Abstract
p53-induced death domain protein (PIDD) facilitates p53-dependent apoptosis through the interaction with components of the death receptor signaling pathways. However, the role of PIDD in hepatocellular carcinoma (HCC) development remains unknown. In this study, we investigated the expression pattern of PIDD in clinical HCC samples and adjacent non-cancerous tissues using immunohistochemistrical and Western blot analyses. The results showed that PIDD was lowly expressed in HCC tissues and HCC cell lines, compared with the adjacent non-tumorous tissues and LO2 normal hepatocytes. In addition, clinicopathological analysis showed that the expression of PIDD was closely related with multiple clinicopathological variables, such as American Joint Committee on Cancer (AJCC) stage, AFP, and poor prognosis of HCC. Univariate and multivariate survival analyses demonstrated that PIDD could serve as an independent prognostic factor to predict the survival of HCC patients. We used serum starvation-refeeding experiment to explore the involvement of PIDD in HCC cell cycle regulation. We found that PIDD was accumulated in growth-arrested HCC cells and was progressively decreased when cells entered into S phase. Moreover, flow cytometry and cell counting kit-8 (CCK-8) assays indicated that depleting the expression of PIDD could facilitate cell cycle progression and accelerate cell proliferation in HepG2 cells, while overexpression of PIDD could result in cell cycle arrest at G1 phase and hinder the cell proliferation in Hep3B cells. Finally, flow cytometry revealed that overexpression of PIDD slightly increased the apoptosis of HCC cells. Taken together, we concluded that PIDD may be a valuable prognostic marker and promising therapeutic target of HCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is a major health concern and the third cause of cancer-related death worldwide, especially in eastern Asia [1]. The number of Chinese patients with HCC accounts for more than 53 % of the total HCC patients in the world [2]. Approximately 90 % HCC cases are attributed to several major risk factors, such as chronic hepatitis B, chronic hepatitis C, alcohol abuse, nonalcoholic steatohepatitis (NASH), and aflatoxin exposure [3]. Despite the fact that considerable effort has been made by both basic scientists and clinical practitioners, the underlying mechanisms of the pathogenesis of HCC remain poorly understood [4]. Therefore, it is significant to identify novel molecules that take part in the regulation of the development of HCC.
Many signaling pathways have been documented to be involved in the occurrence of liver cancer, such as MAPK signaling, NF-κB signaling, and p53 signaling pathways [5]. p53-induced death domain protein (PIDD) was initially identified as a protein involved in cell apoptosis in a p53-dependent manner [6]. Full-length PIDD (PIDD-FL, 910 aa) contains an N-terminal leucine-rich repeats (LRR), two intermediate ZU5 domains, and a C-terminal death domain (DD). PIDD is constitutively auto-processed, giving rise to a 48-kDa N-terminal fragment (PIDD-N) and a 51-kDa C-terminal fragment (PIDD-C). The latter undergoes further cleavage resulting in a 37-kDa fragment (PIDD-CC) [7]. Furthermore, PIDD has four alternative splicing isoforms in compose, PIDD1-4 [8]. PIDD1-3 all contain DD structure domain but PIDD4 without the LRR domain [8, 9]. Serving as a p53-responsive protein, PIDD1 plays a key role in the initiation of p53-dependent apoptosis [10]. PIDD1 functions as a molecular switch to control the translation of the cellular survival pathway and the apoptosis pathway under stress conditions, such as DNA damage and oxidative stress [11]. PIDD2 can prevent PIDD1 from inducing cells apoptosis through direct interaction with FADD and MADD, which are also death domain-containing proteins. PIDD3 exerts a pro-apoptotic role when co-expressed with PIDD1. All of the three isoforms can negatively regulate cell apoptosis via the activation of NF-κB pathway [9]. However, PIDD4 can induce cell apoptosis independently [8]. Recently, several studies have showed that PIDD could induce cell apoptosis through the modulation of multiple pathways, such as p53 signaling pathway [10], NF-κB signaling pathway [12], and ATM/ATR-caspase-2 pathway [13]. In addition, mounting studies have showed that PIDD is involved in the pathology of various diseases, such as colon cancer [14], renal cell carcinomas (RCCs) [15], renal tubular epithelial cell injury (RTEC) [16], oral squamous cell carcinomas (OSCC) [17], and lung cancer [18]. Although PIDD is highly expressed in murine liver [19], the detail role of PIDD in HCC development remains unclear. Thus, it is important to explore the precise role of PIDD in HCC development.
