Abstract
The most diverse and versatile endophytic actinobacteria are relatively unexplored potential sources of bioactive metabolites useful for different medical, agricultural, and other commercial applications. Their diversity in symbiotic association with traditionally utilized medicinal plants of northeast India is scantly available. The present investigation assessed the genetic diversity of endophytic actinobacteria (n = 120) distributed around the root, stem, and leaf tissues of six selected medicinal plants (Emblica officinalis, Terminalia chebula, T. arjuna, Murraya koenigii, Rauwolfia serpentina, and Azadirachta indica) from three different protected areas of evergreen forest—the Gibbon Wildlife Sanctuary (GWS), the Kaziranga National Park (KNP), and the North East Ecological Park (NEEP) of Assam, India. The samples were collected in two seasons (summer and winter). The overall phylogenetic analysis showed significant genetic diversity with 18 distinct genera belonging to 12 families. Overall, the occurrence of Streptomyces genus was predominant across all three sampling sites (76.66%), in both the sampling season (summer and winter). Shannon’s and Simpson’s diversity estimates showed their presence at A. indica (1.496, 0.778), R. serpentina (1.470, 0.858), and E. officinalis (0.975, 0.353). Among the site sampled, GWS had the most diverse community of actinobacteria (Shannon = 0.86 and Simpson = 0.557). The isolates were antagonistically more active against the investigated plant pathogenic bacteria than fungal pathogens. Further analysis revealed the prevalence of polyketide synthase genes (PKS) type II (84%) and PKS type I (16%) in the genome of the antimicrobial isolates. The overall findings confirmed the presence of biosynthetically active diverse actinobacterial members in the selected medicinal plants which offer potential opportunities towards the exploration of biologically active compounds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
There is a need for novel antimicrobial drugs/candidate molecules for the effective treatment of multi-drug-resistant pathogens as well as for protecting against new infections, which are emerging rapidly. Microbial biodiversity associated with traditional Indian medicinal plants is considered as an important reservoir of bioactive compounds which needs due attention and exploration to generate a library of effective molecules against clinical manifestation (Gohain et al. 2015). About 28% of new chemical entities and 42% of anti-cancer drugs that have been introduced into the global market are natural products and their derivatives (Newman and Cragg 2007). Medicinal plants encompass a diverse endophytic actinobacterial population of economic importance (Bulgarelli et al. 2013). They are prospective sources of novel natural products, having an exploitable application in the medical and agricultural industries (Strobel et al. 2004). About 80% of the world’s antibiotics arsenal is derived from the phylum actinobacteria (Bacon and White 2000) with a significant major contribution (50%) from the genus Streptomyces (Mehdi et al. 2006). A number of antimicrobial compounds have been discovered from endophytic actinobacteria, apart from the actinobacteria of soil or water origin. The bioactive compounds like kakadumycins, coronamycin, multicyclic indolsesquiterpenes, spoxazomicins A-C etc. were derived from endophytic actinobacteria species (Castillo et al. 2003; Ezra et al. 2004; Ding et al. 2011; Inahashi et al. 2011). Therefore, endophytic actinobacteria are expected to be potential resources of new bioactive agents in the near future (Duangmal et al. 2008).
Medicinal plants with an ethno-botanical history are considered valuable bio-resources for the isolation of endophytic actinobacteria, which play a vital role in regulating the physiology, health, and fitness of the medicinal plants (Yu et al. 2010). The tropical and temperate rainforests harbor excellent biodiversity which are considered for novel endophytes and their products (Strobel et al. 2004). The tropical rainforests of Northeast India are best-known for their rich floral and faunal diversity (Myers et al. 2000). To date, around 13,500 vascular plants have been discovered in this region, out of which 7000 are considered to be endemic (Pawar et al. 2007). Furthermore, with a high density of ethnic races and tribes, the region has a long ethno-botanical history (Tiwari et al. 1993), which can guide in maximizing the chances of harnessing novel endophytic actinobacterial isolates from the medicinal plants found in the tropical forests of this region. Our previous study reported the antimicrobial biosynthetic potential and genetic diversity of endophytic actinobacteria associated with medicinal plants in the Gibbon Wildlife Sanctuary (GWS) in Assam, India (Gohain et al. 2015). The investigation reported the dominance of the genus Streptomyces with the prevalence of polyketide synthase (PKS) type II gene clusters. However, since the distribution and relative abundance of medicinal plant-associated actinobacteria in general and Streptomyces in particular were not clear, the present study aims to understand their variation across different forested ecosystems in distinct geographical locations. Hence, the present investigation was designed to estimate the endophytic actinobacterial diversity of six ethnobotanically and pharmaceutically important plants (Emblica officinalis, Terminalia chebula, T. arjuna, Murraya koenigii, Azadirachta indica, and Rauwolfia serpentine) from distinct forested ecosystems located at GWS, North East Ecological Park (NEEP) and Kaziranga National Park (KNP) in Assam, India (Supplementary Table S1). Previously, Passari et al. (2015) reported 42 genera of endophytic actinobacteria isolated from medicinal plants in Phawngpuii National Park and Dampa Tiger Reserve Forest in Mizoram, but studies on endophytic actinobacterial diversity in KNP or the NEEP were not carried out till now.
Materials and methods
Sampling and isolation of endophytic actinobacteria
The healthy plant parts (roots, stems, and leaves) of six medicinal plants (E. officinalis, T. chebula, T. arjuna, M. koenigii, R. serpentine, and A. indica) were collected from different forest areas of GWS Jorhat (26°40′ N; 94°20′ E), NEEP Jorhat (26°44′10″N; 94°9′30″E), and KNP Golaghat (26°409′N; 93°21′E) (Supplementary Table S1 and Fig. S1). Samplings were done during the months of December–January (winter) and June–July (summer). The soil pH ranged from 6.5 to 7.4, while the soil moisture content varied from 30.84 to 18.21% (Supplementary Table S2). The samples were processed immediately after reaching the laboratory. The collected samples were washed under running tap water to remove soil and other debris and were cut into small (4 × 4 mm2) pieces. The diced samples were surface-sterilized and air-dried in a laminar airflow cabinet (Taechowisan and Lumyong 2003). The efficacy of surface sterilization was validated (Schulz et al. 1993), and the isolation of endophytic actinobacteria was done in starch casein agar (SCA) medium (Taechowisan and Lumyong 2003) supplemented with antibiotics viz, nalidixic acid (25 mg/L) and nystatin (50 mg/L) (Haque et al. 1992) to inhibit fungal colonies. The plates were incubated at 28 ± 1 °C. Visible colonies were obtained after 5 days of incubation; however, some isolates required a maximum of 22 ± 3days. Purified monocultures were obtained by repeated streaking on SCA medium. The strains were preserved in a 20% glycerol stock and kept at a temperature of − 80 °C for future use. The pH values of the samples were determined by fivefold dilution in sterile distilled water. The pH of the soil samples were also recorded as the soil microenvironment significantly modulates microbial entry and their subsequent colonization within the plant host.
