Abstract
Human hepatocellular carcinoma (HCC) is one of the most common malignant cancers, whose molecular mechanisms is remains largely. PTPN9 has recently been reported to play a critical role in breast cancer development. However, the role of PTPN9 in human HCC remains elusive. The present study aimed at investigating the potential role of PTPN9 in HCC. Western blot and immunohistochemistry were used to examine the expression of PTPN9 protein in HCC and adjacent non-tumorous tissues in 45 patients. Furthermore, Cell Counting Kit-8, flow cytometry and RNA interference experiments were performed to analyze the role of PTPN9 in the regulation of HCC cell proliferation. We showed that the expression level of PTPN9 was significantly reduced in HCC, compared with adjacent non-tumorous tissues. PTPN9 expression was inversely associated with Tumor size (P = 0.014), serum AFP level (P = 0.004) and Ki-67 expression. Low expression of PTPN9 predicted poor survival in HCC patients. Moreover, PTPN9 interference assay that PTPN9 inhibited cell proliferation in HepG2 cells. Cell apoptosis assay revealed that, silencing of PTPN9 expression significantly reduced cell apoptosis, compared with control ShRNA treatment group. Our results suggested that PTPN9 expression was down-regulated in HCC tumor tissues, and reduced PTPN9 expression was associated with worsened overall survival in HCC patients. Depletion of PTPN9 inhibits the apoptosis and promotes the proliferation of HCC cells.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common malignancy and the third most deadly cancer type worldwide [1]. Despite the fact that liver transplantation and surgical resection are effective therapeutic strategies for early-stage HCC, the overall survival for patients with advanced HCCs remains poor because of high frequencies of tumor extrahepatic metastasis, invasiveness, frequent intrahepatic spread, and resistance to chemotherapy [4]. Epidemiologic evidence suggests various risk factors, including HBV and HCV infections and metabolic disorders that lead to chronic liver diseases, along with other etiologies such as gender and dietary aflatoxin, contribute heavily to HCC carcinogenesis [2, 3]. However, the molecular mechanisms underlying HCC pathogenesis remain largely unclear. Therefore, there is an urgent need to identify novel molecular targets for early diagnosis and effective treatment of HCC [5].
Protein-tyrosine phosphatases (PTPs) are a class of enzymes that catalyze the dephosphorylation of proteins on phosphotyrosyl residues [6]. Accumulating evidence has pointed out that PTPs play important roles in the transduction of signaling pathways involved in various cellular processes, including cell apoptosis and proliferation [7]. For example, PTPN13/PTPL1 directly binds to and inactivates c-Src to regulated breast cancer aggressiveness [8]. PTP1B regulates cytokine signaling pathways through dephosphorylating STAT6 and JAK2 [9–11]. Other studies demonstrated that SHP2 [12–14] and PTPN2 [15, 16] modulates cytokine signaling pathways by dephosphorylating STAT3 in the nucleus. These studies suggested that PTPs exerted crucial roles in the regulation of distinctive signaling pathways, and consequently contributed to the development of multiple cancers.
PTPN9 (protein tyrosine phosphatase, nonreceptor type 9), also termed PTP-MEG2, is a cytoplasmic protein tyrosine phosphatase that has been reported to play an important role in promoting intracellular secretary vesicle fusion in hematopoietic cells [17]. Studies have demonstrated that PTPN9 controls the expansion of erythroid cells [18] and regulates embryonic development [19]. It has been also found that PTPN9 promoted the dephosphorylation of ErbB2 and EGFR, leading to impaired activation of STAT5 and STAT3 in breast cancer cells [20, 21]. Recently, PTPN9 was reported to directly dephosphorylate STAT3 at the Tyr705 residue, resulting in resultant inactivation of STAT3 [22]. As such, PTPN9 was reportedly dysregulated in various human cancers, including breast cancer, colorectal cancer and gastric cancer. However, the role of PTPN9 in human HCC remains unclear.
In the present study, we investigated the biological and clinical significance of PTPN9 expression in human HCC. We found that PTPN9 expression was significantly reduced in human HCC specimens, and the downregulation of PTPN9 contributed to uncontrolled proliferation of HCC cells. Our findings indicated that PTPN9 might serve as a tumor suppressor in hepatocarcinogenesis.
