Livak and Schmittgen 2001) with three biological and technical replicates. P value < 0.05 was treated as statistically significant.
Results
ARF annotation in kiwifruit genome
To identify potential ARF genes in kiwifruit genome, 23 Arabidopsis ARF protein sequences were queried against the annotated A. chinensis genome in Ensembl Plants database (http://plants.ensembl.org/index.html). After manual curations, a total of 41 A. chinensis ARFs were identified and named as per their respective chromosomal locations (Table 1). In instances where one chromosome contained two or more than two AcARF genes, chromosome number was used to name the genes, such as AcARFxa and xb, where x represents chromosome number. The number of amino acids (aa) in AcARF proteins ranged from 448 (AcARF28a) to 1115 (AcARF29) with 49.72 to 123.53 kDa MWs, respectively. The PI values ranged from 5.53 (AcARF21c) to 8.1 (AcARF28a), which indicates that different AcARF proteins might work in different microenvironments. All the AcARF proteins were located in the nucleus except AcARF10 and AcARF28a proteins, which were located in the cytosol (Table 1).
Alignment and phylogeny of the AcARFs
All the 41 predicted AcARFs proteins demonstrated characteristic ARF family structure, including a highly conserved B3-like DNA-binding and ARF_resp domain in the N-terminal region as per the sequence alignment and Pfam based protein motif analysis of the predicted proteins (Supplementary Fig. 1). ARFs can function as activators or repressors depending upon the amino acid composition of the MR. Also, according to the previous functional studies on Arabidopsis, ARF proteins containing glutamine (Q)-rich MR function as activators and rest of the ARF proteins function as repressors. Out of the 41 AcARF proteins, AcARF5, 12, 20b, and 29 showed Q-rich MR, which suggests that these proteins might function as transcriptional activators. In contrast, the other 37 AcARF proteins might function as repressors (Supplementary Fig. 1).
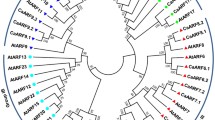
To characterize the evolutionary relationships between kiwifruit and Arabidopsis ARF family members, we carried out an NJ phylogenetic analysis of all the proteins of 41 AcARFs and 23 AtARFs (Fig. 1). As per the phylogenetic analysis, the ARFs were categorized into eight major groups (group 1–8) with well-supported bootstrap values. However, we found only one organism-specific group (group 4) from the phylogenetic analysis. In the common clades, we observed that IAAs were distributed unequally between two organisms. For instance, group 1 contained 11 and 3 members from kiwifruit and Arabidopsis, respectively, and group 2 contained 3 and 11 members from kiwifruit and Arabidopsis, respectively. The Arabidopsis ARF family was overrepresented in this class due to seven genes with tandem duplication, which encodes AtARF12-15 and AtARF20-22 proteins whose orthologous proteins were not found in kiwifruit. All transcriptional activators of AcARF genes were clustered in groups 6 and 7.
Phylogenetic analysis of A. thaliana and A. chinensis ARFs. MEGA V7.0 software was employed to construct the NJ phylogenetic tree with 1000 bootstrap replicates based on the ARF amino acid sequences of A. chinensis and A. thaliana. Different colors represent different AcARFs and AtARFs groups. Black colored dots and stars indicate A. chinensis and A. thaliana, respectively. Branches are drawn to scale; the length of the branch represents the number of substitutions per site
Chromosomal distribution of AcARF genes
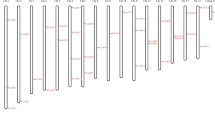
The 41 AcARF genes were distributed across 23 chromosomes as per the in silico chromosomal mapping of the gene loci with 1–4 AcARF genes per chromosome (Fig. 2). These genes were primarily located on chromosomes 1, 4–14, 18–26, 28, and 29, whereas none of the AcARF genes were located on chromosomes 2, 3, 15–17, and 27.