Morphological and biochemical characterization
A preliminary characterization of the purified isolates was done based on the isolate’s morphological characteristics as mentioned in the International Streptomyces Project (Shirling and Gottlieb 1966) and Bergey’s Manual of Determinative Bacteriology (Bergey et al. 1994). Biochemical characterizations, viz, nitrate reduction, catalase, amylase, protease, cellulase, Simmons citrate, H2S production, methyl red test, and indole production, were carried out according to standard protocols in five replications (Shirling and Gottlieb 1966; Hankin and Anagnostakis 1975; Theantana et al. 2007).
Screening for antimicrobial activity
Bacterial and fungal pathogens used in the bioactive assay were obtained from Microbial Type Collection Center, India (MTCC). The test pathogens were Staphylococcus aureus (ATCC 11632™), Escherichia coli (ATCC 11229™), Aspergillus niger (ATCC 16888™), Pseudomonas syringae (MTCC 1604), P. aeruginosa (MTCC 2582), Candida albicans (MTCC 3017), Rhizoctonia solani (MTCC 4633), and Fusarium oxysporum (MTCC 284). The in vitro antimicrobial activities of the actinobacteria were screened against the microbial pathogens using agar well diffusion assay (Saadoun and Muhana 2008) and the zone of inhibition measured. Neomycin (2 mg/mL) and Amphotericin B (3 mg/mL) were used as a control in antibacterial and antifungal tests, respectively.
DNA extraction and genotypic analysis
The genomic DNA of actinobacterial isolates was purified using Ultraclean microbial DNA isolation kit (MO BIO, USA). Genotypic fingerprinting of 120 isolates was carried out using BOXAIR1 primer (5′-CTA CGG CAA GGC GAC GCT GAC G-3′) as described by Pathom-aree et al. (2006) and REP1 (5′-III ICG ICG ICA TCI GGC-3′), REP2 (5′-ICG ITT ATC IGG CCT AC-3′) primers (Smith et al. 2001; Versalovic et al. 1994). Fingerprints were analyzed by hierarchical clustering using CLIQS 1DPRO (http://totallab.com/cliqs-1d-pro/).
Taxonomic characterization of endophytic bacteria
16S rRNA gene was amplified using bacterial universal primers (27f and 1492r). Thermal cycling was performed in the Applied Biosystems® Veriti® 96-Well Thermal Cycler (Applied Biosystems, USA) with an initial denaturation at 95 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, annealing at 45 °C for 1 min, 72 °C for 1 min, followed by final extension at 72 °C for 10 min. The amplified products were purified by a PCR purification Kit (Qiagen, Germany). DNA sequence was determined by fluorescent terminators (Big Dye, Applied Biosystems) and run in an Applied Biosystems ABI Prism automated DNA sequencer (3130 × l).
Detection of biosynthetic genes of polyketide synthase
The conserved regions of biosynthetic genes, PKSI and PKSII, were amplified using two different sets of degenerate primers—viz, K1F/M6R (5′TSA AGTCSA ACATCGGBCA-3′, 5′-CGCAGGTTSCSGTACCAGTA-3′) and KSα/KSβ (5′-TSGCSTGCTTGGAYGCSATC-3′, 5′-TGGAANCCGCCGAABCCTCT-3′). The PCR conditions for both the genes were programmed as initial denaturation at 95 °C for 5 min followed by 35 cycles of 60 s at 95 °C, 90 s at 55 °C for PKSI (K1F/M6R) and 59 °C for PKSII (KSα/KS), 90 s at 72 °C with a final extension of 8 min at 72 °C. The confirmation of the β-ketosynthase domains of PKS (I and II) amplification was done by sequencing and concurrent nr nucleotide database (NCBI GenBank) query using the Blastn tool.
Phylogenetic analysis
For phylogenetic analysis, sequences of 16S rRNA genes from the present study were analyzed for closest homology matches using web-based BLASTn (https://blast.ncbi.nlm.nih.gov) and EzTaxon (https://www.ezbiocloud.net/) tools. Chimeric sequences were processed using Mallard Software Program (version1.02, School of Biosciences, Cardiff University, http://www.bioinformatics-toolkit.org/Mallard/index.html) (Ashelford et al. 2006) and Pintail (version1.0) (Ashelford et al. 2005). Only high-quality trimmed sequences were submitted to NCBI GenBank repository to obtain accession id. MEGA 5.0 software package (Tamura et al. 2011) was used for tree construction and inference. The nucleotide distances were calculated by a Kimura-2-parameter substitution model. Phylogenetic trees were inferred by neighbor-joining (NJ) and UPGMA method on the distance calculated matrix. A bootstrap analysis with 1000 replicates was conducted to estimate the confidence and reproducibility of the particular clade.
The amplified PKS I and PKS II amplicons were sequenced, processed for low-quality bases, and analyzed by BLASTP (blast.ncbi.nlm.nih.gov) program after six frame translations (http://molbiol.ru/eng/scripts/01_13.html). The deduced amino acid sequences were aligned using Clustal Omega. The UPGMA and unrooted NJ tree were then constructed using the Poisson correction model. The consistency of the trees was verified by a bootstrap value consisting of 1000 replications.
Analysis of promoter region of biosynthetic genes
Conserved motifs were predicted using the program MEME (Bailey et al. 2009). Specific motif combinations of both the polyketide genes (type I and type II) were detected using the program GLAM2 (Frith et al. 2008), and sequence logos were constructed.
Statistical data analysis
To calculate relative frequency (RF), the total number of isolates for a genus was divided by the total number of genera cultured from different plant species (El-Shatoury et al. 2013). A diversity index {Shannon’s index (H) and Simpson’s index (Si)}, a richness index (Margalef and Menhinick), and an evenness index (E) were calculated as mentioned previously (Kennedy et al. 1995; Verma et al. 2007, 2009). The variation of positive bioactive isolates within each plant species was also observed with Shannon’s (H) and Simpson’s (Si) diversity indices. RF was expressed as percentages and subjected to ANOVA. All statistical analyses were done with SCaVis2.3 software (https://fossies.org/dox/scavis-2.3/namespacesmtpd.html). TeraPlot 2.0 (http://www.teraplot.com) was used to create the histograms. All experiments were performed in triplicate. Shannon’s index (H) = − ∑ Pi(lnPi); Simpson’s index (Si) = \( 1-\frac{\sum \mathrm{n}\left(\mathrm{n}-1\right)}{\sum \mathrm{N}\left(\mathrm{N}-1\right)} \); Margalef = (S − 1)/lnN; Menhinick = \( \frac{S}{\surd N} \), and Evenness index E = H/lnS were calculated with the formula described by Verma et al. (2007) and Kennedy and Smith (1995), where
- S:
-
numbers of genera encountered,
- n :
-
the total number of isolates of a particular genus,
- N :
-
the total number of isolates of all genera,
- Pi:
-
fraction of entire population made up of particular genus i(Pi = n/N),
- ∑:
-
sum from genus 1 to genus S,
- E:
-
Evenness index,
- H:
-
Shannon diversity index,
- Si:
-
Simpson’s index.
