Abstract
Azo dyes are used as coloring agent in textile industries at larger scale. As a result, large quantity of dye-enriched waste water is generated which subsequently poses environmental problems. Biological tool involving bacteria having azoreductase enzyme has proved to be more effective and efficient in dye effluent treatment. Current work focuses on Staphylococcus caprae (S. caprae) for degradation and decolorization of Reactive Red-195 (RR-195) azo dye. For this purpose, factors such as pH, temperature, inoculums, carbon and nitrogen sources, and dye concentrations have been optimized for maximum decolorization and degradation. S. caprae (4 mg/mL) efficiently resulted into 90% decolorization of RR-195 dye under static condition at 100 µg/mL concentration, 30 °C and pH 7.0 at a 12-h contact period. FTIR analysis has revealed the formation of new functional groups in the treated dye such as O–H stretch at 3370 cm−1, C-H band stretching at 2928 cm−1, and new band at 1608 cm−1 which specify the degradation of aromatic ring, 1382 and 1118 cm−1 represents desulfonated peaks. Biodegraded metabolites of RR-195 dye such as phenol, 3, 5-di-tert-butylphenol, and phthalic acid have been identified respectively that find industrial applications. Phytotoxicity test has shown non-toxic effects of treated dye on germination of Vigna radiata and Triticum aestivum seeds. Further, antibiotic diffusion assay has confirmed the biosafety of S. caprae.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Dye industry contributes heavily to environmental pollution. Synthetic dyes are utilized in different industries such as leather, paper, printer, paint, and textile to color their products (Slama et al. 2021). Azo dye is one of the most commonly used synthetic dyes in the textile industry and its fixation rate varies from 60 to 90%. Unfixed dye effluent (10 to 15%) gets discharged into the environment, leading to water, air, and soil pollution (Yuan et al. 2020; Ikram et al. 2022), and subsequently affecting human health. Therefore, complete mineralization of the azo group is required. Physico-chemical methods are available for the removal of dye effluents (Jargalsaikhan et al. 2021; Al-tohamy et al. 2022). These are highly expensive processes that produce a bulky amount of toxic sludge and need further processing. Bioremediation process, on the other hand, is cost-effective, more viable, and green technology for the decolorization and mineralization of azo dye. Reports are available on the decolorization and degradation of azo dye using algae and fungi such as microalgae (Behl et al. 2019), Lychaete pellucid (Khalaf et al. 2023), Caulerpa lentillifera (Marungrueng and Pavasant 2007; El-Sheekh et al. 2023), and Microcystis (El-Sheekh et al. 2017). However, algae and fungi require prolonged time for decolorization and degradation of dye owing to their long-life cycles. Presence of azoreductase enzyme is reported in various bacterial species, like bacterial consortium (Guo et al. 2020), Pseudomonas aeruginosa (Khan et al. 2023), and Escherichia coli (Ikram et al. 2022), which plays a major role in decolorization and degradation of azo dye. Bacteria require a two-step process for complete detoxification of azo dye. Initially, azo-linked dye gets reduced through the bacterial azoreductase enzyme, and it produces aromatic amines. Aromatic amines are toxic in nature but they get degraded and mineralized in non-toxic form through bacterial action (Amin et al. 2020). Still, there is a need for continuous work to find more suitable and efficient bacteria for dye effluent detoxification in a short time. In the present investigation, S. caprae has been used for the decolorization and degradation of RR-195 dye. S. caprae was isolated from dye effluent, and its mass cultivation was carried out. Most of the reports are available on the detoxification of dye using free cells of bacteria, which are not much feasible to be removed from the waste water. To sort out such problems, beads of S. caprae were prepared and used for decolorization and degradation of RR-195. Various environmental and nutritional factors were optimized for efficient dye decolorization and degradation in a short-time span. Subsequently, the biodegraded product of dye was characterized using UV–visible, FTIR, and GC–MS analysis. A phytotoxicity study was performed to analyze the toxicity range of dye-biodegraded effluent. Antibiotic diffusion assay was also performed to confirm the biosafety of S. caprae using vancomycin and azithromycin antibiotics.
Materials and methods
Dyestuff and chemicals
RR-195 azo dye was obtained through the courtesy of Dream Home Carpet Pvt. Limited, Panipat, India. Stock solution (100 µg/mL) of dye was prepared in sterilized double-distilled water and subsequently solution was filtered using a 0.22-μm pore size filter paper (Millipore filter, India). Agar powder, Mueller–Hinton agar, vancomycin and azithromycin, peptone, yeast extract, nitrogen and carbon sources, and nutritional broth chemicals of analytical grade were purchased from Himedia Laboratory, Delhi, India.