In this study, we aim to conduct a comprehensive analysis of PIDD as a prognostic marker in HCC and determine the physiological relevance of PIDD in HCC development. We found that PIDD was significantly downregulated in HCC tissues, compared with adjacent non-tumorous ones. In addition, Cox regression analysis demonstrated that PIDD could be an independent prognostic factor for the survival of HCC patients. Flow cytometry and cell counting kit-8 (CCK-8) assays indicated that PIDD could regulate cell cycle progression and the proliferation of HCC cells. Finally, we found that overexpression of PIDD could promote cell apoptosis in Hep3B cells. On the basis of these findings, we speculated that PIDD could be a novel prognostic predictor and therapeutic target of HCC.
Materials and methods
Patients and tissue samples
Tissue samples were gained from 99 HCC patients who underwent hepatic curative resection at the Surgery Department, the Affiliated Hospital of Nantong University from 2006 to 2012. The study group consisted of 80 men and 19 women with median age 48 years (21–65 years), and none of them was treated with preoperative systemic chemotherapy. Eight pairs of tissue samples were randomly selected from 99 subjects for using Western blot assays. All HCC specimens were collected in accordance with the protocols approved by the Ethics Committee of Affiliated Hospital of Nantong University, and every patient submitted written informed consent. Histological grades were classified to well (grades I–II; n = 65) and poor (grades III–IV; n = 34) differentiated according to American Joint Committee on Cancer (AJCC) stage. The follow-up time for 99 patients ranged from 1 to 60 months from the date of surgery. The main clinicopathological variables of the patients are shown in Table 1. All specimens were acquired in the operating theater immediately (≤15 min) after tumor resection, snap-frozen in liquid nitrogen, and stored at −80 °C.
Antibodies
The antibodies used for immunohistochemistry and Western blotting were as follows: goat polyclonal anti-PIDD antibody (1:100; Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal anti-Ki-67 antibody (1:100; clone 7B11; Zymed Laboratories, San Francisco, CA, USA), anti-proliferating cell nuclear antigen (PCNA; 1:1000; Santa Cruz Biotechnology), rabbit polyclonal anti-cyclin D1 antibody (1:500, Santa Cruz Biotechnology), rabbit polyclonal anti-cyclin A antibody (1:500, Santa Cruz Biotechnology), rabbit monoclonal anti-GAPDH antibody (glyceraldehyde-3-phosphate dehydrogenase) (1:1000; Santa Cruz Biotechnology), mouse monoclonal anti-β-actin antibody (1:1000, Santa Cruz Biotechnology), and mouse monoclonal anti-caspase-2 antibody (1:1000, Cell Signaling, Danvers, MA, USA). Secondary antibody incubation was performed using horseradish peroxidase-linked IgG (Pierce Biotechnology, Rockford, IL, USA) at a dilution of 1:5000.
Immunohistochemistry and immunohistochemistrical evaluation
In brief, sections were dewaxed twice in xylene for 20 min and rehydrated in graded ethanol. Then, the slides were boiled in 10 mmol/L citrate buffer (pH 6.0) in an autoclave for 3 min to retrieve the antigen. After cooled, sections were blocked by immersion in 3 % methanolic peroxide for 15 min and incubated with primary antibodies: anti-PIDD or anti-Ki-67 antibody overnight at 4 °C, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies. After rinsing in water, slides were counterstained with hematoxylin, dehydrated, and mounted in resin mount.
Three pathologists independently detected all of the sections in a blinded manner without knowledge of the clinical and pathological variables of the patients. For evaluating the expression of PIDD and Ki-67, we randomly selected at least five high-power fields in each specimen and examined the cytoplasm or nuclear staining under a high magnification. Besides, we examined more than 500 cells to determine the mean percentage of signal-positive cells. To determine PIDD expression, the intensity of immunostaining was assessed as 0 (negatively or poorly staining), 1 (moderately staining), and 2 (strongly staining). We divided patients into three groups: low-expression group (<50 %) to be scored 1, moderate-expression group (50–75 %) to be scored 2, and high-expression group (>75 %) to be scored 3 according to PIDD expression ratio (50 and 75 %). Then, according to the average scores (4.2), we multiplied the two scores and divided patients into two groups: high-expression group (>4.2) and low-expression group (≤4.2). In addition, the expression of proliferation marker Ki-67 was scored in a semiquantitative fashion: high expression (≥50 %) and low expression (<50 %).