Results
Isolation and genotypic analysis
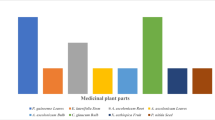
The purification of endophytic isolates and their preliminary characterization yielded a total of 120 morphologically distinct putative endophytic actinobacterial isolates from the medicinal plants. However, some of the isolates retained distinctive morphologies of Streptomyces (Supplementary Table S4). A significant difference [P = 7.22 E-14 and F(21.31) > F(crit = 1.91)] in terms of colonization among the different tissue types (i.e., root, stem, and leaf) was observed. A total of 63 (52.5%) actinobacteria were isolated from the root, 34 (28.33%) from the stem, and 23 (19.16%) from leaves (Fig. 1a and Supplementary Table S5). The actinobacterial proportion differed significantly between the two sampling seasons [P = 1.59E-12 and F = 62 > F(crit = 2.23)] as well as among three sampling sites [(P = 6.59E-07 and F= 6.87 > F(crit = 1.91)] (Fig. 1a, b). Relative abundance was greater in the summer than in the winter. Among the three forest areas, culturability from the plant tissues in GWS was higher than the KNP and NEEP (Supplementary Table S6).
a Endophytic actinomycte assemblages in different plant tissues, i.e., root, stem, and leaf. Highest numbers of actinomycetes were isolated from root followed by stem and leaf. Maximum colonization was recorded in summer than in winter. b Overall colonization of endophytic actinomycetes in three sampling sites. c Relative abundance of endophytic actinomycetes in six medicinal plant species. The genus Streptomyces was found to be common in all the studied plants
Repetitive extragenic palindromic (REP) PCR is one of the most commonly used techniques in bacterial genetic diversity studies to obtain taxonomic resolution. In the present study, the BOX-PCR fingerprinting recorded a broad banding pattern from 500 to 4500 bp while REP-PCR fingerprinting recorded a banding pattern from 500 to 3000 bp (data not shown). The dendrogram obtained from BOX fingerprint resulted in four clusters with three of these clusters concurrently reproduced in the REP-PCR dendrogram; the coefficient of genetic similarity ranged from 0.5 to 1 (Supplementary Fig. S2). The dendrogram of the BOX-PCR fingerprint yielded four major clusters which had similarity coefficient values of > 70%. Furthermore, cluster I was subdivided into four sub-clusters. Sub-cluster I comprised 12 isolates with similarity coefficient values of > 80%; sub-cluster-II had 17 isolates with coefficient values of > 60%; isolates of sub-cluster III and sub-cluster IV had similarity values of approximately 70%. Cluster II was divided into two sub-clusters. Sub-cluster I included 12 isolates having a similarity value of > 60%, and sub-cluster II consisted of 24 isolates with a similarity value of > 70%. Cluster III had two sub-clusters, having 18 isolates with similarity values of > 70%. Cluster IV comprised 15 isolates which had similarity values of > 65%.
The REP-PCR fingerprinting yielded a dendrogram showing three major clusters. Cluster I was composed of 39 isolates with a coefficient value of > 80%. Cluster II, having 34 isolates, was divided into three sub-clusters. Sub-cluster I contained 11 isolates whereas sub-cluster I and sub-cluster III had 16 and 6 isolates, respectively, all sharing a coefficient value > 60%. However, cluster III, with 47 isolates, formed the major cluster and had a coefficient value of > 60%.
Morphological and biochemical characterization of endophytic actinobacterial isolates
Microscopic observations revealed that the isolates were gram-positive, endospore-forming, and had branched networks of hyphae. Most of the isolates showed positive test results for protease, amylase, and catalase activity. However, a few were found to be positive for cellulase, H2S production, HCN production, IAA production, and nitrate reduction (Supplementary Table S4).
Antimicrobial activity of endophytic actinobacteria
Bioassay with test pathogens showed 52 of 120 actinobacterial isolates to possess antimicrobial property with varying intensity. Among the 52 antimicrobial isolates, 44 were found to be active only against bacteria while 3 inhibited only growth of fungi, 6 isolates were found to be active against both bacteria and fungi (Supplementary Table S3). All the 44 antibacterial isolates showed a minimum 10 mm zone of inhibition and maximum zone of 24 mm. Among the antibacterial isolates, EAAG89 was found to be most active against S. aureus (24 mm) and P. syringae (23 mm), followed by the isolate EAAG90, which had inhibition zones of 18 mm and 20 mm against P. syringae and S. aureus, respectively. Additionally, isolate EAAG96 was also found to be active against both P. syringae and S. aureus (Supplementary Table S3). Act30 was the only strain active against E. coli in this study. The isolates EAGB05 and Act60 showed the highest activity against C. albicans (20 mm). The isolate Act60 also showed an inhibitory effect against F. oxysporum (17 mm). However, no activity was found against P. aeruginosa or A. niger.
Taxonomic and phylogenetic placement of endophytes for habitat distribution analysis
A total of 120 distantly related isolates were selected based on genotypic analysis. Based on 16S rRNA sequence analysis, all the 120 actinobacterial strains could be classified into 12 families and 18 different genera (Table 1). The relative frequency of the genera significantly differed among the studied plants [P = 1.84 E-14 and F(10.17) > F(crit = 1.737)]. The genus Streptomyces was evenly distributed among the studied plant species with a high culturability (92 strains across six plant types) while Amycolatopsis, Brevibacterium, Mycobacterium, Nocardia, Leifsonia, Verrucosispora, Isoptericola, Kytococcus, Kitasatospora, Promicromonospora, and Timonella were associated with the single host species (Fig. 1c and Table 1).
The diversity (Shannon’s and Simpson’s) and evenness indices revealed that A. indica (1.496, 0.778), R. serpentina (1.470, 0.858), and E. officinalis (0.975, 0.353) are relatively good habitats for the prospection and isolation of endophytic actinobacteria (Table 2). Based on Margalef’s richness estimator, the highest species richness values were recorded in A. indica, followed by R. serpentina and T. chebula whereas based on Menhinick’s richness index, species richness was high in R. serpentine followed by A. indica. E. officinalis showed the most diverse bioactive strains, having values of 0.358, 0.920, 0.363, and 0.826. Among the sampling sites, plants analyzed from GWS forest ecosystem harbored more diverse communities of actinobacteria compared to KNP and NEEP (Shannon’s = 0.860 and Simpson’s = 0.557) (Table 3).