Isolation and purification of bacteria
Dye effluent was collected from the Bhadohi Carpet Industry (Uttar Pradesh, India) and diluted (10−10 times) in autoclaved double distilled water. Solid agar medium enriched with peptone, lactose, yeast extract (PLY) supplemented with RR-195 dye (100 µg/mL) was prepared under sterilized condition. Dye effluent (10−10 times diluted) was inoculated on solid media containing Petri plates and were incubated for 24 h at 30 °C under aseptic environment using standard method (Singh et al. 2015). Clear zone formations were observed after 12 h. Bacteria were picked up from three clear zones of dye-treated plates. Isolated bacteria were introduced into the PLY (100 mL) broth medium and their growth was measured as absorbance of bacterial suspension at 600 nm (A600) using spectrophotometer (Hitachi U-2900/2910). Each unique bacterial strain was treated with RR-195 dye (100 µg/mL) and kept separately under shaking (100 rpm) and static condition for 12 h. After, every 2-h intervals, 3-mL samples were drawn to centrifuge (Sigma GmbH, Model 1–15 pk, Germany), for 10 min at 5000 rpm. The absorbance of the supernatant was determined using UV–Vis spectrophotometer (Hitachi U-2900) at λ max 520 nm (A520). The most efficient bacterial strain was selected for further study. The decolorization (%) was calculated as per the formula given below.
Decolorization % = (A520 of control – A520 of treated sample × 100) / A520 of control.
Culture identification
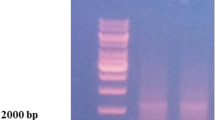
Isolated and purified bacterial culture was identified according to its 16S rRNA gene sequences using Big Dye Terminator v3.1 Cycle Sequencing Kit’s, Bengaluru, Karnataka 560,043. Genomic DNA of the strain was extracted using DNA extraction kit as per the manufacturer’s manual. The 16S rRNA gene was elaborated by PCR using the 16S forward primer (5′-GGATGAGCCCGCGGCCTA-3′) and 16S reverse primer (5′-CGGTGTGTACAAGGCCCGG-3′). The detailed program was set as following: 3 min at 94 °C, followed by 30 cycles for 1 min at 94 °C, 1 min at 50 °C, 2 min at 72 °C, and final extension at 72 °C for 7 min. The PCR products were purified for sequencing. The provided samples were recorded with data of 1433 bp Aligned Sequence. All reagents were of analytical grade.
Preparation of bacterial beads
The exponential phase S. caprae biomass was obtained through centrifugation at 5000 rpm. To generate the beads, the bacterial biomass (100–400 mg) of S. caprae was mixed separately with 50 mL of sodium alginate (4% w/v) prepared in PLY medium. Subsequently, mixture was introduced drop wise into 0.2 mol/L calcium chloride solution with the help of 0.45 × 13 mm gauge syringe under aseptic condition. The produced beads were collected and suspended in (100 mL) growth medium.
Optimization factors for S. caprae for dye decolorization
Dye solution (100 µg/mL) treated with S. caprae beads (100 mg biomass) were incubated for 12 h at pH 5 under static and shaking (120 rpm) condition at 30 °C. Three milliliters of the test sample was drawn at 1-h intervals for 12 h. Subsequently, test samples were centrifuged at 5000 rpm for 10 min to obtain supernatant. The supernatant’s absorbance was measured at 520 nm (A520). Same experiments were incubated at pH 7 and 9 respectively to know the role of pH for decolorization of dye using S. caprae. Dye removal was maximum at pH 7 under static condition at 30 °C (Ikram et al. 2022; Singh et al. 2022).
Size of the inoculums (100–400 mg) and temperature (25–40 °C) were optimized under static condition for dye removal (Singh et al. 2015, 2022). The medium was supplemented with 0.3% w/v carbon sources (glucose, sucrose, fructose, and starch) to increase the dye decolorization by S. caprae (Ikram et al. 2022; Kamal et al. 2022). Nitrogen sources (0.15% w/v concentration) were added in the form of ammonium chloride, ammonium sulfate, ammonium nitrate, and urea to the medium to improve dye decolorization (Singh et al. 2022). Increased dye concentrations from 100 to 500 µg/mL were used to evaluate the efficiency of bacteria for maximum dye decolorization.
Investigation of degraded metabolites by UV–visible scanning, FT-IR, and GC–MS
Aliquots of the 3.0 mL samples from RR-195 dye treated with bacteria and control (100 µg/mL synthetic dye solution) were drawn at 1-h regular time intervals up to 12 h. Samples were centrifuged (Sigma GmbH, Model 1–15 pk, Germany) at 5000 rpm for 10 min to obtain supernatant. Resultant supernatant was scanned at 200–800 nm (λ) against dye solution using UV–Vis spectrophotometer. Dye decolorization was carried out at 30 °C under static condition at pH 7 (Singh et al. 2022).
S. caprae treated dye degraded metabolites were examined using FTIR spectroscopy to know functional groups following standard method (Singh et al. 2018). Freeze-dried state of RR-195 treated supernatant was mixed with KBr (Lyophilizer Christ Alpha 1–2) to make pellets and were used for FT-IR analysis (Perkin Elmer Spectrum version 10.03.05 spectrometer) to know functional groups. GC–MS analysis was used to determine the structure and the m/z (mass spectra) ratio of the newly formed product after dye biodegradation following standard method as discussed by Singh et al. (2022).