Genomic localization of AcARF genes.The numbers at the head of each chromosome represent chromosome serial numbers. 41 AcARF genes were unevenly located on the chromosomes, and these genes were mapped based on the kiwifruit genome database using MapChart V2.2. The length of chromosomes is on the scale (Mb). The ARFs gene location is mentioned on the left while name of the right side
Gene duplication and synteny analyses of the AcARF genes
To determine potential gene duplication within the kiwifruit genome, the duplication events including segmental and whole genome duplications throughout the evolutionary process of ARF gene family were analyzed. A total of 35 duplicated AcARF gene pairs were observed in A. chinensis genome (Fig. 3 and Supplementary Table 1). Out of 35 duplicated gene pairs, 33 were located on different chromosomes, which might be due to whole-genome duplication events, whereas two pairs (AcARF23b & AcARF23c, AcARF8a & AcARF8b) were located on the same chromosomes, which might be due to segmental duplication events. To gain a detailed understanding of the evolutionary constraints on ARF gene family, the Ka/Ks ratios of the AcARF gene pairs were analyzed (Table 2). The outcomes showed Ka/Ks < 1 for 15 duplicated AcARF gene pairs, which suggests that the kiwifruit ARF gene family had undergone selective pressure during evolution. Thus, we hypothesized that these gene duplication events in kiwifruit led to an increase in the AcARF gene family members with higher functional diversity. To further infer the phylogenetic mechanisms of A. chinensis ARF genes family, we constructed a comparative syntenic map of A. chinensis and A. thaliana (Fig. 4). It showed that a total of 18 AcARF genes have a syntenic relationship with ARF genes of Arabidopsis (Supplementary Table 2). It suggests that these genes may have played a crucial role in the evolution of the ARF gene family.
The collinear correlation of the Aux/IAA between A. thaliana and A. chinensis. The green color represents 29 A. chinensis chromosomes, and the red color represents 5 A. thaliana chromosomes. Gray lines in the background indicate the collinear blocks between A. thaliana and A. chinensis, whereas the red lines indicate the syntenic ARF gene pairs between A. thaliana and A. chinensis
Conserved motifs and exon-intronic structures
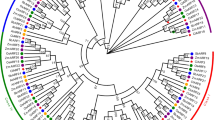
Phylogeny and conserved motif analysis of AcARFs were performed to identify conserved regions (Fig. 5a). A total of 41 AcARFs with 8–15 conserved motifs were detected through this analysis (Fig. 5b and Supplementary Table 3). As per the conserved domains (Supplementary Fig. 1), motif 1 and 3 were identified as B3 conserved domain, motif 6 and motif 7 as Auxin_resp domain, and motif 10 as PB1 domain. Moreover, all the 41 AcARFs contain motifs 1, 3, 6, and 7. And, most of the AcARFs (30 / 41) proteins contain PB1 (AUX_IAA) domain except AcARF8b, 10, 11, 18a, 21a, 22a, 22b, 23c, 26b, 26c, and 28a. The structural analysis demonstrated that AcARFs contain 2–14 exons and 1–13 introns (Fig. 5c). Notably, the number of introns between the AcARF genes was different; however, the genes present in one cluster showed highly identical distributions of exons and introns. Motif composition, arrangements, and gene structures were in line with the phylogenetic tree (Fig. 5a). Furthermore, an in-depth structural analysis of exon–intron can unravel the evolution of the AcARFs gene family.
Phylogeny, motifs, and exon-intronic structures of AcARFs. (a) Unrooted NJ phylogeny of AcARFs. MEGA V7.0 was employed to construct the phylogenetic tree based on full-length AcARFs amino acid sequences (bootstrap = 1000, Poisson model) with a length corresponding to the number of substitutions per site. (b) MEME was used to determine the conserved motifs of AcARFs. Motifs and their positions (1–15) are represented by the colored boxes at the bottom. The scale bar denotes the length of the amino acid sequence (Supplementary Table 1). (c) GSDS was used to depict the structure of AcARF genes. Exons (yellow box); UTR (green); scale bar represents the length of the respective DNA sequences
Putative cis-regulatory elements in the promoters of AcARFs
The interaction between cis-elements and the corresponding trans-acting factors modulates gene regulation. PlantCARE was used to predict the cis-regulatory elements in the promoter region of the AcARF genes to unravel the AcARFs mediated gene regulation. We found that promoter sequences of all the 41 AcARF genes contain several light-responsive elements. It indicates that AcARFs play a crucial role in kiwifruit morphogenesis. Besides, we found cis-regulatory elements related to hormonal stress (due to JA, SA, ABA, and GA), stress-responsive elements (anaerobic induction, defense, drought, and low temperature) in the promoter regions of the majority of the AcARF genes (Table 3). However, few AcARF genes showed tissue-specific elements (endosperm, meristem, and cell cycle), and circadian control elements.