Materials and Methods
Patients and Tissue Samples
In this study, the paraffin-embedded pathologic specimens from 45 patients with liver cancer were obtained from the Surgery Department, at the Affiliated Hospital of Nantong University. All patients undergoing hepatic surgical resection without postoperative systemic chemotherapy between 2004 and 2005. The cases selected have been examined by experienced pathologists through histological examination of H&E stained biopsy sections. Clinicopathologic characteristics of these patients, including Gender, Age, Histological grade, Metastasis, Vein invasion, Tumor size, No. of tumor nodes, Capsular formation, HBsAg, Cirrhosis, and serum AFP level are presented in Table 1. Their ages range from 21 to 63 years, with an average age of 42 years. The male: to female ratio was 36:9. Histological grades were classified to well differentiated (grade I; n = 8), moderately differentiated (gradeII; n = 35), and poorly differentiated (grade III; n = 2). The follow-up time was 5 years for 45 patients after surgery ranging from 1 to 80 months .
Immunohistochemistry
The TMA slides were dried overnight at 60 °C, deparaffinized through graded alcohol, and quenched endogenous peroxidase activity was blocked by 0.3 % hydrogen peroxide for 30 min, and antigen was recovered by boiling for 3 min in 10 mM sodium citrate buffer (pH 6.0). The slides then were blocked with rabbit normal serum at room temperature for 45 min to reduce nonspecific reaction. Then, the slides were incubated with rabbit polyclonal anti-human PTPN9 antibody (1:50 dilution, Santa Cruz Biotechnology, Santa Cruz, CA) for 2 h at room temperature. The slides were sequentially incubated with secondary antibodies (biotin-labeled; Santa Cruz Biotechnology) and third antibodies (peroxidase-labeled; Santa Cruz Biotechnology) and stained with DAB. Finally, the sections were counterstained with hematoxylin, and dehydrated, and mounted. For the assessment of PTPN9 and Ki-67, 5 high-power fields in each specimen were randomly chosen, and at least 500 cells were counted to determine the Labeling index (LI), which represented the percentage of immunostained cells related to the total number of cells [23]. The scoring of PTPN9 and Ki-67 were carried out both the staining intensity and the percentage of positively stained tumor cells. The percent positivity of PTPN9 was scored as follows: 0 (<10 % tumor cells stained); 1 (10–30 % tumor cells stained); 2 (31–50 % tumor cells stained); 3 (51–70 % tumor cells stained); 4 (>70 % tumor cells stained). The percent positivity of Ki67 was scored as: 0 (<5 % tumor cells stained); 1 (5–25 % tumor cells stained); 2 (26–45 % tumor cells stained); 3 (46–60 % tumor cells stained); 4 (>60 % tumor cells stained). The staining intensity (0 for negative, 1 for weak, 2 for moderate, 3 for strong). The immunostaining score was evaluated using the percentage positive score × the staining intensity score, and the immunostaining score ranged from 0 to 12. For statistical analysis, 0–4 were counted as low expression, while 5–12 were counted as high expression [24].
Western Blot Analysis
Western blot assay was performed as reported previously [25]. Tissues and cell samples were lysed in RIPA lysis buffer (50 mM Tris⋅HCl pH 7.5, 150 mM NaCl, 1 % NP-40, 1 % sodium deoxycholate, 0.1 % SDS, 1 mM EDTA and Complete protease inhibitor cocktail (Roche Diagnostics, Mannheim, Germany)), followed by centrifugation at 11, 000g for 30 min at 4 °C to collect the supernatant. An equivalent amount of total proteins were separated by 10 % SDS-polyacrylamide gel, and transferred to polyvinylidine difluoride filter (PVDF) membranes (Millipore, Bedford,MA, USA). The membranes were blocked with 5 % low-fat milk in TBS-T, followed by incubation with the primary antibodies. Antibodies used in Western blot were as follows: anti-PTPN9 (1:1, 000; Santa Cruz Biotechnology); anti-STAT3 (1:1, 000; Santa Cruz Biotechnology); anti-pSTAT3 (1:1, 000; Santa Cruz Biotechnology); anti-Bcl-xL (1:1, 000; Santa Cruz Biotechnology); anti-cyclin D1 (1:1, 000; Santa Cruz Biotechnology); anti-GAPDH (1:1, 000; Sigma). After washing with TBS-T, horseradish peroxidase (HRP)-conjugated goat anti-rabbit and anti-mouse IgG were used as the secondary antibodies (diluted 1:5000 in TBS-T). The membranes were visualized using an enhanced chemiluminescent (ECL) detection reaction (NEN Life Science Products, Boston, MA, USA).