Toxicity assessment
Phytotoxicity assessment
Phytotoxicity test was carried out to analyze the toxicity of bio-degraded metabolite (Singh et al. 2017). T. aestivum (wheat) Var. HUW 510 and V. radiata (Moong) Var. HUM-12 seeds were procured from the Institute of Agriculture, BHU, Varanasi, India. Phytotoxicity test was followed up to assess the effect of bacterial treated and untreated (100 µg/mL RR-195) dye on T. aestivim and V. radiata. Control sample (distilled water) was used for the comparative study. In the beginning of experimentation, 0.1% sodium hypochlorite solutions were used to sterilize seeds for 1 min. Subsequently, seeds were washed and then soaked for 10 min in sterilized distilled water. After that, each sterilized Petri plate was covered with two layers of sterile distilled water (10 mL) moistened wet tissue paper (Maxwin, India). After that, 10 seeds were distributed on each plate and sprinkled every day at a fix time with a 5-mL supernatant of bacterial-treated dye. As stated by the Organization for Economic Co-Operation Development OECD’s (OECD 2003) for the seed germination test, the growth of the plumule (shoot), radicle (root), and germination (%) were measured every 24 h up to 4 days.
Antibiotic resistant test
Antibiotic resistant testing is required to authenticate susceptibility to selected antimicrobial agents or to identify resistance in isolates. In the present investigation, an antibiotic diffusion assay was used for (vancomycin and azithromycin) antibiotic resistance testing in S. caprae. Initially solid agar plates were prepared using 3.8% Mueller–Hinton agar medium in sterilized double-distilled water under aseptic condition. Bacterial culture (100 µL) was spread over the solid media containing Petri plates. A well was created in the center of plate using pipette. The two antibiotics (5 µg/mL) were added in two Petri plates in a well using the micropipette and were incubated for 16 h at 30 °C under aseptic environment (Bauer et al. 1966). After 16 h, a clear zone of inhibition was observed confirming the susceptibility of the isolate against these antibiotics (Fig. 6).
Quality assurance and control
Glass wares were systematically cleaned and retained in 2 mol/L HNO3 for 20 h before being washed and dried. Quality control (QC) was affirmed for each test using one control sample and one triplicate.
Statistical analysis
Results of triplicate experiments were represented as mean value ± standard error (SE). When ANOVA was significant (p ≤ 0.05), means were segregated using Turkey’s numerous test. For statistical analysis, SPSS v. (SPSS 16.0; IL, Chicago, USA) has been used.
Results and discussion
Culture identification
Isolated microbe was identified as Staphylococcus caprae strain 116,030,216 on the basis of 16S ribosomal RNA gene sequencing. This strain showed 99.86% similarity with Staphylococcus sp. strain 92,738. Although, 98% similarity is examined as an edge for prokaryotic species identification. Phylogenetic (maximum likelihood) tree of the isolated bacteria was constructed using MEGA software (Hall 2013) with Kimura 2 parameter model (Fig. 1). Identified bacteria S. caprae strain 116,030,216 is distinct with bold bullet accession number. The partial sequence of the identified strain was subsequently deposited to NCBI under the accession number, OP522362. Reports are available on several other Staphylococcus strains, such as S. capitis JCM2420, S. caprae ATCC35538, S. capitis LK499, S. caprae DSM 20608, S. hominis RMLRT03, S. saprophyticus BHUSSX5, Staphylococcus K2204, Pseudomonas putida R43, and Enterobactor which are used for dye degradation and are isolated from dye effluents (Zahoor and Rehman 2009; Basharat et al. 2023; Stingley et al. 2010; Singh et al. 2014; Kumari et al. 2015; Velayutham et al. 2018; Garg et al. 2012; Singh et al. 2022). These strains were included in phylogenetic tree.
Effect of optimization factors on S. caprae for RR-195 dye decolorization
pH is essential factor for the optimum physiological performance of microbial cultures for dye decolorization. It has an impact on several biochemical and enzymatic processes, including cell development. Nutrients as well as dye molecules are transported inside the bacterial cell across the membrane at specific pH, which help in decolorization of dye (Bekhit et al. 2020; Lade et al. 2015; Carolin et al. 2021). Effects of pH (5 to 9) were observed on the RR-195 dye decolorization efficiency of S. caprae. Beads of 100 mg biomass of S. caprae was found effective to decolorize 54%, 60%, and 52% RR-195 dye of 100 µg/mL concentration at pH 5, 7, and 9 respectively after 12 h of incubation. Maximum RR-195 dye decolorization was observed at pH 7 (Fig. 2a). Avci et al. (2023) also observed a maximum decolorization rate (89.8%) of Reactive Orange (RO 13) at pH 7 in case of Bacillus sp. Bacillus stratosphericus SCA 107 showed its efficiency to decolorize RO 16 at pH 7 (Akansha et al. 2022). Ewida et al. (2019) have reported optimal decolorization of Acid Red 337 dye by Bacillus megaterium at pH 7. Carolin et al. (2021) have also reported maximum methyl orange dye decolorization using M. yunnaenensis at neutral pH 7. However, the ability of decolorization decreased at pH values less than 7. This is attributed to negative effect of acidic condition on the transport of azo dye inside the bacterial cells (Guo et al. 2020). At the same time under acidic condition, azo dye got converted into protonated form which could not be identified by the microbes for their action and decolorization rate decreases (Du et al. 2015). A similar result was observed by Singh et al. (2022) for 86.34% RY-145 decolorization using mixed consortia at pH 7. Roy et al. (2018) have also stated maximum decolorization of Crystal Violet dye using bacteria at pH 7. However, decolorization trend decreased under acidic and alkaline pH. Basharat et al. (2023) have also reported 96.0% Congo red dye decolorization by S. caprae at pH 7.