Cell culture and cell cycle analysis
HCC cell lines (Huh7, HepG2, Hep1B, Hep3B, and MHCC-97H) and LO2 normal hepatocytes were obtained from the Shanghai Institute of Cell Biology, Academic Sinica, and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma, St. Louis, MO, USA) supplemented with 10 % fetal bovine serum (HyClone, South America), 100 U/ml penicillin–streptomycin mixture at 37 °C, and 5 % CO2.
Flow cytometric analysis was performed in accordance with previous reports by Wan et al. [20]. In brief, cells were fixed with 70 % methanol for 24 h at −20 °C. Then, the cells were washed for three times with 1 ml phosphate buffered saline (PBS), and incubated with 1 mg/ml RNase A for 30 min at 37 °C. Subsequently, cells were incubated with 50 μg/ml propidium iodide (PI; Becton Dickinson, San Jose, CA, USA) for staining in 0.5 % Tween-20 in PBS. Finally, we used a BD FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA) coupled with Cell Quest acquisition and analysis programs to analyze the distribution of cell cycle.
Western blot analysis
Briefly, tissues and cell samples were immediately homogenized in a lysis buffer containing 1 M Tris–HCl (pH 7.5), 1 % Triton X-100, 1 % NP-40 (Nonidet P-40), 10 % sodium dodecyl sulfate (SDS), 0.5 % sodium deoxycholate, 0.5 M EDTA, 10 μg/mL leupeptin, 10 μg/mL aprotinin, and 1 mM PMSF and then centrifuged at 13,000×g, 4 °C for 20 min to collect the supernatant. Protein concentrations were determined using a BCA protein assay kit (Bio-Rad, Hercules, CA, USA). Subsequently, the supernatant was diluted in 2× SDS loading buffer and boiled for 15 min. The protein samples were separated with 10 % SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and transferred to polyvinylidene difluoride filter (PVDF) membranes (Millipore, Bedford, MA, USA). Next, the membranes were blocked with 5 % no-fat milk in TBST (150 mM NaCl, 20 mM Tris and 0.05 % Tween-20) for 2 h and then incubated with primary antibodies overnight at 4 °C. Thereafter, the membranes were incubated with the secondary antibody horseradish peroxidase-linked IgG for 2 h. The detection of chemiluminescent signals was performed by ECL method (ZhongShan Biotech Company, China).
Cell transfection
Small interference RNAs (siRNA) were purchased from Genechem (Shanghai). Expression of human PIDD was knocked down with siRNA duplexes targeting the sequence: 5′-GCTTTGTCTTCTACTCGCA-3′, 5′-CCTCAGATTTGGACAGCTT-3′, and 5′-CTGTTCCTGACCTCAGATT-3′. The pCMV-HA-PIDD plasmid was purchased from Public Protein/Plasmid Library. HepG2, Hep3B, and LO2 cells were seeded the day before transfection using DMEM with 10 % fetal bovine serum (FBS) without antibiotics. Cells were transfected using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. Transfected cells were used for the subsequent experiments 48 h after transfection.
Cell proliferation assay
Cell counting kit-8 (CCK-8) assays was used to evaluate the cell proliferation in accordance with the manufacturer’s protocol. Briefly, cells were plated at a density of 2 × 104 cells/well in a 96-well plate in a volume of 100 μl and grown overnight. Then, Cell Counting Kit-8 (Dojindo, Kumamoto, Japan) reagent was added to a subset of wells under different treatments. After the cells were incubated for 2 h at 37 °C, we quantified the absorbance on an automated plate reader.