Cell Culture and Cell-Cycle Analysis
HepG2, SK-Hep-1, Hep3B, Huh7, L02 cells were obtained from Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM medium supplemented with 10 % fetal bovine serum, streptomycin 100 ug/mL, penicillin 100 U/mL in 5 % CO2 astrosphere at 37 °C. Cell-cycle analysis was performed by methods analogous to those reported previously [26].
Cell Proliferation Assay
The HepG2 cells were transfected with PTPN9 ShRNA or NC ShRNA, and seeded on 96-well plates at a density of 1000 cells/well in cluster (Corning Inc., Corning, NY, USA) in 100 uL medium. Cell proliferation was measured with the Cell Counting Kit-8 (CCK-8) assay kit (Dojindo, Kumamoto, Japan) in accordance with the manufacturer’s instrument. Briefly, 10 μL CCK-8 was added into each well and the cells were incubated with CCK-8 reagent for 1 h at 37 °C. The relative optical density (OD)/well was determined at a test wavelength of 490 nm and a reference wavelength of 630 nm on an automated plate reader (BioTek Instruments, Winooski, VT).
Plasmid Constructs ShRNA and Transfection
The RNAi species for the PTPN9 knockdown were synthesized by Genechem (Shanghai, China). The target sequences of PTPN9 ShRNAs were as follows: ShRNA1: 5′-ATACTCAGACAGATTACAT-3′; ShRNA2: 5′-TATCAAACTGTGCCTTATT-3′; ShRNA3: 5′-AGGCGTGGAGAACATGAAT-3′; ShRNA4: 5′-AAGGAAGGAAGGCATTGTA-3′; The sequence of control ShRNA is: 5′-UUCUCCGAACGUGUCACGU-3′. Cell transfection was performed with Super-Fectin (Qiagen, Valencia, CA) according to the manufacturer’s instruction.
Annexin-V/PI Apoptotic Assay
48 h after transfection with PTPN9 or NC ShRNA, the cells were collected up and washed in ice-cold PBS. Thereafter, the cells were resuspended in 100 uL of 1 × binding buffer, and incubated with Annexin V-FITC (Bestbio, China) at room temperature for 15 min in the dark. Subsequently, the cells were incubated with propidium iodide and 1 μg/ml RNase A at room temperature for 5 min in the dark. Then, the samples were analyzed using a flow cytometer (Beckman, USA).
Statistical Analysis
Statistical analysis was carried out using the SPSS 16.0 (standard version 16.0; SPSS, Chicago, IL). The correlations among Ki-67and PTPN9 expression and patients clinicopathologic features were evaluated using the x 2 test. The correlation between Ki-67 and PTPN9 expression were studied using the Spearman rank correlation test. A Cox proportional-hazards analysis was used in univariate and multivariate analyses to explore the effects of PTPN9 expression and HCC clinicopathological factors on survival. Differences were considered to be statistically significant if the P value from a 2-tailed test was <0.05.
Results
PTPN9 was Downregulated in HCC Specimens
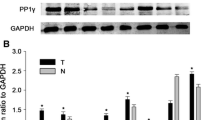
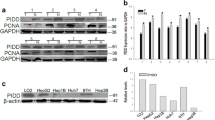
The expression profile of PTPN9 in eight paired adjacent non-tumorous tissues and HCC biopsy samples was first determined. PTPN9 was significantly downregulated in tumor samples, compared with the adjacent non-tumorous tissues (Fig. 1a, b). We next examined the expression of PTPN9 protein in a panel of 4 HCC cell lines and L02 normal liver cells. In this way, we found that PTPN9 protein was highly expressed in L02 cells, compared with HCC cell lines (Fig. 1c, d). These findings suggested that the expression of PTPN9 was decreased in HCC specimens and cell lines.