An optimum temperature for the viability of bacterial cells is required. It regulates the bacterial metabolism and their enzymatic properties. Therefore, experiments were performed at various temperatures. S. caprae was incubated with RR-195 dye at 25, 30, 35, and 40 °C for 12 h. Bacteria were found efficient in 65% decolorization of RR-195 dye at 30 °C (Fig. 2b). A decrease in decolorization and degradation of RR-195 dye was observed with further increase or decrease in temperature. Shah et al. (2013) also recorded maximum decolorization of methyl orange by Pseudomonas sp. at 30 °C. A maximum (91%) decolorization of AR 337 dye was obtained by B. megaterium at 30 °C (Ewida et al. 2019). Carolin et al. (2021) have described a decrease in dye decolorization at high temperature. This was mainly due to negative impact of high temperatures on viability of bacterial cells and subsequently on their enzymatic properties.
Decolorization and degradation of dyes are also governed by anaerobic and aerobic conditions (Garget al. 2020). Biodegradation of azo dyes may be inhibited or favored in the presence of oxygen, which depends on oxygen sensitive and insensitive enzymes present inside the bacterial cells used in the experiment (Singh et al. 2015). In the current investigation, 70% and 65% RR-195 dye decolorization by the S. caprae BM8 was achieved under static and shaking culture conditions, respectively, after 12 h of incubation (Fig. 2c). Results showed that under shaking conditions, more oxygen was available, which favored better bacterial cell biomass production as compared to static condition. However, maximum RR-195 dye decolorization was noted under static rather than shaking culture conditions. Enzyme azoreductase under shaking conditions becomes inactive due to being oxygen sensitive, and dye decolorization and degradation get hampered (Kumar et al. 2019; Haque et al. 2021). Singh et al. (2022) have also reported maximum azo dye decolorization (95.73%) using mixed consortia within 12 h under static rather than (90.57%) shaking condition.
Inoculum size also plays a major role in dye decolorization and degradation. When RR-195 dye of 100 mL having 100 µg/mL concentrations was treated with bacterial bead biomass of 100 to 400 mg, there was increase in the decolorization percentage (Fig. 2d). The highest decolorization of 90% was obtained using 4 mg/mL inoculum, which was followed successfully by that of 88%, 78%, and 70% using 3 mg/mL, 2 mg/mL, and 1 mg/mL biomass. There was no further increase in decolorization percentage on further increasing bacterial bead biomass by more than 4 mg/mL. Bonugli-Santos et al. (2016) and Roy et al. (2018) also reported that the decolorization potential increases in inoculum size to a certain extent, beyond which it decreases, as enzyme production is reduced due to nutritional depletion caused by increased biomass. This results in a decrease in the metabolic activity of microorganisms. Singh et al. (2022) have reported that an initial increase in inoculum favors the rate of decolorization. However, further increase in the size of inoculum causes overlapping of their surface area. Subsequently, dye decolorization is inhibited due to lesser availability of bacterial surface area.
The ability of S. caprae for RR-195 dye decolorization was studied in the presence of co-substrates such as various forms of carbon and nitrogen (Fig. 2e, f). Generally, azo dyes have a less carbon content therefore in the absence of carbon as co-substrate the degradation of azo dyes by bacteria gets inhibited (Al-Tohamy et al. 2020; Garg et al. 2020).
To investigate the effect of carbon on S. caprae for dye decolorization, experiments were performed with various forms of carbon sources (glucose, sucrose, fructose, and starch) at 0.3% (w/v) concentration while keeping other parameters constant (pH 7, 100 µg/mL dye concentration, 30 °C temperature, 4% inoculum size). Dye degradation was 95% with glucose as a co-substrate (Fig. 2e). However, other forms of carbon sources showed the least dye degradation as compared to glucose. Guadie et al. (2017) reported 95 to 100% decolorization of azo dye using Bacillus sp. in presence of glucose as a carbon source. Glucose is the simplest form of carbon, which could be easily metabolized and subsequently transported inside the bacterial cells, where it helps with decolorization and degradation of dye (Al-Tohamy et al. 2020; Carolin et al. 2021). Similar results were recorded by Singh et al. (2022) who used glucose as carbon source for decolorization and biodegradation of azo dye with the help of mixed consortia.
Decolorization of dye was observed under different nitrogen sources like ammonium sulfate, ammonium chloride, ammonium nitrate, and urea at 0.15% (w/v) concentration while keeping other parameters constant. In the present study, ammonium sulfate showed 97% decolorization of RR-195 dye (Fig. 2f). Decolorization was increased in ammonium sulfate supplemented media because it was assimilated easily by bacteria as compared to other nitrogen sources (Shi et al. 2021). A nitrogen source also generates NADH, which acts as an electron donor for the azoreductase enzyme, which reduces azo dyes (Guo et al. 2020; Carolin et al. 2021). Similar results were observed by Seyedi et al. (2020).
At a 100-µg/mL dye concentration, decolorization was observed 97% while 45% decolorization was recorded at 500 µg/mL within a 12-h incubation period (Fig. 2g). With increasing dye concentrations, the efficiency of dye decolorization decreases. Increasing concentrations of dye cause toxicity to microbial growth and thus reduce the ability of the bacterial biomass to catalyze the degradation process (Prasad and Ram 2020). Avci et al. (2023) have observed that with increase of initial concentration of dye, there was concomitant decrease in decolorization of dye Reactive Orange 13 (RO-13) by Bacillus sp. SBT8. Dye inside the bacterial cells acts as redox active, and causes univalent reduction of molecular oxygen and consequently produces H2O2, and hydroxyl radical (Farias et al. 2021). During the dye degradation process, there is formation of more toxic intermediate compounds like aromatic amines (Balapure et al. 2015) which causes oxidative stress. Similar result was reported by Cabiscol et al. (2000).