Expression analyses of the ARF Genes in Actinidia chinensis organs
We investigated the spatial-specific expression pattern of the 41 AcARFs in six different organs, including roots, stems, leaves, flowers, small green fruit (SF) and ripe fruits (RF), to investigate the physiological functions of AcARFs (Fig. 6). All the AcARFs were detected in different organs and showed tissue-specific expression patterns in Actinidia chinensis. Most AcARFs showed higher stem and flower specific expression compared with other organs, except AcARF7 and AcARF29, whose expression levels were upregulated in leaf. AcARF10 showed root-specific expression. AcARF23a and AcARF23b showed SF-specific expression, while AcARF26d showed RF-specific expression. Transcriptional analysis of the AcARFs showed tissue-specific expression in Actinidia chinensis, suggesting that AcARF genes might play distinct roles in different organs during kiwi fruit development.
Heatmap of AcARFs’ transcriptional profiles in different organs. The log2 values of the relative expression data were used for the heatmap visualization. The relative expressions of individual AcARFs were normalized to ACT 2 gene in different tissues, including root, stem, leaf, flower, small green fruit (SF, approximately 30 days after flower), and ripe fruit (RF, approximately 120 days after flower). Green blocks and red blocks represent downregulated and upregulated transcription levels, respectively
The AcARFs expression patterns under various environmental and hormonal stress
To test the responsiveness of AcARs to exogenous hormonal stimuli, we choose 12 ARF genes from different clusters and no collinear relationship between them as far as possible for expression profiling. So, we analyzed expression patterns of AcARF genes such as AcARF1a, AcARF4, AcARF5, AcARF6a, AcARF7, AcARF10, AcARF18b, AcARF19a, AcARF23a, AcARF26a, AcARF28a, AcARF28b in the kiwifruit leaves, 1, 6 and 12 h after IAA, ABA, GA, SA, MeJA treatment using qRT-PCR (Fig. 7 and Supplementary Fig. 2). The outcome of this analysis demonstrated that most of these AcARF genes were responsive to auxin except for AcARF1a and AcARF23a. Post-IAA treatment, AcARF4, AcARF5, AcARF6a, AcARF10, AcARF19a, AcARF28a, AcARF28b genes were down-regulated at different time-points, while AcARF7, AcARF26a and AcARF18b were up-regulated. Interestingly, the AcARF7 and AcARF26a were up-regulated by IAA, but they were down-regulated by ABA treatment. In contrast, the AcARF4 gene was up-regulated by ABA at all the time-points and down-regulated by IA. AcARF1a gene was overexpressed 6 h post ABA treatment, and later its expression level decreased to the baseline level. The transcript levels of most of the genes such as AcARF6a, AcARF7, AcARF23a and AcARF28b increased significantly post GA treatment, whereas AcARF26a and AcARF28a gene expression was first down-regulated and later reverted to normal. SA treatment induced the down-regulation of AcARF1a and AcARF10 genes at all time points. JA treatment induced the upregulation of AcARF6a and AcARF28a after 6 h post treatment, while AcARF18b was up-regulated at all time points. The diverse AcARF gene expression pattern in response to different hormonal treatments indicates the complexity of auxin-regulated gene expression.
Heatmap showing differential expression patterns of 12 AcARFs under hormonal stress. Transcription levels at 0 h (untreated plant) were taken as an internal control (mean value = 0) to determine the relative mRNA abundance of each gene. The ACT 2 gene was used as an internal standard. The experiments were performed in triplicates, and the mean expression values from three independent experiments were used for the heatmap construction
To explore the expression patterns of AcARF genes in kiwifruit under drought and salt stress, the expression pattern of 12 AcARF genes was investigated through qRT-PCR (Fig. 8 and Supplementary Figs. 3 and 4). The outcome of this analysis showed that drought stress induces the upregulation of AcARF1a, AcARF5, AcARF7, AcARF19a, AcARF26a, AcARF28a, and AcARF28b genes. As demonstrated in Fig. 8 and Supplementary Fig. 3, AcARF1a, AcARF19a, AcARF26a, AcARF28a, and AcARF28b genes were up-regulated throughout different time points, while the transcription levels of AcARF5 and AcARF7 increased only after 168 h post-drought stress induction. Besides, once the water supply was restored, the expression level of these genes reverted to normal. Under salt stress, the AcARF26a gene was up-regulated across all the time-points, while AcARF4, AcARF5, AcARF23a, and AcARF28a genes were up-regulated 12 h, 24 h and 48 h post salt stress induction (Fig. 8 and Supplementary Fig. 4).