Further, to observe the relationships among the isolates, 16S rRNA gene sequences were aligned with sequences from types strain retrieved from NCBI-GenBank. Most of the actinobacterial strains were grouped into the family Streptomycetaceae (77.5%). The rest were grouped into Micrococcaceae (5%); Streptosporagiaceae (4.16%); Micromonosporaceae, Microbacteriaceae, and Pseudonocardiaceae (2.5% each); Promicromonosporaceae (1.66%); and Brevibacteriaceae, Intrasporangiaceae, Mycobacteriaceae, Nocardiaceae, and Sanguibacteraceae (1.31% each). Since the Streptomyces was proven to be the dominant genus within the host studied and they conferred broad range activity against most of the pathogens, their intra-generic variation was determined by NJ and UPGMA methods. Further, due to highly divergent alignment nature of the 120 sequences with the reference sequences lead to cluster instability, which was resolved by carrying out separate phylogenetic analysis of the remaining 28 genera (other than Streptomyces) (Fig. 2a, b). For the Streptomyces strains, both NJ and UPGMA methods yielded the same type of tree topology, with two major clusters (Supplementary Fig. S3). The larger cluster I consisted of 88 strains along with all the reference sequences, while the four strains of cluster II (Act10, Act26, Act21, Act44) phylogenetically bifurcated from the rest. The UPGMA method also showed a similar type of clustering pattern, which confirmed the correct phylogenetic placement of each strain.
a NJ phylogenetic tree of rest 28 endophytic actinomycetes strains (black triangle) on 16S rRNA gene sequence alignments rooted with Pseudomonas aeruginosa and b UPGMA phylogenetic tree. Bootstrap values greater than 50% are shown at the nodes and are based on 1000 replicates. Sequence of type strains and related isolates were retrieved from GenBank, and accession numbers appear in parentheses. Scale bar indicates 0.02 substitutions per nucleotide
Similarly, two major clusters were evident among the rest of the actinobacterial isolates, as analyzed separately by both NJ and UPGMA clustering methods (Fig. 2a, b). Cluster I was the major cluster with 26 actinobacterial strains along with reference sequences. The majority of the members of cluster I shared similarities with the Micromonospora and Micrococcus groups. Cluster II, which had only two actinobacterial strains (EAAG52 and EAAG81), was similar to the Mycobacterium group. The topology of the clades and the relative phylogenetic positions of strains obtained with NJ clustering corroborated with UPGMA clustering, which confirmed the correct phylogenetic placement of each strain.
Detection of biosynthetic genes in the active strains and in vitro antimicrobial activity
The actinobacteria were further studied for the presence of genes encoding polyketide synthase I (PKSI) and polyketide synthase II (PKSII). Of the strains analyzed, 84% produced the expected bands for PKSII, while PKSI candidate amplicons were detected only in six strains (16%). The genus Streptomyces had broad activity against most of the pathogens, and the highest detection rates for PKSI and PKSII was found in this genus. Out of 52 active strains, PKSI and PKSII were detected in 33 strains. However, the strains Act19, Act58, and Act74 did not show any antagonistic activity even though PKS I or PKS II was present in these strains. In the same way, strains EAAG89 and EAAG91 showed antimicrobial activity despite the non-detection of PKS I and PKS II genes. Among the isolates studied, only two actinobacterial strains (EAAG95 and EAAG96) showed the prevalence of both PKSI and PKS II.
Phylogeny of ketosynthase domains of the detected polyketide type I and type II
In this study, the keto synthase (KS) domains of PKSI were detected in six isolates and shared 60–70% homology with the KS domains retrieved from GenBank. The phylogenetic tree of PKSI showed two distinct clusters (Fig. 3a). Cluster I consisted three isolates viz. EAAG95 (KX790329), EAAG96 (KX925851), and Act44 (KX925850) with similarity values of > 80%. The EAAG95 and EAAG96 isolates were retrieved from the GWS site while Act44 was from KNP. Similarly, cluster II also had three isolates, Act52 from KNP and EAAG33, Act19 from GWS. These isolates also had similarity values of > 80%.
a An unrooted neighbor-joining tree constructed using aligned KS domain amino acid sequence from type I PKSs (black triangle). Bootstrap values calculated from 1000 replicates. Scale bar represents 0.2 substitutions per amino acid position and b an unrooted neighbor joining tree constructed using aligned KS domain amino acid sequence from type II PKSs (black triangle). Scale bar represents 0.05 substitutions per amino acid position. Evolutionary distances were calculated using Poisson correction model
The phylogeny of PKSII was found to be more diverse than that of PKSI (Fig. 3a, b). The study included 32 strains which had the biosynthetic gene PKS II. Three major clusters (cluster I, cluster II, and cluster III) were observed (Fig. 3b). Cluster I was divided into two sub-clusters. Sub-cluster I had six strains while sub-cluster II had four strains with a similarity value of > 50%. Cluster II consisted of ten strains with a similarity value of > 70%. As shown in Fig. 3b, cluster III had 12 isolates with a similarity value of > 60%. The phylogenetic analysis inferred that PKSII is more widespread than PKSI. It is interesting to note that all the KS domains retrieved from the isolates belonged to all the three different sampling sites.
Motif analysis
GLAM2 accurately predicted the motifs for PKS types I and II. GLAM2 reported nine motifs for the domains of both PKSI and PKSII with a definite score. The best motif for PKSI (highest score = 311.061) and PKSII (highest score = 4087.99) is shown in Supplementary Fig. S4a and 4b. Observed insertion (gray letters) and deletion (dots) in the motif of PKSII were, however, not included in the final sequence logo.
Discussion
Plants are hotspots for diverse groups of endophytic microorganisms. Many of them are responsible for the production of a wide variety of natural bioactive products like antibiotic, antitumor, and anti-infection agents. This study investigated the diversity of endophytic actinobacteria associated with six traditional medicinal plants in three different habitats, as well as the coding potential for the putative antibiotic biosynthetic gene. To our knowledge, this is the first report on the assessment of genetic diversity of medicinal plant-associated endophytic actinobacterial strains and evaluation of their bioactivity in KNP and NEEP in Northeast India. The present study revealed a significant diversity of endophytic actinobacteria among the six medicinal plants considered. There were significant variations in the abundance of actinobacterial isolates from the plant parts that were considered for isolation. The maximum endophytic actinobacteria population was observed in the roots (52.5%), followed by the stems (28.33%) and leaves (19.16%). This corroborated with our previous results (Gohain et al. 2015) and with similar studies from other researchers (Taechowisan and Lumyong 2003; Verma et al. 2009). Since actinobacteria is a common soil-dwelling bacteria and roots are the main source of nutrient and water uptake in plants, endophytic actinomycetes may colonize in the roots of plants more easily than in leaves and stems (Janso and Carter 2010).
A comparative genotypic analysis was performed on the endophytic actinobacterial isolates from six medicinally important plants from three different habitats. The PCR-based fingerprinting techniques (i.e., BOX and REP-PCR) were employed to examine genetic variability among the cultured actinobacterial isolates. There was observable variation in the molecular data. Genomic profiling using BOX-PCR fingerprinting and clustering grouped EAAG94, Act21, EAAG98, EAAG97, Act08, EAAG95, Act35 into the same cluster; however, they did not reveal similar grouping pattern in REP-PCR fingerprinting. Thus, the grouping obtained from BOX and REP-PCR techniques revealed poor correspondence among the 120 isolates. Earlier studies provided similar results (Davelos et al. 2004; Naik et al. 2008). This may be due to the probable chromosomal rearrangements which results in instability of the actinobacteria genome (Dharmalingam and Cullum 1996; Fischer et al. 1997) and is reflected in the genomic fingerprint.