Investigation of metabolites of treated and untreated RR- 195 dye by UV–visible scanning, FT-IR, and GC–MS
Treated and untreated samples were monitored under UV–Vis absorbance, and the results are shown in (Fig. 3). It was seen that the intensity of visible band in a bacterial-treated dye sample disappeared, due to cleavage of the azo bond of the dye. Zhang et al. (2007) have also reported reduction in UV band intensity at 289 nm after bacterial treatment of dye sample.
The FT-IR spectra of untreated and bacterial treated RR-195 azo dye are presented in (Fig. 4). Untreated dye showed specific bands at 3447, 1622, 1555, 1493, 1195, 1123, 1050, and 993 cm−1. While in treated dye, the peaks were observed at 3370, 2928, 2371, 1608, 1382, and 1118 cm−1 (Table 1). Bands at 3447 cm−1 in untreated dye attributes to the − NH2 groups. In treated dye, there is change in the peak from 3447 to 3370 cm−1, and it showed broad appearance which could be due to overlap with the stretching vibration of the OH groups (Hasanain et al. 2011). Spectrum bands at 1555 cm−1 signify the presence of azo band in dye. In bacterial-treated dye samples, azo band peak gets to disappear which strongly evidenced for dye degradation. Peak at 1622 cm−1 represent C = C stretching of aromatic double bond. However, in treated samples shifting of band to 1608 cm−1 indicates the degradation of aromatic ring. Peak at 1493 cm−1 displays N–H stretching of secondary amide. This band was found to disappear in treated dye sample due to degradation of dye by bacteria. In an untreated dye, the peaks at 1123, 1050, 1195, and 993 cm−1 represent sulfonated group. Bacteria were found efficient to desulfonate. Therefore, respective sulfonated peaks disappeared and subsequently there is sifting of new peak at 1118 and 1382 cm−1 (Sudha et al. 2018). Peak at 2928 cm−1 represents asymmetric stretching of C-H band. Jain et al. (2012) have also reported similar stretching at 2928 cm−1.
The GC–MS analysis results were used to determine the probable metabolites of RR-195 generated after treatment with S. caprae strain BM8. These metabolites were identified as 3,5-di-tert-butylphenol and phthalic acid (Fig. 5). Naphthalene is a bicyclic aromatic hydrocarbon which acts as a precursor for the production of phthalic acid. Desulfonation and oxidative deamination of 1,5-naphthalene disulfonic acid and napthylamine -3hydroxy-5,9-disulfonic acid produce phthalic acid, which acts as precursor for the synthesis of dye. Desulfonation of benzene sulfonate ethane sulfonic acid gives the 3,5-di-tert-butylphenol. Similar product was obtained by Guo et al. (2021) during the degradation of azo dye using halo thermophilic bacterial consortium. Peaks in the chromatogram at 9.96, 14.01 min with a charge ion mass standard equal to 206, 418.6093 m/z respectively were present in bacterial treated RR-195 dye. As per report, phthalic acid is used in perfume, nail varnish, hairsprays, and other personal/cosmetic uses. 3,5-di-tert-butylphenol has anticariogenic potential (Kumar et al. 2019).
Phytotoxicity assay of untreated and treated RR-195 dye
To assess the impact of RR-195 dye on the seeds of T. aestivum and V. radiata, a phytotoxicity test was carried out. The plumule and radical length as well as the germination rate of V. radiata and T. aestivum were adversely affected by RR-195 azo dye. However, control (distilled water) and treated (100 µg/mL) showed identical growth of V. radiata and T. aestivum (Table 2). Kuberan et al. (2011) have also carried out phytotoxicity study using degraded metabolites of azo dye. They used Listeria sp. for this study and observed that germination of seeds and growth of plants were arrested when soaked in azo dye. However, phytotoxicity test of present investigation specifies that the biodegradable products were not hazardous when compared with original dye. These products do not restrict plant seeds germination. Data of phytotoxicity test will help to analyze environmental pollution (Singh et al. 2022).
Susceptibility of the S. caprae against antibiotics and bio-safety information
Antibiotic diffusion assay confirms the susceptibility of the S. caprae against vancomycin and azithromycin antibiotics (Fig. 6). A susceptible result indicates that the infection of S. caprae could be treated with vancomycin and azithromycin. Vancomycin antibiotic is a glycopeptide antibiotic which is used for the treatment of multiple bacterial infections especially caused by multi drug resistant S. aureus and S. epidermis. It is also known as “last resort of antibiotics.” Azithromycin is also used for the treatment of multiple bacterial infections such as pneumonia (caused by S. pneumoniae), intestinal infections, and ear infections (Zeng et al. 2016).
Ogura et al. (2022) have reported that the S. caprae (coagulase-negative) inhibits other Staphylococcus spp. such as S. aureus and S. epidermidis and contributes significantly in skin lesion healing. Besides, hemolytic activity of rabbit blood cells of S. aureus is suppressed by S. caprae. While, hemolytic activity of S. caprae was suppressed by S. aureus. Thus, each of the two Staphylococcus spp. suppresses the pathogenicity of the other and this inequity between the two is linked through recurrent pressure injuries (RPI).