Apoptosis assay
Hep3B cells transfected with pCMV-HA-PIDD and a control plasmid were cultured for 48 h and harvested, then washed three times, resuspended, and incubated with Annexin V-FITC (Bestbio, China) for 15 min at 4 °C in the dark, according to the manufacturer’s instructions. After staining, the cells were incubated with propidium iodide for 5 min at 4 °C in the dark and then analyzed using a flow cytometer (Beckman, USA).
Statistical analysis
All data were analyzed by SPSS 17.0 software and presented as mean ± SD. χ2 test was used to analyze the association between PIDD and Ki-67 expression and clinicopathological variables. For the analysis of survival data, Kaplan–Meier curves were constructed, and the log-rank test was performed. Multivariate analysis was performed using Cox proportional hazards model. The hazard ratio and its 95 % confidence interval were recorded for each variable. P < 0.05 was considered statistically significant.
Results
PIDD was lowly expressed in HCC tissues and cell lines
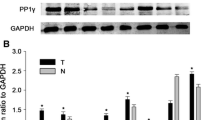
To explore the role of PIDD in HCC development, we first investigated the expression pattern of PIDD in eight pairs of matched HCC tissues and adjacent normal liver tissues. Western blotting analysis showed that PIDD was lowly expressed in HCC tissues compared with adjacent normal liver tissues (Fig. 1a, b). Then, we examined the expression profile of PIDD in human embryo liver LO2 cell and several HCC cell lines, such as Huh7, HepG2, Hep1B, Hep3B, and MHCC-97H. We found that PIDD had a lower expression in HCC cell lines than in LO2 cells (Fig. 1c, d). In addition, immunohistochemistry assay was performed to further examine the expression of PIDD in 99 HCC samples. As expected, we found that PIDD was lowly expressed in most tumorous tissues, whereas highly expressed of PIDD was observed in adjacent non-tumorous liver tissues (Fig. 2a, c). However, the expression of Ki-67 was higher in most tumorous tissues, compared with the adjacent non-tumorous tissues (Fig. 2b, d). These findings together implicated that PIDD was lowly expressed in HCC tissues and cell lines.
PIDD was lowly expressed in HCC tissue and cell lines. a The protein level of PIDD was low in eight representative paired samples of HCC tissues (T) compared with non-tumorous adjacent tissues (N). GAPDH was used as a loading control. The experiment has been repeated for three times. b Quantification graphs (relative optical density) of the intensity of staining of PIDD and PCNA to GAPDH. c The expression of PIDD was low in HCC cell lines compared with LO2 normal hepatocytes. d The bar chart of the ratio of PIDD protein to β-actin by densitometry in the indicated cell lines. Mean ± SD of three independent experiments. *P < 0.01 compared with control non-tumorous adjacent tissues (N)
Correlation between PIDD expression and clinicopathological variables in HCC patients
To further explore the pathophysiological significance of PIDD expression with tumor characteristics, the clinicopathological data were summarized in Table 1. The results showed that the expression of PIDD was significantly related with AFP (P = 0.039), AJCC stage (P = 0.002) as well as Ki-67 (P = 0.001), while PIDD expression was not correlated with other prognostic factors, such as gender and age. Moreover, we also investigated the correlation between Ki-67 expression and patients’ clinicopathological parameters, through which we observed significant correlations of Ki-67 expression with AFP (P = 0.001) and AJCC stage (P = 0.012), but not with other factors. In addition, we used the Spearman’s correlation coefficient to analyze the correlation between PIDD and Ki-67 expression in the examined HCC tissues. The results indicated that there was a remarkable inverse correlation between PIDD and Ki-67 (Fig. 3a). Combined with these results together, we consider that downregulated expression of PIDD might contribute to the progression of HCC.