The analysis of 16S rRNA sequences revealed that 77% of the strains share a close relationship to the genus Streptomyces. This is consistent with the findings of earlier studies (Janso and Carter 2010; Passari et al. 2015). The 92 isolates representing 37 different species reflected significant intra-generic diversity of endophytes. A reconstruction of the molecular phylogeny grouped the 92 Streptomyces isolates into two major clusters. Interestingly, all four Streptomyces isolates in cluster II were isolated from the stem of T. chebula, although the host habitats were different. Streptomyces strains Act10 and Act21 were isolated from the GWS while other two strains (Act26 and Act44) were isolated from KNP. The members of cluster II also showed a similar pattern of host dependency, rather than habitat variation. Thus, the actinobacterial strains were host-specific rather than habitat-specific (Holliday 1998). Similarly, intergeneric diversity was noticed in the remaining 28 strains, representing 16 different genera.
The highest species richness index was recorded in A. indica.. The next most species-rich samples were those of R. serpentina and E. officinalis. Again, an even distribution was recorded in A. indica followed by R. serpentina. Genetic diversity was also recorded in T. arjuna and M. koenigii. Although all the plant species have some medicinal values of their own, the highest numbers of bioactive isolates were isolated from E. officinalis and T. chebula. This might be due to differences in plant-specific factors, such as the chemical metabolisms of the plants (Reinhold-Hurek and Hurek 2011).
Furthermore, the existing climatic conditions and seasonal variations are key factors in the colonization of endophytic microorganisms (Barka et al. 2016). We have previously reported the endophytic nature of Promicromonospora thailandica (isolated from E. officinalis), although it was previously reported to be the inhabitant of marine sediments (Thawai and Kudo 2012). Likewise, the habitat variability of Microbacterium testaceum was first observed during this investigation. This is the first report of the presence of Microbacterium testaceum in the root interior of E. officinalis, although its predominance in nature has been described in agronomic and prairie plants (Zinniel et al. 2002).
Moreover, higher endophytic colonization rates are observed in summer than in the winter. This finding is consistent with those of previous reports (Silva et al. 2013; Gohain et al. 2015). The genus Streptomyces was frequently isolated in both seasons while the rest of the 18 genera were recovered during summer only. The greater availability of moisture content during the summer is likely a major driver for this increased colonization (Barka et al. 2016).
Habitat-specific variation was also noticed among the endophytic actinobacterial strains. Altogether, the study revealed the highest endophytic actinobacterial diversity in the GWS. This might be due to environmental and edaphic factors that govern different study sites (Hou et al. 2009). As described earlier, soil pH and moisture content have tremendous effects on variation in actinobacterial populations (Silva et al. 2013; Barka et al. 2016). The soil pH of the sampling sites of the GWS was 6.5 in both the seasons while, in KNP and the NEEP, soil pH levels were 7.4 and 7.2, respectively. Similarly, there was variation in terms of soil moisture contents in the selected study sites. Indeed, the moisture content of these sites varied seasonally. The moisture content of soil at the GWS was 30.84% in summer and 18.21% in winter. These values for KNP soil were 30.31% in the summer and 17.10% in the winter. For NEEP soil, the values were 18.11% in the summer and 14.10% in the winter (Supplementary Table S2). Effect of a moderately acidic soil environment in promoting richer endophytic actinobacterial diversity in the GWS needs to be studied and explored further. A similar type of predisposition was also observed from an earlier investigation (Kim et al. 2003). Similarly, endophytic actinobacterial growth in higher moisture conditions is reported by Barka et al. (2016).
The functional characterization of endophytic actinobacterial isolates was further confirmed by their antimicrobial activities against plant and animal pathogens. The strains possessed higher antibacterial than antifungal activity. Other studies have shown the pronounced antifungal nature of endophytic actinomycetes (Bascom-Slack et al. 2009; Zhao et al. 2011). There is a discrepancy between antimicrobial metabolite profiles and their biological activity, even metabolic profiles of the same species may vary depending upon the chemical properties of the host plant (Paulus et al. 2006). Thus, the complex chemical properties of host plants may influence the process of endophytic metabolite production directly or indirectly.
The bacterial polyketide synthases are multifunctional group of enzymes. The polyketide gene clusters have the capability to produce varied pharmaceutically important metabolites that help the bacteria compete with others in colonizing a niche. Among the bacterial genera, Streptomyces are known as prolific producers of several clinically active compounds. Different biosynthetic gene clusters in the Streptomyces genome are responsible for the production of bioactive compounds. The modular polyketide synthases are one of the important Streptomyces gene cluster responsible for the production of bioactive metabolites, like antibiotics, antiparasitics, insecticides etc. Therefore, the bioactive strains in the present study were screened for the presence of the PKS gene cluster to understand the basis of observed antagonism against tested pathogens.
The studied actinobacterial strains showed a dominance of PKSII (84% of the active strains contained this gene) over PKSI genetic system. The high detection rate of the biosynthetic genes was recorded with Streptomyces. This is supported by previous reports which have found Streptomyces to be one of the major genera of actinomycetes, having the ability to produce nearly 80% of the world’s antibiotics (Li et al. 2008; Passari et al. 2017). However, the observed antimicrobial activity sometimes do not depend on the presence of PKS I and PKS II fragments in the bacterial genome (Zhao et al. 2011; Gohain et al. 2015) as additional biosynthetic genes (such as PKSIII, PKSE, NRPS, phzE, CYP-450, dTGD etc.) can confer antimicrobial properties in these strains (Yuan et al. 2014). Similar predisposition was noticed in some of the actinobacterial strains from the present study. Two possibilities that may be accounted for are (i) endophytic actinobacteria within the plant samples do not harbor PKS biosynthetic genetic cluster or (ii) the divergent nature of the gene sequences resulted in non-priming of target primers during PCR amplification (Miller et al. 2012). Furthermore, some strains did not show significant antimicrobial activity but had positive PCR reactions for PKSI or PKSII genes. In this case, the concentration of active fraction of the secondary metabolites could be too low to inhibit the growth of the pathogens (Zhao et al. 2011; Passari et al. 2015) or due to cryptic nature of the gene cluster. The pathways for encoding these biosynthetic genes may not be active under the studied environmental conditions but could be activated under modified conditions (Passari et al. 2015).
Moreover, the occurrence of PKSI and PKSII genes, taken together with the detection of bioactivity and the diversity of actinobacteria in the present study, clearly explains the nature of endophytic actinobacterial strains as promising sources of natural bioactive agents (Wu and Jiang 2012). The sequence analysis of the biosynthetic gene and phylogenetic analysis provide an insight into the activity of the KS domains of PKS type I and type II genes. Phylogenetic trees were constructed to recount the structural and functional connections in these genes. Two distinct phylogenetic clades were obtained from the detected KS domains. This indicates that the genes associated with these organisms have not been isolated so far or that, until now, these organisms were not studied for their PKS gene pathways (Li et al. 2008).