Paharik et al. (2017) have stated that S. caprae competes with S. aureus (methicillin-resistant) by inhibiting its quorum sensing and suppresses skin colonization and infection. Thus, the application of S. caprae for dye removal will help in reducing the public health trouble of pathogenic S. aureus-mediated skin infection and the environmental dissemination of antimicrobial resistance.
Conclusion
S. caprae was found highly effective for 90% decolorization and degradation of RR-195 dye. FTIR analysis of a S. caprae-treated dye shows the disappearance of 1555 cm−1 band which signifies the degradation of azo group. Carbon (glucose) and nitrogen (ammonium sulfate) were found most favorable nutrient source to enhance (95 and 97%) decolorization and degradation of RR-195 dye respectively. Commercially, useful metabolites of bacterial treated dye were identified through GC–MS analysis. Phytotoxicity study also reveals non-toxic effect of bacterial-treated dye on seed growth of V. radiata and T. aestivum. Antibiotic diffusion test also reveals that S. caprae is susceptible against vancomycin and azithromycin antibiotics. Therefore, S. caprae is recommended for dye removal which also takes care of public health from a S. aureus-mediated skin infection and the environmental spreading of antimicrobial resistance.
References
Akansha K, Yadav AN, Kumar M, Chakraborty D, Sachan SG (2022) olorization and degradation of Reactive Orange 16 by Bacillus stratosphericus SCA1007. Folia Microbiol 67:91–102
Al-Tohamy R, Sun J, Fareed MF, Kenawy ER, Ali SS (2020) Ecofriendly biodegradation of Reactive Black 5 by newly isolated Sterigmatomyces halophilus SSA1575, valued for textile azo dye wastewater processing and detoxification. Sci Rep 10(1):12370
Al-tohamy R, Ali SS, Li F, Okasha K, Mahmoud YAG, Elsamahy T, Jiao H, Fu Y, Sun J (2022) A critical review on the treatment of dye-containing wastewater: ecotoxicological and health concerns of textile dyes and possible remediation approaches for environmental safety. Ecotoxicol Environ Saf 231:113160
Amin S, Rastogi RP, Chaubey MG, Jain K, Divecha J, Desai C, Madamwar D (2020) Degradation and toxicity analysis of a reactive textile diazo dye-Direct Red 81 by newly isolated Bacillus sp. DMS2. Front Microbiol 11:576680
Avci A, Yıldırım A, Cerit İ, Yılmazer Keskin S, Hamk M, Keskin CS, Demirkol O (2023) Influence of culture age and environmental conditions on the decolorization and biodegradation of Reactive Orange 13 by Bacillus sp. SBT8. Biomass Convers Biorefin 13(5):4353–4363
Balapure K, Bhatt N, Madamwar D (2015) Mineralization of reactive azo dyes present in simulated textile waste water using down flow microaerophilic fixed film bioreactor. Bioresour Technol 175:1–7
Basharat Z, Asghar S, Yasmin A (2023) Leveraging molecular docking to understand Congo red degradation by Staphylococcus caprae MB400. Arch Microbiol 205(6):250
Bauer AW, Kirby WMM, Sherris JC, Turck M (1966) Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45(4):493–496
Behl K, Joshi M, Sharma M, Tandon S, Chaurasia AK, Bhatnagar A, Nigam S (2019) Performance evaluation of isolated electrogenic microalga coupled with graphene oxide for decolorization of textile dye wastewater and subsequent lipid production. J Chem Eng 375:121950
Bekhit F, Farag S, Attia AM (2020) Decolorization and degradation of the Azo dye by bacterial cells coated with magnetic iron oxide nanoparticles. Environ Nanotechnol Monit Manag 14:100376
Bonugli-Santos RC, Vieira GA, Collins C, Fernandes TCC, Marin-Morales MA, Murray P, Sette LD (2016) Enhanced textile dye decolorization by marine-derived Basidiomycete Peniophora sp. CBMAI 1063 using integrated statistical design. Environ Sci Pollut Res 23:8659–8668
Cabiscol E, Tamarit J, Ros J (2000) Oxidative stress in bacteria and protein damage by reactive oxygen species. Int Microbiol 3(1):3–8
Carolin CF, Kumar PS, Joshiba GJ (2021) Sustainable approach to decolourize methyl orange dye from aqueous solution using novel bacterial strain and its metabolites characterization. Clean Technol Environ Policy 23:173–181
Du LN, Li G, Zhao YH, Xu HK, Wang Y, Zhou Y, Wang L (2015) Efficient metabolism of the azo dye methyl orange by Aeromonas sp. strain DH-6: characteristics and partial mechanism. Int Biodeterior Biodegrad 105:66–72
El-Sheekh MM, Abou-El-Souod GW, El Asrag HA (2017) Biodegradation of some dyes by the cyanobacteria species Pseudoanabaena sp. and Microcystis aeruginosa Kützing. Egypt J Exp Biol (zoo) 13:233–243
El-Sheekh MM, El Shafay SM, El-Shanshoury AERR, Hamouda R, Gharieb DY, Abou-El-Souod GW (2023) Impact of immobilized algae and its consortium in biodegradation of the textile dyes. Int J Phytoremediation 25:687–696
Ewida AYI, El-Sesy ME, Abou Zeid A (2019) Complete degradation of azo dye acid red 337 by Bacillus megaterium KY848339. 1 isolated from textile wastewater. Water Sci Technol 33(1), 154–161.