Correlation between PIDD expression and Ki-67 index in HCC specimens. a The correlation between PIDD and Ki-67 expression in HCC was further evaluated by Spearman rank correlation test (P = 0.001). b Kaplan–Meier survival curves for low versus high PIDD expression in 99 HCC patients showed a significant difference between the curves (P < 0.001, log-rank test). Overall survival, probability of survival of all patients with HCC: high expression, n = 37; low expression, n = 62, P < 0.001. Patients with high expression of PIDD had a longer survival than those with low expression. c Kaplan–Meier analysis of patients’ survival with low versus high Ki-67 expression in the total 99 HCC patients. Patients within the high Ki-67 expression group show worse overall survival (P = 0.002)
Prognostic significance of PIDD expression
In order to explore the correlation between PIDD expression and patients’ survival, Kaplan–Meier analysis was performed in the total of 99 patients with follow-up data. The survival curves revealed that HCC patients with low PIDD expression had poorer overall survival than those with high PIDD expression (Fig. 3b). However, patients with high Ki-67 expression predicted significantly shortened overall survival (Fig. 3c). Moreover, we evaluated the association of patient’s survival status with clinicopathological parameters using univariate analysis. We found that there was a significant correlation between patients’ survival status and clinicopathological factors such as AFP (P < 0.001), AJCC stage (P < 0.001), the expression of Ki-67 (P = 0.002), and the expression of PIDD (P < 0.001) (Table 2). Multivariate analysis showed that tumor size (P = 0.010), AJCC stage (P < 0.001), and the expression of PIDD (P = 0.040) were independent prognostic factors in HCC patients (Table 3). These data highlighted that PIDD could be a valuable prognostic indicator for the prediction of patients’ survival.
Expression of PIDD in proliferating HCC cells
Our aforementioned data have demonstrated the physiological significance of PIDD as a prognostic indicator to predict the survival of HCC patients. Next, we employed HepG2 cells to further investigate the role of PIDD in HCC development. Because the expression of PIDD was associated with the level of AFP, AJCC stage, and Ki67 expression in HCC specimens, we speculated that PIDD might influence the proliferation of HCC cells. We therefore analyzed the expression of PIDD in different proliferating statuses of HCC cells. HepG2 cells were serum starved for 72 h and then refed with 10 % FBS-containing medium. Flow cytometry analysis revealed that HepG2 cells were arrested at the G0/G1 phase after serum depletion for 72 h. After serum refeeding, HepG2 cells were released from the G1 phase and entered into S and G2/M phases (Fig. 4a). Western blotting results displayed that the expression of PIDD was decreased after serum stimulation in HepG2 cells, whereas the expression of cyclin D1 was coincidentally upregulated (Fig. 4b). These results suggested PIDD might work as a negative regulator of HCC cell proliferation.
The expression of PIDD and cell cycle-related proteins in serum-starved and serum-refed HCC cells. a, c Flow cytometry analysis of cell cycle distribution in serum-starved and serum-released HepG2 cells. Cells were accumulated at G1 phase after serum starvation for 72 h and then progressively entered into cell cycle upon serum refeeding. b Western blot analysis of PIDD and cyclin D1 expression in serum-starved and serum-refed HepG2 cells. β-actin was used as a loading control. d Quantitative analysis of the intensities of PIDD, and cyclin D1 versus β-actin at different time points in HepG2 cells. The data are mean ± SEM of three independent experiments. (*, ∧, # P < 0.05 compared with the cells starved for 72 h). S serum starvation, R serum refeeding
The role of PIDD in cell cycle and cell proliferation
To further study the role of PIDD in cell proliferation, we used siRNA to interfere the expression of PIDD in LO2 and HepG2 cells. The results showed that the expression of PIDD was dramatically decreased in LO2 cells transfected with PIDD siRNA, especially in the cells transfected with PIDD siRNA#2, compared with the cells transfected with control siRNA (Fig. 5a). However, the expression of cyclin A, cyclin D1, and PCNA were significantly increased in HepG2 cells transfected with PIDD siRNA#2 (Fig. 5b). In addition, flow cytometry analysis showed that interference of PIDD led to a significantly increased population in S phase, and a decreased population in G1 phase, compared with control siRNA-transfected cells (Fig. 5c). Finally, cell counting kit-8 (CCK-8) assay showed that the proliferation of cells was significantly increased in PIDD siRNA#2-transfected cells, compared with control siRNA transfected (Fig. 5d). However, overexpression of PIDD remarkably decreased the expression of cyclin A, cyclin D1, and PCNA in Hep3B cells, indicating suppressed cell proliferation (Fig. 5e). In addition, Hep3B cells were arrested in the G1 phase in the presence of ectopic PIDD (Fig. 5f). Furthermore, CCK-8 assay indicated that the proliferation of Hep3B cells was significantly decreased after PIDD overexpression (Fig. 5g). Taken these results together, we confirmed that PIDD could modulate cell cycle and cell proliferation.