PTPN9 is downregulated in HCC specimens, compared with paired adjacent non-tumorous tissues. a Western blot analysis of 8 representative paired samples of HCC tissues (T) and adjacent non-tumorous tissues (N) using an anti-PTPN9 antibody. GAPDH was used as a control for protein load and integrity. b The bar chart demonstrates the ratio of PTPN9 protein to GAPDH by densitometry. Results are presented as mean ± SD from three independent experiments (*P < 0.05). c The PTPN9 protein expression was lowly expressed in the human hepatocellular carcinoma cell lines, compared with the non-tumorous liver cell line L02. d The bar chart indicates the relative expression of PTPN9 to GAPDH in the cell lines
Immunohistochemical Analysis of PTPN9 Expression in HCC Clinical Samples and Its Relationship to Clinicopathological Parameters
Next, we investigated the correlation between PTPN9 expression and clinicopathological parameters in HCC patients. Immunohistochemical analysis of PTPN9 expression was carried out on a tissue microarray containing 45 HCC samples. PTPN9 and Ki-67 expression in HCC was scored as low expression (<5), and high expression (≥5) (Fig. 2). PTPN9 expression in HCC was scored as positive when the immunoreactivity was found in the cytoplasm. The percentage of PTPN9 positive cells ranged from 6.39 to 80.39 % in HCC samples. The mean percentage of positive cells was 43.39 %. Ki-67 expression in HCC was scored as positive when displaying strong nuclear staining. The percentage of Ki-67 positive cells ranged from 2.29 to 62. 29 %. The mean percentage of Ki-67 positive cells was 32.29 %.
Immunohistochemical analysis of PTPN9 and Ki-67 expression. a The representative image of HCC specimen with low PTPN9 expression. b The representative image of HCC specimen with high PTPN9 expression. c The image showed HCC specimen that exhibited low Ki-67 expression. d The image showed HCC specimen that exhibited high Ki-67 expression (SP × 200)
The relationship between PTPN9 expression and clinicopathologic parameters was further analyzed. The clinicopathologic data of these HCC cases were summarized in Table 1. We evaluated the association between PTPN9 expression and clinicopathological variables. PTPN9 expression was significantly correlated with Tumor size (P = 0.014), and serum AFP level (P = 0.004). However, there was no statistical association between PTPN9 expression and the remaining clinicopathologic parameters, such as Age, Histological grade, Vein invasion, No. of tumor nodes, Capsular formation, HBsAg, Cirrhosis, Metastasis, and Gender (P > 0.05). Furthermore, in most specimens, it was found that tumors with high expression of PTPN9 exhibited low levels of Ki-67. A negative correlation between PTPN9 and Ki-67 expression was identified (P < 0.01; Fig. 3).
Low PTPN9 Expression was Associated with Damal Prognosis in HCC Patients
The prognostic value of PTPN9 was investigated by comparing patients with high PTPN9 expression and those with low PTPN9 expression. According to the Kaplan–Meier survival analysis, patients with low PTPN9 expression were likely to have a significantly worsened overall prognosis. HCC patients with low level of PTPN9 predicted significantly shortened overall survival compared with those with high PTPN9 expression (Fig. 4, P < 0.05).
Next, we performed Cox proportional-hazards analysis to further clarify the prognostic significance of PTPN9 expression and other clinical pathological parameters in HCC patients. Using univariate analysis, it was found that PTPN9 and Ki-67 expression exhibited prognostic merits (Table 2). The prognostic merit of PTPN9 and Ki-67 expression were further examined using multivariate analysis. Results showed that PTPN9 was an independent predictor. Thus, PTPN9 expression would be closely correlated with the overall survival (P < 0.05, Table 2).
PTPN9 was Lowly Expressed in Proliferating HCC Cells
Based on the finding that PTPN9 was associated with tumor size and Ki-67 expression in HCC, we speculated that PTPN9 might be involved in the regulation of HCC cell growth. As such, we detected the expression of PTPN9 during cell cycle progression in HCC cells. HepG2 cells were arrested in G1 phase by serum deprivation for 48 h, released from G1 phase and reentered into S phase through serum refeeding (Fig. 5a). As predicted, the expression of cyclin D1, a marker protein of cell proliferation, was also up-regulated (Fig. 4b). Moreover, western blot analysis showed that the expression of PTPN9 was down- regulated after serum refeeding. These results indicated that PTPN9 was down-regulated during HCC cell cycle progression.