Heatmap showing differential expression of 12 AcARFs under drought and salt stresses. The annotation is the same as Fig. 6
Discussion
Auxin plays a crucial role in the growth and development of plants. ARFs are vital components of the auxin signaling pathway; thus, they can regulate the transcription of auxin-responsive genes involved in the majority of plant-growth stages, development, and stress responses (Tiryaki 2009). Thus, to elucidate the role of kiwifruit’ ARFs in specific auxin responses, we used A. chinensis genome and performed a genome-wide comprehensive survey of ARF gene family in kiwifruit. In this study, 41 kiwifruit ARF genes were identified and designated as per their respective chromosomal location (Fig. 2). The number of AcARF genes in kiwifruit was more as compared to other species, such as Arabidopsis (23), tomato (22), and rice (25) due to a higher number of gene duplication events (Supplementary Tables 4). Synteny analysis showed that 35 AcARF gene pairs underwent gene duplication events via the segment and whole genome duplication mechanism (Fig. 3), and 15 duplicated AcARF gene pairs had Ka/Ks < 1. Comparative genomic analysis of the kiwifruit and Arabidopsis AcARF genes validated gene duplication events (whole genome duplication) in at least 18 orthologous AcARF genes (Fig. 4). Segmental and whole-genome duplication events are vital for the gene family’s expansion (Li et al. 2015b). The phylogenetic analysis of kiwifruit and Arabidopsis showed that kiwifruit AcARFs have orthologs in Arabidopsis except for two specific genes, AcARF13 and AcARF23a, in Group 4 (Fig. 1).
In this study, protein domain analysis revealed that kiwifruit AcARFs contain highly conserved DNA binding domain along with plant-specific B3-type and an ARF subdomain, which binds explicitly to auxin response elements (AuxRE: TGTCTC) in the promoter region of auxin response genes (Fig. 5). ARFs were categorized into two groups: transcriptional activators and repressors, as per the amino acid composition of MR. The average activator/repressor ratio of AcARFs was 0.1, which was less than that of Arabidopsis (0.59), rice (0.56), tomato (0.27). The CTD showed the presence of two motifs, domains III and IV, which are also found in Aux/IAA. These motifs facilitate the formation of homodimers and heterodimers between ARFs and Aux/IAAs. The percentage of CTD-truncated AcARF (13.67%) was similar to that of Arabidopsis (17.39%), but it was lower than other species, such as tomato (28.57%), rice (24%), and sweet orange (21.05%). The AcARFs without CTD suggest that some auxin-responsive genes in kiwifruit are regulated in an auxin independent manner. Also, the cis-element analysis confirmed that most of the ARF gene promoter sequences contain hormonal stress response-related cis-regulatory elements, tissue-specific, and stress response elements (Table 3).
Expression patterns of AcARFs were analysed in different organs using qRT-PCR to study their physiological functions (Fig. 6). Most AcARFs showed higher expression levels in the leaf and stem, which indicated their differential roles during kiwifruit development. AcARF23a and AcARF23b showed preferential expression in small fruit, suggesting their important roles during early fruit development. Also, we observed higher transcription levels of AcARF26b from the small fruit to the ripe fruit stage and higher expression throughout the fruit ripening. This study showed AcARFs have organ-specific expression patterns, and the function of them also needs to be further studied.