The role of PIDD in cell cycle and cell proliferation. a Western blot analysis of the expression of PIDD in LO2 cells transfected with control siRNA and different PIDD-targeting siRNA. b, e Western blot analysis explored the expression of cyclin A, cyclin D1, and PCNA in HepG2 and Hep3B cells transfected with siRNA and overexpressing plasmid. c, f Flow cytometry analysis explored the role of PIDD in cell cycle in HepG2 and Hep3B cells. d, g CCK-8 assay detected the role of PIDD in cell proliferation in HepG2 and Hep3B cells. The data are mean ± SEM of three independent experiments. (*P < 0.05, compared with the control group)
Overexpression of PIDD slightly augmented the apoptosis in HCC cells
Studies have showed that PIDD could induce cell apoptosis [6]. Thus, we investigated whether PIDD was involved in the apoptosis of HCC cells. Hep3B cells were transiently transfected with pCMV-HA-PIDD plasmid or control plasmid for 72 h and then subjected to Annexin-V/PI apoptotic analysis. Flow cytometry results showed that the apoptosis of cells transfected with PIDD-overexpressing plasmid was increased significantly (Fig. 6a). Therefore, these results showed that overexpression of PIDD in Hep3B cells promoted cell apoptosis.
Overexpression of PIDD promoted cell apoptosis in Hep3B cells. a Hep3B cells were transiently transfected with pCMV-HA-PIDD plasmid or control plasmid and cultured in a humidified incubator. At 72 h post-infection, cells were harvested and measured for apoptosis using Annexin V-FITC apoptosis detection kit followed by flow cytometry analysis. b Cells were lysed and analyzed by Western blot. GAPDH was used as loading control
To further investigate the mechanisms underlying PIDD-induced apoptosis in Hep3B cells, we also examined the protein expression which might involve in the apoptotic pathway. As shown in Fig. 6b, the level of activation of cleaved caspase-2 was increased in Hep3B cells transfected with PIDD overexpressing plasmid, compared with the cells transfected with control plasmid. These results indicated that overexpression of PIDD could promote HCC cells apoptosis via the initiation of caspase-2 apoptotic pathway.
Discussion
Despite the improvements in surveillance and clinical treatment strategies in HCC, the prognosis of HCC patients with high recurrence and metastasis remains poor [21]. Therefore, it is a huge challenge to find new molecular markers related with the prognosis and development of HCC for clinical practitioner and basic scientists. In this study, we depicted the potential role of PIDD in the development of HCC. We found that PIDD was lowly expressed in the majority of clinical HCC samples and HCC cell lines, compared with the adjacent non-tumorous tissues and LO2 normal hepatocytes. Moreover, we also found that the expression of PIDD was significantly correlated with AJCC stage, serum level of AFP, the expression of Ki-67 as well as poor prognosis. Univariate and multivariate analyses indicated that PIDD could be an independent prognostic indicator of patients’ survival. In addition, we used serum starvation-refeeding experiment to explore the potential role of PIDD in HCC cell cycle. In particular, we found that interference of PIDD in HepG2 cells led to accelerate cell growth, whereas overexpression of PIDD in Hep3B cells resulted to cell cycle arrest and decrease cell proliferation. Finally, the overexpression of PIDD could promote apoptosis in Hep3B cells. Taken together, these findings validated that PIDD may be a novel prognostic marker and therapeutic target of HCC.
Cell cycle is the basic process of cell expansion and plays an important role in tumorigenesis. Recent studies have showed that aberrant expression of PIDD led to uncontrolled cell proliferation and carcinogenesis [22]. Trudy et al. have showed that overexpression of PIDD hindered cell growth and increased the percentage of cells in G1 phase [7]. Oliver et al. have showed that interference of PIDD resulted in an increase in the percentage of cells in G2/M phase [18]. In addition, G Bradley et al. have showed that the expression of PIDD was associated with cell proliferation in OSCC [17]. Taken these date together, we speculated that PIDD might modulate HCC cell cycle progression and be involved in HCC cell proliferation. In this study, we found that depleting the expression of PIDD led to an increased population in S phase in LO2 and HepG2 cells, whereas overexpression of PIDD resulted in cell cycle arrest at G1 phase. We also found that the expression of PIDD was related with Ki-67, a nuclear antigen that has been widely used as a marker for cell proliferation in HCC and is only expressed during cell cycle progression [23]. There was a remarkable negative correlation between PIDD and Ki-67 expression in the examined 99 HCC tissues using the Spearman’s correlation coefficient. In addition, we found that interference of PIDD accelerated cell proliferation in HepG2 cells, whereas overexpression of PIDD inhibited cell proliferation in Hep3B cells. These results indicated that PIDD could regulate HCC cell cycle progression and cell proliferation. However, the mechanism detail remains undefined.