The expression of PTPN9 and cell cycle-related molecules in proliferating HCC cells. a Flow cytometry quantitation of cell cycle progress in HepG2 cells. Cells were arrested in G1 phase by serum deprivation for 48 h, then entered into the cell cycle following serum stimulation for 0, 4, 8, 12, or 24 h. Following serum refeeding, a significant proportion of cells entered into S phase. b Cells were serum-starved for 48 h in HepG2, and then subjected to serum refeeding for the indicated period of time. Cell lysates were prepared and subjected to Western blot analysis. GAPDH was used as a control for protein load and integrity. c The bar chart demonstrates the ratio of PTPN9, cyclin D1, protein to GAPDH for each time point by densitometry. Results are presented as mean ± SD from three independent experiments (#, * P < 0.05, compared with control: 0 h). R serum relea
PTPN9 Inhibits Cell Proliferation in HCC Cell Lines
To further investigate the potential involvement of PTPN9 in HCC cell proliferation, ShRNA were employed to knockdown PTPN9 expression in HepG2 cells. HepG2 cells were transiently transfected with control ShRNA or four different PTPN9-ShRNA. 48 h after transfection, the knockdown efficiencies of the ShRNA were determined using western blot analysis. PTPN9 ShRNA4 exhibited the best knockdown efficiency among the four different PTPN9 ShRNA (Fig. 6a). Therefore, PTPN9 ShRNA4 was used for subsequent experiments. To assess the impact of PTPN9 depletion on cell proliferation, PTPN9-ShRNA4 and control ShRNA were transfected into HepG2 cells. Westernblotgand CCK-8 assays were conducted to analyze the influence of PTPN9 interference on HCC cell proliferation. CCK-8 assay showed that interference of PTPN9 resulted in significantly increased cell proliferation in HepG2 cells, compared with control ShRNA (Fig. 6b). In addition, it was found that PTPN9 knockdown also led to up-regulated level of p-STAT3, cyclin D1 and Bcl-xL in HepG2 cells (Fig. 6c). To determine whether the cell-cycle distribution was following after depletion of PTPN9 in HCC cells, flow cytometrical analysis was performed and found that HepG2 cells showed an apparently elevated population of cells in S phase, whereas cells in G1 phase was markedly reduced after the transfection of PTPN9 ShRNA (Fig. 6d). These results highlighted the importance of PTPN9 in regulating HCC cell proliferation.
Interference of PTPN9 promoted the proliferation of HCC cells. a 48 h after transiently transfected with PTPN9-ShRNAs or control ShRNA, the interference efficiencies of PTPN9 ShRNA were examined using Western blot analysis. The bar chart indicates the ratio of PTPN9 protein to GAPDH by densitometry. b Cell proliferation was measured using CCK-8 assay. HepG2 cells that have been transfected with PTPN9 ShRNA4 exhibited enhanced cell proliferation. c Western blot analysis of STAT3, p-STAT3, cyclin D1, Bcl-xL and in control and PTPN9-depleted HepG2 cells. d Depletion of PTPN9 resulted in decreased cell population in G1 phase and increased proportion of cells in S phase, in comparison with cells transfected with control ShRNA
Suppression of PTPN9 Inhibits Apoptosis via Dephosphorylation of STAT3
Finally, we analyze the role of PTPN9 in the regulation of apoptotic cell death in HCC cells. HepG2 cells were transiently transfected with control ShRNA or PTPN9 ShRNA4. 48 h after transfection, annexin-V/PI apoptotic assay revealed that, depletion of PTPN9 significantly reduced spontaneous apoptosis of HepG2 cells, compared with control ShRNA treated group (Fig. 7a). These results suggested that depletion of PTPN9 decreased the apoptosis of HCC cells. Moreover, western blot analysis was performed to examine the potential involvement of PTPN9 inhibition on signaling pathways associated with apoptosis. As shown in Fig. 7b, depletion of PTPN9 expression led to up-regulation of p-STAT3 (Tyr705). These results suggested that the regulatory role of PTPN9 in HCC cell survival viability might involve the dephosphorylation of STAT3.
Depletion of PTPN9 inhibits the apoptosis of HepG2 cells. a HepG2 cells were transfected with PTPN9 ShRNA4 or control ShRNA and cultured in a humidified incubator. 48 h after transfection, the cells were subjected to annexin-V/PI apoptotic assay. b Cells were lysed and analyzed using Western blot assay to determine the level of phosporylated STAT3 (p-STAT3). GAPDH was used as a control for protein load and integrity (*P < 0.05)
Discussion
Despite the fact that major pathological factors leading to HCC have been clarified, the molecular mechanisms underlying hepatocarcinogenesis remains largely obscure. In the current study, we showed that PTPN9 was remarkably downregulated in HCC specimens. Additionally, we found that PTPN9 expression was inversely correlated with tumor size and Ki-67 expression in HCC tissues. Moreover, univariate and multivariate survival analyses demonstrated that PTPN9 expression could be an independent prognostic factor to predict the survival of HCC patients. Using HCC cells, we identified that PTPN9 participated in the regulation of HCC proliferation, which involves the activity of STAT3 transcription factor. These findings inferred that PTPN9 downregulation might be a key determinant in the pathogenesis of HCC.