Kiwifruits are recurrently exposed to abiotic stresses, such as drought, cold, salinity, defense, and hormonal stimulation during the fruiting and various developmental stages. Previous studies have reported that ARF as transcriptional regulators are directly involved in Auxin signal transposition (Ljung 2013; Guilfoyle and Hagen 2007), and stress response, and hormonal cross-talks (Guilfoyle and Hagen 2007; McAtee et al. 2013). In the current study, we selected 12 AcARF candidate genes to determine their response in stresses induced by five distinct phytohormones along with salt and drought stress. In the present study, we found that all the 12 candidate ARF genes were responsive to exogenous hormone and abiotic stresses in a time-dependent manner. According to the cis-elements, we found that some ARF-induced expression results confirmed the results of promoter element analysis, for example, AcARF1a, 5, 7,10, 28b, induced by exogenous hormones. However, some ARF-induced expression patterns were in line with promoter element analysis; for instance, AcARF4, 28a, lacking IAA promoter response elements were significantly down-regulated by IAA; 18b, 19a, 23a promoters containing ABA elements were not affected by the ABA treatment. The differential expression patterns of 12 AcARF genes indicated that all these genes respond to abiotic stress, which suggests their distinct functions during the kiwi fruit development as far as the stress response is concerned. The current study provides a foundation for an in-depth investigation of ARF functions, specifically the ARF-mediated hormonal cross-talks during the fruit growth, development, and stress responses.
Conclusions
This is the first study of which we are aware to have comprehensively profiled and annotated ARF expression profiles in kiwifruit in response to different stresses. Herein, we were able to identify a total of 41 AcARFs in the kiwifruit genome, which allowed us to explore the functional significance and evolutionary development of this gene family. Notably, the AcARFs expression patterns suggested organ-specific expression patterns and the complex inducible involvements of these genes in abiotic stresses and signaling. These findings present new prospects for a detailed study of the precise functions of AcARFs. It highlights the correlations between the AcARFs expression and stress response; besides, the data could facilitate the screening of ARFs for in-depth functional characterization and genetic improvement of kiwifruit’s agronomic traits.
References
Aloni R, Aloni E, Langhans M, Ullrich CI (2006) Role of auxin in regulating Arabidopsis flower development. Planta 223:315–328. https://doi.org/10.1007/s00425-005-0088-9
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–208. https://doi.org/10.1093/nar/gkp335
Berendzen KW, Weiste C, Wanke D, Kilian J, Harter K, Droge-Laser W (2012) Bioinformatic cis-element analyses performed in Arabidopsis and rice disclose bZIP- and MYB-related binding sites as potential AuxRE-coupling elements in auxin-mediated transcription. Bmc Plant Biol. https://doi.org/10.1186/1471-2229-12-125
Bouzroud S, Gouiaa S, Hu N, Bernadac A, Mila I, Bendaou N, Smouni A, Bouzayen M, Zouine, (2018) Auxin Response Factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS ONE. 13(2):e0193517. https://doi.org/10.1371/journal.pone.0193517
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500
de Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH (2009) The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57:160–170. https://doi.org/10.1111/j.1365-313X.2008.03671.x
de Jong M, Wolters-Arts M, Garcia-Martinez JL, Mariani C, Vriezen WH (2011) The Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7) mediates cross-talk between auxin and gibberellin signalling during tomato fruit set and development. J Exp Bot 62:617–626. https://doi.org/10.1093/jxb/erq293
Esmon CA, Tinsley AG, Ljung K, Sandberg G, Hearne LB, Liscum E (2006) A gradient of auxin and auxin-dependent transcription precedes tropic growth responses. Proc Natl Acad Sci USA 103:236–241. https://doi.org/10.1073/pnas.0507127103
Finn RD, Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–285. https://doi.org/10.1093/nar/gkv1344
Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10:453–460. https://doi.org/10.1016/j.pbi.2007.08.014
Hao Y, Hu G, Breitel D, Liu M, Mila I, Frasse P, Fu Y, Aharoni A, Bouzayen M, Zouine M (2015) Auxin response factor SlARF2 is an essential component of the regulatory mechanism controlling fruit ripening in tomato. PLoS GENET 11:e1005649. https://doi.org/10.1371/journal.pgen.1005649