Since Lin et al. have found that overexpression of PIDD could suppress cell viability in a p53-responsive manner in 2000 [6], more and more researches in succession have indicated that PIDD was involved in the regulation of cell apoptosis in recent years. For instance, pPIDDThr788 plays a critical role in the activation of caspase-2 (an apoptosis-related protease) through forming an activating platform (PIDDosome) with receptor interacting protein (RIP)-associated ICH-1/CED-3 homologous protein with a death domain’ (RAIDD) and pro-caspase-2 [6, 10, 13, 16, 24]. Thus, we speculated that PIDD might modulate HCC cell viability by inducing cell apoptosis via caspase-2 signaling pathway. We found that the apoptosis of Hep3B cells transfected with PIDD-overexpressing plasmid was increased compared with cells transfected with control plasmid. Meanwhile, we found that the activation of cleaved caspase-2 was increased in Hep3B cells transfected with PIDD overexpressing plasmid. However, although our data is informative to suggest a potential role of PIDD in HCC cell viability, the increased rate of apoptosis in PIDD-overexpressing HCC cells was moderate, probably because of low apoptotic rate in unstimulated HCC cells. Further studies are needed to convincingly determine whether PIDD was involved in the viability and chemoresistance of HCC cells.
To summarize, this study for the first time showed that PIDD was lowly expressed in HCC tissues and correlated with AFP, AJCC stage, Ki-67 as well as poor prognosis, which suggested a prognostic value of PIDD in HCC. In addition, Cox regression analysis demonstrated that PIDD could be an independent prognostic factor for the survival of HCC patients. Interestingly, we found that interfering the expression of PIDD could promote cell proliferation, whereas overexpression of PIDD could hinder cell cycle progression and suppress cell proliferation. Finally, overexpression of PIDD promoted cell apoptosis in Hep3B cells via caspase-2 signaling pathway. Our findings indicated that PIDD may be a novel potential target for the development of diagnostic and therapeutic strategies for HCC.
References
Bruix J, Gores GJ, Mazzaferro V. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 2014;63:844–55.
Wu Q, Qin SK. Features and treatment options of Chinese hepatocellular carcinoma. Chin Clin Oncol. 2013;2:38.
Moudgil V, Redhu D, Dhanda S, Singh J. A review of molecular mechanisms in the development of hepatocellular carcinoma by aflatoxin and hepatitis b and c viruses. J Environ Pathol Toxicol Oncol Off Organ Int Soc Environ Toxicol Cancer. 2013;32:165–75.
Finn RS. Development of molecularly targeted therapies in hepatocellular carcinoma: where do we go now? Clin Cancer Res Off J Am Assoc Cancer Res. 2010;16:390–7.
Fornaro L, Vivaldi C, Caparello C, Sacco R, Rotella V, Musettini G, et al. Dissecting signaling pathways in hepatocellular carcinoma: new perspectives in medical therapy. Future Oncol. 2014;10:285–304.
Lin Y, Ma W, Benchimol S. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet. 2000;26:122–7.
Tinel A, Tschopp J. The piddosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–6.
Huang L, Han D, Yang X, Qin B, Ji G, Yu L. PIDD4, a novel PIDD isoform without the LRR domain, can independently induce cell apoptosis in cytoplasm. Biochem Biophys Res Commun. 2011;407:86–91.
Cuenin S, Tinel A, Janssens S, Tschopp J. P53-induced protein with a death domain (PIDD) isoforms differentially activate nuclear factor-kappab and caspase-2 in response to genotoxic stress. Oncogene. 2008;27:387–96.