Increasing evidence indicated that Protein-tyrosine phosphatases (PTPs) played important roles in human tumorigenesis. Studies have shown that several PTPs, such as PTPN6, had oncogenic properties, however several other PTPs, including PTPN9 had tumor suppressive functions [27, 28]. PTPN9 directly mediates the dephosphorylation of p-STAT in breast cancer, and negatively regulates STAT3 activity [22]. However, the role of PTPN9 in human HCC development has not yet been documented. In this study, we detected PTPN9 protein expression in HCC tissue specimens and HCC cell lines using Western blotting and immunohistochemical analyses. We found that PTPN9 was decreased in most primary HCC tumor tissues. We also found that low expression of PTPN9 protein was significantly associated with poor patient survival, compared to the patients with high PTPN9 expression. In addition, multivariate analysis suggested that PTPN9 protein was an independent prognostic factor for overall survival. These findings implied that PTPN9 could be a valuable predictor of the prognosis of patients with HCC.
HCC progression involves complex dynamics in various cellular signaling pathways, including both oncogenic and tumor-suppressive pathways [29]. Although the molecular mechanisms underlying the initiation of many oncogenic pathways have been intensively investigated, the regulatory role of signaling pathway termination in cancer development attracted less attention. STAT3 transcription factor has been critically implicated in HCC development [30]. Persistent STAT3 activation has been frequently observed in HCC tissues, while deletion of STAT3 led to the attenuation of HCC progression [31]. STAT3 mainly regulates the proliferation of HCC cells via the transcription of various cell cycle regulators, such as Cyclin D1 [32] and c-Myc [33]. The activation of STAT3 works through tyrosine phosphorylation by upstream Janus Kinases, including JAK2 and JAK3 [34]. Serving as a protein-tyrosine phosphatase, PTPN9 dephosphorylates and inactivates STAT3 through direct dephosphorylation of STAT3 at residue Try705 [22]. However, whether PTPN9 played a role in the regulation of STAT3 dephosphorylation during HCC progression remain unclear. In the current study, we showed that interference of PTPN9 was associated with hyperactivation of STAT3, which consequently led to enhanced proliferation of HCC cells. Therefore, we speculated that the downregulation of PTPN9 in HCC specimens might contribute to persistent activation of STAT3 transcription factor, and resultant HCC progression. Of great intrigue, we found that the level of PTPN9 was apparently downregulated in proliferating HCC cells, whose mechanism remains unknown. Because the regulation of PTPN9 expression in cancer cells has been little investigated, the mechanism by which PTPN9 expression was decreased in serum-refed HCC cells remains to be elucidated. Some studies have shown that PTPN9 was regulated by miRNAs in cancer cells. WW Du et al. reported that miR-24 facilitated breast cancer development through targeting PTPN9 and PTPRF [35]. PTPN9 may also be potentially regulated by miR-126 [36]. In addition, studies have demonstrated that PTPN9 overexpression reduces invasion in the breast cancer [21]. Therefore, whether the PTPN9 overexpression reduces invasion in HCC will be an interesting question in future studies.
In conclusion, we demonstrated that the downregulation of PTPN9 was associated with enhanced proliferation and poor prognosis in human HCC. Our study suggested that PTPN9 is a tumor suppressor in HCC carcinogenesis, especially in tumor growth and progression. Our results implicated that restoring PTPN9 expression may exert beneficial effects in the treatment of human HCC.