Hardtke CS, Berleth T (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J 17:1405–1411. https://doi.org/10.1093/emboj/17.5.1405
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587. https://doi.org/10.1093/nar/gkm259
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015a) GSDS 20: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Hu W, Zuo J, Hou X, Yan Y, Wei Y, Liu J, Li M, Xu B, Jin Z (2015b) The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front Plant Sci 6:742. https://doi.org/10.3389/fpls.2015.00742
Huang S, Jian D, Deng D, Tang W, Liu Y (2013) Draft genome of the kiwifruit Actinidia chinensis. Nat Commun 4(4):2640. https://doi.org/10.1038/ncomms3640
Jain M, Khurana JP (2009) Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. Febs J 276:3148–3162. https://doi.org/10.1111/j.1742-4658.2009.07033.x
Ji C, Jiang L (2015) Molecular characterization of ARF family proteins in Arabidopsis. Mol Biol Cell 26:862–875
Jing Z, Liu Z (2018) Genome-wide identification of WRKY transcription factors in kiwifruit (Actinidia spp.) and analysis of WRKY expression in responses to biotic and abiotic stresses. Genes Genomics 40:429–446. https://doi.org/10.1007/s13258-017-0645-1
Kalluri UC, Difazio SP, Brunner AM, Tuskan GA (2007) Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol 7:59. https://doi.org/10.1186/1471-2229-7-59
Kalve S, Sizani BL, Markakis MN, Helsmoortel C, Vandeweyer G, Laukens K, Sommen M, Naulaerts S, Vissenberg K, Prinsen E, Beemster GTS (2020) Osmotic stress inhibits leaf growth of Arabidopsis thaliana by enhancing ARF-mediated auxin responses. New Phytol 226:1766–1780. https://doi.org/10.1111/nph.16490
Kazan K (2013) Auxin and the integration of environmental signals into plant root development. Ann Bot 112:1655–1665. https://doi.org/10.1093/aob/mct229
Keller O, Odronitz F, Stanke M, Kollmar M, Waack S (2008) Scipio: using protein sequences to determine the precise exon/intron structures of genes and their orthologs in closely related species. BMC Bioinform 9:278. https://doi.org/10.1186/1471-2105-9-278
Kumar R, Tyagi AK, Sharma AK (2011) Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol Genet Genomics 285:245–260. https://doi.org/10.1007/s00438-011-0602-7
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. https://doi.org/10.1093/nar/30.1.325
Li SB, OuYang WZ, Hou XJ, Xie LL, Hu CG, Zhang JZ (2015a) Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Front Plant Sci 6:119. https://doi.org/10.3389/fpls.2015.00119
Li Q, Yu H, Cao PB, Fawal N, Mathe C, Azar S, Cassan-Wang H, Myburg AA, Grima-Pettenati J, Marque C (2015b) Explosive tandem and segmental duplications of multigenic families in eucalyptus grandis. Genome Biol Evol (4):1068–1081. https://doi.org/10.1093/gbe/evv048
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Ljung K (2013) Auxin metabolism and homeostasis during plant development. Development 140:943–950. https://doi.org/10.1242/dev.086363
Mattsson J, Ckurshumova W, Berleth T (2003) Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol 131:1327–1339. https://doi.org/10.1104/pp.013623
McAtee P, Karim S, Schaffer R, David K (2013) A dynamic interplay between phytohormones is required for fruit development, maturation, and ripening. Front Plant Sci 4:79. https://doi.org/10.3389/fpls.2013.00079
Mun JH, Yu HJ, Shin JY, Oh M, Hwang HJ, Chung H (2012) Auxin response factor gene family in Brassica rapa: genomic organization, divergence, expression, and evolution. Mol Genet Genomics 287:765–784. https://doi.org/10.1007/s00438-012-0718-4
Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS (2005) Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132:4107–4118. https://doi.org/10.1242/dev.01955
Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17:444–463. https://doi.org/10.1105/tpc.104.028316
Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19:118–130. https://doi.org/10.1105/tpc.106.047761
Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Hong Q (2005) Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17:3282–3300. https://doi.org/10.1105/tpc.105.036723
Pekker I, Alvarez JP, Eshed Y (2005) Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17:2899–2910. https://doi.org/10.1105/tpc.105.034876
Schultz J, Milpetz F, Bork P, Ponting CP (1998) SMART, a simple modular architecture research tool: Identification of signaling domains. Proc Natl Acad Sci USA 95:5857–5864. https://doi.org/10.1186/gb-2000-1-1-reports234