The study could not find any distinctive motif from the isolate EAAG33, though all the sequences were selected for motif analysis. The study determined the best motif by comparing it with the rest of the sequences (EAAG95, EAAG96, Act19, Act44, and Act52). In addition to this, the isolate EAAG33 was found to diverge only slightly from cluster II in the phylogenetic tree. Therefore, this may be considered a unique KS (Li et al. 2008). Similarly, the tree topology of PKSII depicted three distinct clusters, and the best motif was found to have a score of 5363.97 and utilized in sequence logos construction. These logos graphically represent sequence conservation rates of amino or nucleotide sequences and also depict the diversity of sequences (Bailey et al. 2009). Stronger motifs are always indicated by higher scores (Frith et al. 2008).
The overall investigation verified the diversity of endophytic actinobacteria as well as the novelty of the genes responsible for synthesizing novel bioactive compounds. Most of the bioactive endophytic actinobacterial isolates and their active KS domains were retrieved in the samples collected from the GWS, providing information about the possibility of harnessing novel, diverse bioactive compounds from the plant resources for future studies. Most of the microbial communities produce bioactive secondary metabolites in their natural habitats under different environmental conditions (temperature, pH, moisture content, etc.) (Barka et al. 2016). Moreover, the study has shown that the investigated ethnobotanical medicinal plants are a reservoir of bioactive and antagonistic endophytic actinobacteria, which could be an ideal bioresource for the extraction of novel bioactive compounds in the near future.
A comparative genotypic analysis was performed on the endophytic actinobacterial isolates from six medicinally important plants from three different habitats. The PCR-based fingerprinting techniques (i.e., BOX and REP-PCR) were employed to examine genetic variability among the cultured actinobacterial isolates. There was observable variation in 16S gene molecular data. Genomic profiling using BOX-PCR fingerprinting and clustering grouped EAAG94, Act21, EAAG98, EAAG97, Act08, EAAG95, Act35 into the same cluster; however, they did not reveal similar grouping pattern in REP-PCR fingerprinting. Thus, the grouping obtained from BOX and REP PCR techniques revealed poor correspondence among the 120 isolates. Earlier studies provided similar results (Davelos et al. 2004; Naik et al. 2008). This may be due to the probable chromosomal rearrangements in the genome and the DNA amplification which reflects the instability of the actinobacteria genome (Dharmalingam and Cullum 1996; Fischer et al. 1997).
The analysis of 16S rRNA sequences revealed that 77% of the strains share a close relationship to the genus Streptomyces. This is consistent with the findings of earlier studies (Janso and Carter 2010; Passari et al. 2015). The 92 isolates representing 37 different species reflected significant intra-generic diversity of endophytes. A reconstruction of the molecular phylogeny grouped the 92 Streptomyces isolates into two major clusters. Interestingly, all four Streptomyces isolates in cluster II were isolated from the stem of T. chebula, although the host habitats were different. Streptomyces strains Act10 and Act21 were isolated from the GWS while other two strains (Act26 and Act44) were isolated from KNP. The members of cluster II also showed a similar pattern of host dependency, rather than habitat variation. Thus, the actinobacterial strains were host-specific rather than habitat-specific (Holliday 1998). Similarly, intergeneric diversity had been noticed in the remaining 28 strains, representing 16 different genera.
The highest species richness index recorded in A. indica. Accordingly, A. indica has diverse types of actinobacteria. The next most species-rich samples were those of R. serpentina and E. officinalis. Again, an even distribution was recorded in A. indica followed by R. serpentina. Genetic diversity was also recorded in T. arjuna and M. koenigii. Although all the plant species have some medicinal values of their own, the highest numbers of bioactive isolates were isolated from E. officinalis and T. chebula. This might be due to differences in plant-specific factors, such as the chemical metabolisms of the plants (Reinhold-Hurek and Hurek 2011).
Furthermore, the existing climatic conditions and seasonal variations are key factors in the colonization of endophytic microorganisms (Barka et al. 2016). We have previously reported the endophytic nature of Promicromonospora thailandica (isolated from E. officinalis), although it was previously reported to be the inhabitant of marine sediments (Thawai and Kudo 2012). Likewise, the habitat variability of Microbacterium testaceum was first observed during this investigation. This is the first report of the presence of Microbacterium testaceum in the root interior of E. officinalis, although its predominance in nature has been described in agronomic and prairie plants (Zinniel et al. 2002).
Moreover, higher endophytic colonization rates are observed in summer than in the winter. This finding is consistent with those of previous reports (Silva et al. 2013; Gohain et al. 2015). The genus Streptomyces was frequently isolated in both seasons while the rest of the 18 genera were recovered during summer only. The greater availability of moisture content during the summer is likely a major driver for this increased colonization (Barka et al. 2016).
Moreover, habitat specific variation was noticed among the endophytic actinobacterial strains. Overall, the study revealed the highest endophytic actinobacterial diversity in the GWS. This might be due to environmental and edaphic factors that govern different study sites (Hou et al. 2009). As described earlier, soil pH and moisture content have tremendous effects on variation in actinobacterial populations (Silva et al. 2013; Barka et al. 2016). The soil pH of the sampling sites of the GWS was 6.5 in both the seasons while, in KNP and the NEEP, soil pH levels were 7.4 and 7.2, respectively. Similarly, there was variation in terms of soil moisture contents in the selected study sites. Indeed, the moisture content of these sites varied seasonally. The moisture content of soil at the GWS was 30.84% in summer and 18.21% in winter. These values for KNP soil were 30.31% in the summer and 17.10% in the winter. For NEEP soil, the values were 18.11% in the summer and 14.10% in the winter (Supplementary Table S2). Thus, the highest endophytic actinobacterial diversity in the GWS indicates the effect of a moderately acidic soil environment. A similar type of predisposition was also observed from an earlier investigation (Kim et al. 2003). Similarly, endophytic actinobacterial growth in higher moisture conditions is reported by Barka et al. (2016).
The functional characterization of endophytic actinobacterial isolates was further confirmed by their antimicrobial activities against plant and animal pathogens. The strains possessed higher antibacterial activity than antifungal activity. However, previous studies have shown the pronounced antifungal nature of endophytic actinomycetes (Bascom-Slack et al. 2009; Zhao et al. 2011). There is a discrepancy between antimicrobial metabolite profiles and their biological activity, even when isolates of the same species are related to the chemical properties of the host plant (Paulus et al. 2006). Thus, the complex chemical properties of host plants may influence the process of endophytic metabolite production directly or indirectly.
The bacterial polyketide synthases are multifunctional group of enzymes. The polyketide gene clusters have the capability to produce varied pharmaceutically important antimicrobial, immunosuppressive or anticancer compounds that make the bacteria potentially active against various infectious diseases. Among the bacterial genera, Streptomyces are known for the prolific producers of several clinically active compounds. Different biosynthetic gene clusters in the Streptomyces genome are responsible for the production of bioactive compounds. The modular polyketide synthases are one of the important Streptomyces gene cluster responsible for the production of bioactive metabolites, like antibiotics, antiparasitics, insecticides etc. Therefore, the bioactive strains of the present study were screened for the presence of the PKS gene cluster to understand the basis of the observed antagonistic activity of the selected strains.