Farías R, Norambuena J, Ferrer A, Camejo P, Zapata C, Chávez R et al (2021) Redox stress response and UV tolerance in the acidophilic iron-oxidizing bacteria Leptospirillum ferriphilum and Acidithiobacillus ferrooxidans. Res Microbiol 172:103833
Garg N, Garg A, Mukherji S (2020) Eco-friendly decolorization and degradation of reactive yellow 145 textile dye by Pseudomonas aeruginosa and Thiosphaera pantotropha. J Environ Manage 263:110383
Garg SK, Tripathi M, Singh SK, Tiwari JK (2012) Biodecolorization of textile dye effluent by Pseudomonas putida SKG-1 (MTCC 10510) under the conditions optimized for monoazo dye orange II color removal in simulated minimal salt medium. Int Biodeter Biodegr 74:24–35
Guadie A, Tizazu S, Melese M, Guo W, Ngo HH, Xia S (2017) Biodecolorization of textile azo dye using Bacillus sp. strain CH12 isolated from alkaline lake. Biotechnol Rep 15:92–100
Guo G, Hao J, Tian F, Liu C, Ding K, Xu J, Guan Z (2020) Decolorization and detoxification of azo dye by halo-alkaliphilic bacterial consortium: Systematic investigations of performance, pathway and metagenome. Ecotoxicol Environ Saf 204:111073
Guo G, Liu C, Hao J, Tian F, Ding K, Zhang C, Yang F, Liu T, Xu J, Guan Z (2021) Development and characterization of a thermophilic bacterial consortium for decolorization of azo dye. Chemosphere 272:129916
Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30(5):1229–1235
Haque MM, Haque MA, Mosharaf MK, Marcus PK (2021) Decolorization, degradation and detoxification of carcinogenic sulfonatedazo dye methyl orange by newly developed biofilm consortia. Saudi J Biol Sci 793–804
Hasanain ASAM, Ahmad AYA, Hussain KA (2011) The preparation, characterization and the study of the linear optical properties of a new azo compound. J Basrah Res 37:2
Ikram M, Naeem M, Zahoor M, Hanafiah MM, Oyekanmi AA, Ullah R, Gulfam N (2022) Biological degradation of the azo dye basic orange 2 by Escherichia coli: A sustainable and ecofriendly approach for the treatment of textile wastewater. Water 14:2063
Jain K, Shah V, Chapla D, Madamwar D (2012) olorization and degradation of azo dye–Reactive Violet 5R by an acclimatized indigenous bacterial mixed cultures-SB4 isolated from anthropogenic dye contaminated soil. Environ Dev Sustain 213:378–386
Jargalsaikhan M, Lee J, Jang A, Jeong S (2021) Efficient removal of Azo dye from waste water using the non-toxic potassium ferrate oxidation–coagulation process. Appl Sci 11:6825
Kamal IM, Abdeltawab NF, Ragab YM, Farag MA, Ramadan MA (2022) Biodegradation, decolorization, and detoxification of di-azo dye direct Red 81 by halotolerant, alkali-thermo-tolerant bacterial mixed cultures. Microorganisms 10(5):994
Khalaf HA, El-Sheekh MM, Makhlof ME (2023) Lychaete pellucida as a novel biosorbent for the biodegradation of hazardous azo dyes. Environ Monit Assess 195(8):929
Khan Ullah A, Zahoor M, Ur Rehman M, Ikram M, Zhu D, Naveed Umar M, Ali EA (2023) Bioremediation of Azo Dye Brown 703 by Pseudomonas aeruginosa: an effective treatment technique for dye-polluted wastewater. Microbiol Res 14(3):1049–1066
Kuberan T, Anburaj J, Sundaravadivelan C, Kumar P (2011) Biodegradation of azo dye by Listeria sp. Int J Environ Sci 1(7):1760–1769
Kumar N, Sinha S, Mehrotra T, Singh R, Tandon S, Thakur IS (2019) Biodecolorization of azo dye Acid Black 24 by Bacillus pseudomycoides: process optimization using Box Behnken design model and toxicity assessment. Bioresour Technol Rep 8:100311
Kumari L, Verma AK, Tiwary D, Giri DD, Nath G, Mishra PK (2015) Biodegradation of Navy N5RL1 carpet dye by Staphylococcus saprophyticus strain BHUSS X3. 3 Biotech 5:775–782
Lade H, Kadam A, Paul D, Govindwar S (2015) Biodegradation and detoxification of textile azo dyes by bacterial consortium under sequential microaerophilic/aerobic processes. EXCLI J 14:158
Marungrueng K, Pavasant P (2007) High performance biosorbent (Caulerpa lentillifera) for basic dye removal. Bioresour Technol 98(8):1567–1572
Nicoletti G, Scarpetta S (2003) Regulation, productivity and growth: OECD evidence. Econ Policy 8(36):9–72
Ogura K, Furuya H, Takahashi N, Shibata K, Endo M, Watanabe S, Cui L, Miyoshi-Akiyama T, Okamoto S, Ogai K, Sugama J (2022) Interspecies regulation between Staphylococcus caprae and Staphylococcus aureus colonized on healed skin after injury. Front Microbiol 13:818398
Paharik AE, Parlet CP, Chung N, Todd DA, Rodriguez EI, Van Dyke MJ, Horswill AR (2017) Coagulase-negative staphylococcal strain prevents Staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe 22(6):746–756