Berube C, Boucher LM, Ma W, Wakeham A, Salmena L, Hakem R, et al. Apoptosis caused by p53-induced protein with death domain (PIDD) depends on the death adapter protein RAIDD. Proc Natl Acad Sci U S A. 2005;102:14314–20.
Wu ZH, Mabb A, Miyamoto S. PIDD: a switch hitter. Cell. 2005;123:980–2.
Janssens S, Tinel A, Lippens S, Tschopp J. PIDD mediates NF-kappab activation in response to DNA damage. Cell. 2005;123:1079–92.
Ando K, Kernan JL, Liu PH, Sanda T, Logette E, Tschopp J, et al. PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival piddosome signaling. Mol Cell. 2012;47:681–93.
Oskouian B, Sooriyakumaran P, Borowsky AD, Crans A, Dillard-Telm L, Tam YY, et al. Sphingosine-1-phosphate lyase potentiates apoptosis via p53- and p38-dependent pathways and is down-regulated in colon cancer. Proc Natl Acad Sci U S A. 2006;103:17384–9.
Heikaus S, Pejin I, Gabbert HE, Ramp U, Mahotka C. Piddosome expression and the role of caspase-2 activation for chemotherapy-induced apoptosis in RCCs. Cell Oncol Off J Int Soc Cell Oncol. 2010;32:29–42.
Seth R, Yang C, Kaushal V, Shah SV, Kaushal GP. P53-dependent caspase-2 activation in mitochondrial release of apoptosis-inducing factor and its role in renal tubular epithelial cell injury. J Biol Chem. 2005;280:31230–9.
Bradley G, Tremblay S, Irish J, MacMillan C, Baker G, Gullane P, et al. The expression of p53-induced protein with death domain (Pidd) and apoptosis in oral squamous cell carcinoma. Br J Cancer. 2007;96:1425–32.
Oliver TG, Mercer KL, Sayles LC, Burke JR, Mendus D, Lovejoy KS, et al. Chronic cisplatin treatment promotes enhanced damage repair and tumor progression in a mouse model of lung cancer. Genes Dev. 2010;24:837–52.
Wang J, Huo K, Ma L, Tang L, Li D, Huang X, et al. Toward an understanding of the protein interaction network of the human liver. Mol Syst Biol. 2011;7:536.
Wan C, Hou S, Ni R, Lv L, Ding Z, Huang X, et al. MIF4G domain containing protein regulates cell cycle and hepatic carcinogenesis by antagonizing CDK2-dependent p27 stability. Oncogene. 2015;34:237–45.
Inagawa S, Itabashi M, Adachi S, Kawamoto T, Hori M, Shimazaki J, et al. Expression and prognostic roles of beta-catenin in hepatocellular carcinoma: correlation with tumor progression and postoperative survival. Clin Cancer Res Off J Am Assoc Cancer Res. 2002;8:450–6.
Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70.
Kitamoto M, Nakanishi T, Kira S, Kawaguchi M, Nakashio R, Suemori S, et al. The assessment of proliferating cell nuclear antigen immunohistochemical staining in small hepatocellular carcinoma and its relationship to histologic characteristics and prognosis. Cancer. 1993;72:1859–65.
Ren J, Shi M, Liu R, Yang QH, Johnson T, Skarnes WC, et al. The Birc6 (Bruce) gene regulates p53 and the mitochondrial pathway of apoptosis and is essential for mouse embryonic development. Proc Natl Acad Sci U S A. 2005;102:565–70.
Acknowledgments
This work was supported by the National Natural Scientific Foundation of China (no. 81272708).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
None
Ethics approval and consent to participate
All HCC specimens were collected in accordance with the protocols approved by the Ethics Committee of Affiliated Hospital of Nantong University, and every patient submitted written informed consent.
Additional information
Weidong Shi and Wei Huang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(GIF 119 kb)
Rights and permissions
About this article
Cite this article
Shi, W., Huang, W., Chen, Y. et al. Low expression of PIDD is associated with cell proliferation and apoptosis in hepatocellular carcinoma. Tumor Biol. 37, 10447–10457 (2016). https://doi.org/10.1007/s13277-015-4556-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-4556-y