The studied actinobacterial strains showed a dominance of the PKSII biosynthetic gene (84% of the active strains contained this gene) over PKSI genetic system. The high detection rate of the biosynthetic genes was recorded with Streptomyces. This is supported by previous reports which have found Streptomyces to be one of the major genera of actinomycetes, having the ability to produce nearly 80% of the world’s antibiotics (Li et al. 2008; Passari et al. 2017). However, the observed antimicrobial activity sometimes does not depend on the presence of active segments of PKS I and PKS II in the bacterial genome (Zhao et al. 2011; Gohain et al. 2015) or the antimicrobial activity in these strains may have been caused by some other biosynthetic genes such as PKSIII, PKSE, NRPS, phzE, CYP-450, dTGD etc. (Yuan et al. 2014). Similar predisposition was noticed in some of the actinobacterial strains from the present study. Two possibilities that may be accounted for are (i) endophytic actinobacteria within the plant samples do not harbor PKS biosynthetic genetic cluster or (ii) the divergent nature of the gene sequences resulted in non-priming of target primers during PCR amplification (Miller et al. 2012). Furthermore, some strains did not show significant antimicrobial activity but had positive PCR reactions for PKSI or PKSII genes. In this case, the concentration of secondary metabolites of actinobacterial origins could be too low to inhibit the growth of the pathogens (Zhao et al. 2011; Passari et al. 2015). Thus, it is evident that some of the pathways for encoding these biosynthetic genes may not be active under the studied environmental conditions but could be activated under modified conditions (Passari et al. 2015).
Moreover, the occurrence of PKSI and PKSII genes, taken together with the detection of bioactivity and the diversity of actinobacteria in the present study, clearly explains the nature of endophytic actinobacterial strains as promising sources of natural bioactive agents (Wu and Jiang 2012). The sequence analysis of the biosynthetic gene and phylogenetic analysis provide an insight into the activity of the KS domains of PKS type I and type II genes. Phylogenetic trees were constructed to recount the structural and functional connections of these genes. Two distinct phylogenetic clades were obtained from the detected KS domains. This indicates that the genes associated with these organisms have not been isolated so far or that, until now, these organisms were not studied for their PKS gene pathways (Li et al. 2008).
Again, the study could not find any distinctive motif from the isolate EAAG33, though all the sequences were selected for motif analysis. The study determined the best motif by comparing it with the rest of the sequences (EAAG95, EAAG96, Act19, Act44, and Act52). In addition to this, the isolate EAAG33 was found to diverge only slightly from cluster II in the phylogenetic tree. Therefore, this may be considered a unique KS (Li et al. 2008). Similarly, the tree topology of PKSII depicted three distinct clusters, and the best motif was found to have a score of 5363.97, and sequence logos were constructed. These logos graphically represent sequence conservation rates of amino or nucleotide sequences and also depict the diversity of sequences (Bailey et al. 2009). Stronger motifs are always indicated by higher scores (Frith et al. 2008).
The overall investigation verified the diversity of endophytic actinobacteria as well as the novelty of the genes responsible for synthesizing novel bioactive compounds. Most of the bioactive endophytic actinobacterial isolates and their active KS domains were retrieved in the samples collected from the GWS, providing information about the possibility of harnessing novel, diverse bioactive compounds from the plant resources for future studies. Most of the microbial communities produce bioactive secondary metabolites in their natural habitats under different environmental conditions (temperature, pH, moisture content, etc.) (Barka et al. 2016). Moreover, the study has shown that the investigated ethnobotanical medicinal plants are a reservoir of bioactive and antagonistic endophytic actinobacteria, which could be an ideal bioresource for the extraction of novel bioactive compounds in the future.
Abbreviations
- PKS1:
-
Polyketide synthase type I
- PKSII:
-
Polyketide synthase type II
- GWS:
-
Gibbon Wildlife Sanctuary
- KNP:
-
Kaziranga National Park
- NEEP:
-
North East Ecological Park
References
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2005) At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71:7724–7736
Ashelford KE, Chuzhanova NA, Jones AJ, Weightman AJ (2006) New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72:5734–5741
Bacon CW, White JF Jr (2000) Microbial endophytes. Marcel Dekker, New York
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Barka EA, Vatsa P, Sanchez L, Vaillant NG, Jacquard C, Klenk HP, Clément C, Ouhduch Y, van Wezel GP (2016) Taxonomy, physiology, and natural products of Actinobacteria. Microbiol Mol Biol Rev 80:1–43
Bascom-Slack CA, Ma C, Moore E, Babbs B, Fenn K, Greene JS, Hann BD, Keehner J, Kelley-Swift EG, Kembaiyan V, Lee SJ, Li P, Light DY, Lin EH, Schorn MA, Vekhter D, Boulanger LA, Hess WM, Vargas PN, Strobel GA, Strobel SA (2009) Multiple, novel biologically active endophytic actinomycetes isolated from upper Amazonian rainforests. Microb Ecol 58:374–383
Bergey DH, Holt JG, Krieg NR, Sneath PHA (1994) Bergey’s manual of determinative bacteriology, 9th edn. Williams & Wilkins Co., Baltimore, pp 1860–1937
Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P (2013) Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838
Castillo U, Harper JK, Strobel GA, Sears J, Alesi K, Ford E, Lin J, Hunter M, Maranta M, Ge H, Yaver D, Jensen JB, Porter H, Robison R, Millar D, Hess WM, Condron M, Teplow D (2003) Kakadumycins, novel antibiotics from Streptomyces sp. NRRL 30566, an endophyte of Grevillea pteridifolia. FEMS Microbiol Lett 224:183–190
Davelos AL, Xiao K, Samac DA, Martin AP, Kinkel LL (2004) Spatial variation in Streptomyces genetic composition and diversity in a prairie soil. Microb Ecol 48:601–612
Dharmalingam K, Cullum J (1996) Genetic instability in Streptomyces. J Biosci 21:433–444
Ding L, Maier A, Fiebig HH, Lin WH, Hertweck C (2011) A family of multi cyclic indol osesquiterpenes from a bacterial endophyte. Org Biomol Chem 9:4029–4031
Duangmal K, Thamchaipenet A, Ara I, Matsumoto A, Takahashi Y (2008) Kineococcus gynurae sp. nov., isolated from a Thai medicinal plant. Int J Syst Evol Microbiol 58:2439–2442