References
Wenzh El-Serag HB (2011) Hepatocellular carcinoma. N Engl J Med 365:1118–1127
El-Serag HB, Rudolph KL (2007) Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132:2557–2576
But D-Y-K, Lai C-L, Yuen M-F (2008) Natural history of hepatitis-related hepatocellular carcinoma. World J Gastroenterol 14:1652–1656
Poon RT, Ng IO, Fan ST, Lai EC, Lo CM, Liu CL et al (2001) Clinicopathologic features of long-term survivors and disease-free survivors after resection of hepatocellular carcinoma: a study of a prospective cohort. J Clin Oncol 19:3037–3044
Frau M, Biasi F, Feo F et al (2010) Prognostic markers and putative therapeutic targets for hepatocellular carcinoma. Mol Asp Med 31:179–193
Tonks NK (2006) Nat Rev Mol Cell Biol 7:833–846
den Hertog J, Ostman A, Bohmer FD (2008) Protein tyrosine phosphatases:regulatory mechanisms. FEBS J 275:831–847
Glondu-Lassis M, Dromard M, Lacroix-Triki M, Nirde P, Puech C, Knani D, Chalbos D, Freiss G (2010) PTPL1/PTPN13 regulates breast cancer cell aggressiveness through direct inactivation of Src kinase. Cancer Res 70:5116–5126
Aoki N, Matsuda T (2002) A nuclear protein tyrosine phosphatase TC-PTP is a potential negative regulator of the PRL-mediated signaling pathway: dephosphorylation and deactivation of signal transducer and activator of transcription 5a and 5b by TC-PTP in nucleus. Mol Endocrinol 16:58–69
Johnson KJ, Peck AR, Liu C, Tran TH, Utama FE, Sjolund AB, Schaber JD, Witkiewicz AK, Rui H (2010) PTP1B suppresses prolactin activation of Stat5 in breast cancer cells. Am J Pathol
Lu X, Malumbres R, Shields B, Jiang X, Sarosiek KA, Natkunam Y, Tiganis T, Lossos IS (2008) PTP1B is a negative regulator of interleukin 4-induced STAT6 signaling. Blood 112:4098–4108
Tebbutt NC, Giraud AS, Inglese M, Jenkins B, Waring P, Clay FJ, Malki S, Alderman BM, Grail D, Hollande F, Heath JK, Ernst M (2002) Reciprocal regulation of gastrointestinal homeostasis by SHP2 and STAT-mediated trefoil gene activation in gp130 mutant mice. Nat Med 8:1089–1097
Baron M, Davignon JL (2008) Inhibition of IFN-gamma-induced STAT1 tyrosine phosphorylation by human CMV is mediated by SHP2. J Immunol 181:5530–5536
Tsai CC, Kai JI, Huang WC, Wang CY, Wang Y, Chen CL, Fang YT, Lin YS, Anderson R, Chen SH, Tsao CW, Lin CF (2009) Glycogen synthase kinase-3beta facilitates IFN-gamma-induced STAT1 activation by regulating Src homology-2 domain-containing phosphatase 2. J Immunol 183:856–864
ten Hoeve J, de Jesus I-SM, Fu Y, Zhu W, Tremblay M, David M, Shuai K (2002) Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol 22:5662–5668
Meyer T, Hendry L, Begitt A, John S, Vinkemeier U (2004) A single residue modulates tyrosine dephosphorylation, oligomerization, and nuclear accumulation of stat transcription factors. J Biol Chem 279:18998–19007
Huynh H, Bottini N, Williams S, Cherepanov V, Musumeci L, Saito K, Bruckner S, Vachon E, Wang X, Kruger J, Chow CW, Pellecchia M, Monosov E, Greer PA, Trimble W, Downey GP, Mustelin T (2004) Nat Cell Biol 6:831–839
Xu MJ, Sui X, Zhao R, Dai C, Krantz SB, Zhao ZJ (2003) Blood 102:4354–4360
Wang Y, Vachon E, Zhang J, Cherepanov V, Kruger J, Li J, Saito K, Shannon P, Bottini N, Huynh H, Ni H, Yang H, McKerlie C, Quaggin S, Zhao ZJ, Marsden PA, Mustelin T, Siminovitch KA, Downey GP (2005) J Exp Med 202:1587–1597
Furth PA, Nakles RE, Millman S, Diaz-Cruz ES, Cabrera MC (2011) Signal transducer and activator of transcription 5 as a key signaling pathway in normal mammary gland developmental biology and breast cancer. Breast Cancer Res 13:220
Yuan T, Wang Y, Zhao ZJ, Gu H (2010) Protein-tyrosine phosphatase PTPN9 negatively regulates ErbB2 and epidermal growth factor receptor signaling in breast cancer cells. J Biol Chem 285:14861–14870
Su F, Ren F et al (2012) Protein tyrosine phosphatase Meg2 dephosphorylates signal transducer and activator of transcription 3 and suppresses tumor growth in breast cancer. Breast Cancer Res. doi:10.1186/bcr3134