Sessions A, Nemhauser JL, McColl A, Roe JL, Feldmann KA, Zambryski PC (1997) ETTIN patterns the arabidopsis floral meristem and reproductive organs. Development 124:4481–4491
Silva AE, Villanueva WJ, Knidel H, Bonato VC, Reis SF, Von Zuben FJ (2005) A multi-neighbor-joining approach for phylogenetic tree reconstruction and visualization. Genet Mol Res 4:525–534
Singh VK, Rajkumar MS, Garg R, Jain M (2017) Genome-wide identification and co-expression network analysis provide insights into the roles of auxin response factor gene family in chickpea. Sci Rep 7:10895. https://doi.org/10.1038/s41598-017-11327-5
Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L (2008) The arabidopsis information resource (TAIR): gene structure and function annotation. Nucleic Acids Res 36:D1009–D1014. https://doi.org/10.1093/nar/gkm965
Tippmann HF (2004) Analysis for free: comparing programs for sequence analysis. Brief Bioinform 5:82–87
Tiryaki I (2009) Biosynthesis metabolism and signaling pathway of Auxin in plants Philipp. Agric Sci 92:243–253
Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543
Ulmasov T, Hagen G, Guilfoyle TJ (1999) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96:5844–5849
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang D, Pei K, Fu Y, Sun Z, Li S, Liu H, Tang K, Han B, Tao Y (2007) Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394:13–24. https://doi.org/10.1016/j.gene.2007.01.006
Wang YC et al (2018) Auxin regulates anthocyanin biosynthesis through the Aux/IAA-ARF signaling pathway in apple. Hortic Res-Engl. https://doi.org/10.1038/s41438-018-0068-4
Wang SX, Shi FY, Dong XX, Li YX, Zhang ZH, Li H (2019) Genome-wide identification and expression analysis of auxin response factor (ARF) gene family in strawberry (Fragaria vesca). J Integr Agr 18:1587–1603. https://doi.org/10.1016/S2095-3119(19)62556-6
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Tae-ho L, Jin H, Barry M, Guo H (2012) Mcscanx: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40(7):e49–e49. https://doi.org/10.1093/nar/gkr1293
Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112:531–552
Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW (2005) NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43:118–130. https://doi.org/10.1111/j.1365-313X.2005.02432.x
Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot 95:707–735. https://doi.org/10.1093/aob/mci083
Xing H, Pudake RN, Guo G, Xing G, Hu Z, Zhang Y, Sun Q, Ni Z (2011) Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 12:178. https://doi.org/10.1186/1471-2164-12-178
Yu H, Soler M, Mila I, San Clemente H, Savelli B, Dunand C, Paiva JA, Myburg AA, Bouzayen M, Grima-Pettenati J (2014) Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in Eucalyptus grandis. PLoS ONE 9:e108906. https://doi.org/10.1371/journal.pone.0108906
Yuan YJ, Xu X, Gong ZH, Tang YW, Deng W (2019) Auxin response factor 6A regulates photosynthesis, sugar accumulation, and fruit development in tomato. Hortic Res-Engl. https://doi.org/10.1038/s41438-019-0167-x
Zhou P, Fatima M, Ma XY, Liu J, Ming R (2019) Auxin regulation involved in gynoecium morphogenesis of papaya flowers. Hortic Res Engl. https://doi.org/10.1038/s41438-019-0205-8
Acknowledgements
We gratefully thank for funding from Shaanxi Engineering Research Center for Kiwifruit, Shaanxi Aerospace Breeding Engineering Research Center and Key Disciplines of Botany of Xi'an City.
Funding
This research was funded by National Natural Science Foundation of China (31701935), Agricultural technology R & D project of Xi’an City (20NYYF0037), Key Industry Chain Project of Shaanxi Province (2018TSCXL-NY-01–03; 2020ZDLNY03-01; S2021-YF-YBNY-0248) and Natural Science Foundation of Chongqing (cstc2020jcyj-msxmX1064).
Author information
Authors and Affiliations
Contributions
Conceptualization, QL and LS; methodology, MX, YL, JD, YW and LS; software, LS; formal analysis, LS; resources, LS; data curation, LS. and QL; writing—original draft preparation, LS; writing—review and editing, QL; funding acquisition, LS and QL; All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Su, L., Xu, M., Zhang, J. et al. Genome-wide identification of auxin response factor (ARF) family in kiwifruit (Actinidia chinensis) and analysis of their inducible involvements in abiotic stresses. Physiol Mol Biol Plants 27, 1261–1276 (2021). https://doi.org/10.1007/s12298-021-01011-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-021-01011-4