Prasad SS, Ram RL (2020) Effects of dye toxicity on bacterial growth and its kinetics. Ad Plant Sci 33:131–134
Roy DC, Biswas SK, Saha AK, Sikdar B, Rahman M, Roy AK, Tang SS (2018) Biodegradation of Crystal Violet dye by bacteria isolated from textile industry effluents. Peer J 6:e5015
Seyedi ZS, Zahraei Z, JookarKashi F (2020) olorization of reactive black 5 and reactive red 152 Azo dyes by new haloalkaliphilic bacteria isolated from the textile wastewater. Curr Microbiol 77:2084–2092
Shah MP, Patel KA, Nair SS, Darji AM (2013) Microbial decolourization of methyl orange dye by Pseudomonas spp. Biotechnol Bioprocess Eng 2(1):10
Shi Y, Yang Z, Xing L, Zhang X, Li X, Zhang D (2021) Recent advances in the biodegradation of azo dyes. World J Micro Boil Biotechnol 37:1–18
Singh AL, Chaudhary S, Kayastha AM, Yadav A (2015) olorization and degradation of textile effluent with the help of Enterobacter asburiae. Indian J Biotech 14:101–106
Singh AL, Chaudhary S, Kayastha AM, Yadav A (2017) olourization, degradation and removal of heavy metals of textile effluent with the help of mixed bacterial consortium. Indian J Biotechnol 16:258–264
Singh AL, Chaudhary S, Kumar S, Kumar A, Singh A, Yadav A (2022) Biodegradation of Reactive Yellow-145 azo dye using bacterial consortium: A deterministic analysis based on degradable Metabolite, phytotoxicity and genotoxicity study. Chemosphere 300:134504
Singh AK, Kumar A, Singh PK, Singh AL, Kumar A (2018) Bacterial desulphurization of low-rank coal: a case study of Eocene Lignite of Western Rajasthan, India. Energy Sources a: Recovery Util Environ 40(10):1199–1208
Singh RP, Singh PK, Singh RL (2014) Bacterial decolorization of textile azo dye acid orange by Staphylococcus hominis RMLRT03. Toxicol Int 21(2):160
Slama HB, ChenariBouket A, Pourhassan Z, Alenezi FN, Silini A, Cherif-Silini H, Belbahri L (2021) Diversity of synthetic dyes from textile industries, discharge impacts and treatment methods. Appl Sci 11(14):6255
Stingley RL, Zou W, Heinze TM, Chen H, Cerniglia CE (2010) Metabolism of azo dyes by human skin microbiota. J Med Microbiol 59(1):108–114
Sudha M, Bakiyaraj G, Saranya A, Sivakumar N, Selvakumar G (2018) Prospective assessment of the Enterobacter aerogenes PP002 in decolorization and degradation of azo dyes DB 71 and DG 28. J Environ Chem Eng 6(1):95–109
Velayutham K, Madhava AK, Pushparaj M, Thanarasu A, Devaraj T, Periyasamy K, Subramanian S (2018) Biodegradation of Remazol Brilliant Blue R using isolated bacterial culture (Staphylococcus sp. K2204). Environ Technol 39 (22):2900–2907.
Yuan H, Chen L, Cao Z, Hong FF (2020) Enhanced decolourization efficiency of textile dye Reactive Blue 19 in a horizontal rotating reactor using strips of BNC-immobilized laccase: Optimization of conditions and comparison of decolourization efficiency. Biochem Eng J 156:107501
Zahoor A, Rehman A (2009) Isolation of Cr (VI) reducing bacteria from industrial effluents and their potential use in bioremediation of chromium containing wastewater. J Environ Sci 21(6):814–820
Zeng D, Debabov D, Hartsell TL, Cano RJ, Adams S, Schuyler JA, Pace JL (2016) Approved glycopeptide antibacterial drugs: mechanism of action and resistance. Csh Perspect Med 6(12):a026989
Zhang F, Yediler A, Liang X (2007) omposition pathways and reaction intermediate formation of the purified, hydrolyzed azo reactive dye CI Reactive Red 120 during ozonation. Chemosphere 67(4):712–717
Acknowledgements
ALS thankfully acknowledges the Department of Botany, Banaras Hindu University, for extending the facilities and for the financial support in the form of Incentive Grant received under IoE scheme of the University. MY is thankful to the financial support given by B.H.U. in the form of fellowship and for providing one time support from IOE grant for analysis.
Author information
Authors and Affiliations
Contributions
Asha Lata Singh: conceptualization, study design, methodology, supervision, writing the original draft, figures, tables, reviewing and editing. Monika Yadav: experimentation and data generation, resources and writing, figures and tables formatting.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yadav, M., Singh, A.L. Decolourization and detoxification of Reactive Red-195 azo dye by Staphylococcus caprae isolated from textile effluent. Folia Microbiol (2024). https://doi.org/10.1007/s12223-024-01175-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12223-024-01175-y