Xu X, Yamamoto H, Sakon M et al (2003) Overexpression of CDC25A phosphatase is associated with hypergrowth activity and poor prognosis of human hepatocellular carcinomas. Clin Cancer Res 9:1764–1772
Yu C, Chen K, Zheng H et al (2009) Overexpression of astrocyte elevated gene-1 (AEG-1) is associated with esophageal squamous cell carcinoma (ESCC) progression and pathogenesis. Carcinogenesis 30:894–901
Xue Q, Lv L, Wan C, Chen B, Li M, Ni T et al (2013) Expression and clinical role of small glutamine-rich tetratricopeptide repeat (TPR)-containing protein alpha (SGTA) as a novel cell cycle protein in NSCLC. J Cancer Res Clin Oncol 139(9):1539–1549. doi:10.1007/s00432-013-1474-5
Wan C, Hou S, Shen A et al (2015) MIF4G domain containing protein regulates cell cycle and hepatic carcinogenesis by antagonizing CDK2-dependent p27 stability. Oncogene 34(2):237–245. doi:10.1038/onc.2013.536
Sun T, Aceto N, Meerbrey KL, Kessler JD, Zhou C, Migliaccio I, Nguyen DX, Pavlova NN, Botero M, Huang J, Bernardi RJ, Schmitt E, Hu G, Li MZ, Dephoure N, Gygi SP, Rao M, Creighton CJ, Hilsenbeck SG, Shaw CA, Muzny D, Gibbs RA, Wheeler DA, Osborne CK, Schiff R, Bentires-Alj M, Elledge SJ, Westbrook TF (2011) Activation of multiple proto-oncogenic tyrosinekinases in breast cancer via loss of the PTPN12 phosphatase. Cell 144:703–718
Julien SG, Dube N, Hardy S, Tremblay ML (2010) Inside the human cancer tyrosine phosphatome. Nat Rev Cancer 11:35–49
Hu TH, Huang CC, Lin PR et al (2003) Expression and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular carcinoma. Cancer 97:1929–1940
Sánchez A, Nagy P, Thorgeirsson SS (2003) STAT-3 activity in chemically-induced hepatocellular carcinoma. Eur J Cancer 39:2093–2098
Fuke H, Shiraki K, Sugimoto K, Tanaka J, Beppu T, Yoneda K, Yamamoto N, Ito K, Masuya M, Takei Y (2007) Jak inhibitor induces S phase cell-cycle arrest and augments TRAIL-induced apoptosis in human hepatocellular carcinoma cells. Biochem Biophys Res Commun 363:738–744
Leslie K, Lang C, Devgan G, Azare J, Berishaj M, Gerald W, Kim YB, Paz K, Darnell JE, Albanese C, Sakamaki T, Pestell R, Bromberg J (2006) Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. Cancer Res 66:2544–2552
Bowman T, Broome MA, Sinibaldi D, Wharton W, Pledger WJ, Sedivy JM, Irby R, Yeatman T, Courtneidge SA, Jove R (2001) Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci U S A 98:7319–7324
Yue P, Turkson J (2009) Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs 18:45–56
Du WW, Fang L, Yang BB et al (2013) MicroRNA miR-24 enhances tumor invasion and metastasis by targeting PTPN9 and PTPRF to promote EGF signaling. J Cell Sci 126(6):1440–1453. doi:10.1242/jcs.118299
Huang X, Gschweng E, Witte ON et al (2011) Regulated expression of microRNAs-126/126* inhibits erythropoiesis from human embryonic stem cells. Blood 117(7):2157–2165. doi:10.1182/blood-2010-08-302711
Acknowledgments
This work was supported by the National Natural Scientific Foundation of China (no. 81401985)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
We investigated 45 cases of liver cancer provided by Surgery Department of Affiliated Hospital of Nantong University. All liver cancer tissues were collected using protocols approved by Ethics Committee of Affiliated Hospital of Nantong University, and written informed consent was obtained from every patient.
Conflict of Interest
All authors declare no conflicts of interest
Additional information
Baoying Hu and Xia Yan contributed equally to this work.
Rights and permissions
About this article
Cite this article
Hu, B., Yan, X., Liu, F. et al. Downregulated Expression of PTPN9 Contributes to Human Hepatocellular Carcinoma Growth and Progression. Pathol. Oncol. Res. 22, 555–565 (2016). https://doi.org/10.1007/s12253-015-0038-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-015-0038-1