El-Shatoury SA, El-Kraly OA, Trujillo ME, El-Kazzaz WM, El-Din E-SG, Dewedar A (2013) A generic and functional diversity in endophytic actinomycetes from wild Compositae plant species at South Sinai-Egypt. Res Microbiol 164:761–769
Ezra D, Castillo UF, Strobel GA, Hess WM, Porter H, Jensen JB, Condron MAM, Teplow DB, Sears J, Maranta M, Hunter M, Weber B, Yaver D (2004) Coronamycins, peptide antibiotics produced by a verticillate Streptomyces sp. (MSU2110) endophytic on Monstera sp. Microbiology 150:785–793
Fischer G, Kyriacou A, Decaris B, Leblond P (1997) Genetic instability and its possible evolutionary implications on the chromosomal structure of Streptomyces. Biochimie 79:555–558
Frith MC, Saunders NFW, Kobe B, Bailey TL (2008) Discovering sequence motifs with arbitrary insertions and deletions. PLoS Comput Biol 4(5):e1000071
Gohain A, Gogoi A, Debnath R, Yadav A, Singh BP, Gupta VK, Sharma R, Saikia R (2015) Antimicrobial biosynthetic potential and genetic diversity of endophytic actinomycetes associated with medicinal plants. FEMS Microbiol Lett 362. https://doi.org/10.1093/femsle/fnv158
Hankin L, Anagnostakis SL (1975) The Use of Solid Media for Detection of Enzyme Production by Fungi. Mycologia 67:597–607AAA
Haque SF, Sen SK, Pal SC. (1992) Screening and identification of antibiotic producing strains of Streptomyces. Hindustan Antibiot Bull 34:76–84
Holliday P (1998) A dictionary of plant pathology. Cambridge University Press, Cambridge
Hou BC, Wang ET, Li Y, Jia RZ, Chen WF, Man CX, Sui XH, Chen WX (2009) Rhizobial resource associated with epidemic legumes in Tibet. Microb Ecol 57:69–81
Inahashi Y, Iwatsuki M, Ishiyama A, Namatame M, Tsukashima AN, Matsumoto A, Hirose T, Sunazuka T, Yamada H, Otoguro K, Takahashi Y, Ōmura Kazuro Shiomi S (2011) Spoxazomicins AC, novel antitrypanosomal alkaloids produced by an endophytic actinomycete, Streptosporangium oxazolinicum K070460(T). J Antibiot 64:303–307
Janso JE, Carter GT (2010) Biosynthetic potential of phylogenetically unique endophytic actinomycetes from tropical plants. Appl Environ Microbiol 76:4377–4386
Kennedy AC, Smith KL (1995) Soil bacterial diversity and ecosystem functioning. Plant Soil 170:75–86
Kim SB, Lonsdale J, Seong CN, Goodfellow M (2003) Streptacidiphilus gen. Nov., acidophilic actinomycetes with wall chemotype I and emendation of the family Streptomycetaceae (Waksman and Henrici (1943)) emend. Rainey et al. 1997. Antonie Van Leeuwenhoek 83:107–116
Li J, Zhao G, Chen H, Wang H, Qin S, Zhu W, Xu L, Jiang C, Li W (2008) Antitumour and antimicrobial activities of endophytic streptomycetes from pharmaceutical plants in rainforest. Letters in Applied Microbiology 47:574–580
Mehdi RBA, Sioud S, Fguira LFB, Bejar S, Mellouli L (2006) Purification and structure determination of four bioactive molecules from a newly isolated Streptomyces sp. TN97 strain. Proc Biochem 41:1506–1513
Miller KI, Qing C, Man Yuen Sze D, Neilan BA (2012) Investigation of the biosynthetic potential of endophytes in traditional Chinese anticancer herbs. PLoS One 7(5):e35953
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GA, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858
Naik PR, Sahoo N, Goswami D, Ayyadurai N, Sakthivel N (2008) Genetic and functional diversity among fluorescent pseudomonads isolated from the rhizosphere of banana. Microb Ecol 56:492–504
Newman DJ, Cragg GM (2007) Natural products as sources of new drugs over the last 25 years. J Nat Prod 70(3):461–477
Passari AK, Mishra VK, Saikia R, Gupta VK, Singh BP (2015) Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front Microbiol 6. https://doi.org/10.3389/fmicb.2015.00273
Passari AK, Mishra VK, Singh G, Singh P, Kumar B, Gupta VK, Sharma RK, Saikia R, Donovan AO, Singh BP (2017) Insights into the functionality of endophytic actinobacteria with a focus on their biosynthetic potential and secondary metabolites production. Sci Rep 7:11809. https://doi.org/10.1038/s41598-017-12235-4
Pathom-Aree W, Stach JE, Ward AC, Horikoshi K, Bull AT, Goodfellow M (2006) Diversity of actinomycetes isolated from challenger deep sediment (10,898 m) from the mariana trench. Extremophiles 10, 181–189
Paulus B, Kanowski J, Gadek P, Hyde KD (2006) Diversity and distribution of saprobic micro fungi in leaf litter of an Australian tropical rainforest. Mycol Res 110:1441–1454
Pawar SS, Birand AC, Ahmed MF, Sengupta S, Shankar Raman TR (2007) Conservation biogeography in north-East India: hierarchical analysis of cross-taxon distributional congruence. Divers Distrib 13:53–65
Reinhold-Hurek B, Hurek T (2011) Living inside plants: bacterial endophytes. Curr Opin Plant Biol 14:435–443
Saadoun I, Muhana A (2008) Optimal production conditions, extraction, partial purification and characterization of inhibitory compound(s) produced by Streptomyces ds-104 isolate against multi-drug resistant Candida albicans. Curr Trends Biotechnol Pharm 2:402–420
Schulz B, Wanke U, Draeger S, Aust HJ (1993) Endophytes from herbaceous plants and shrubs: effectiveness of surface sterilization methods. Mycol Res 97:1447–1450
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Silva MS, Sales AN, Magalhães-Guedes KT, Dias DRSS, Schwan RF (2013) Brazilian Cerrado soil actinobacteria ecology. BioMed Res Int. https://doi.org/10.1155/2013/503805
Smith NC, Hennesy J, Stead DE (2001) Repetitive sequence-derived PCR profiling using the BOX-A1 primer for rapid identification of plant pathogen Clavibacter michiganensis ssp. sepedonicus. Eur J Plant Pathol 107:739–748
Strobel G, Daisy B, Castillo U, Harper J (2004) Natural products from endophytic microorganisms. J Nat Prod 67:257–268
Taechowisan T, Lumyong S (2003) Activity of endophytic actinomycetes from roots of Zingiber officinale and Alpinia galangal against phytopathogenic fungi. Ann Microbiol 53:291–298
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thawai C, Kudo T (2012) Promicromonospora thailandica sp. nov, isolated from marine sediment. Int J Syst Evol Microbiol 62:2140–2144
Theantana T, Hyde KD, Lumyong S. (2007) Asparaginase production by endophytic fungi isolated from some Thai medicinal plants. KMITL Science and Technology Journal 7: 13-18
Tiwari KC, Sharma BN, Majumder R, Pandey G (1993) Studies in medicinal plants of Khasi and Jayantiya hills in north east region of India. J Econ Bot Res 17:275–281
Verma VC, Gond SK, Kumar A, Kharwar RN, Strobel G (2007) The endophytic mycofflora of bark, leaf, and stem tissues of Azadirachta indica a. Juss (neem) from Varanasi (India). Microb Ecol 54:119–125
Verma VC, Gond SK, Kumar A, Mishra A, Kharwar RN, Gange AC (2009) Endophytic actinomycetes from Azadirachta indica a. Juss.: isolation, diversity, and anti-microbial activity. Microb Ecol 57:749–756
Versalovic J, Schneider M, de Brulin FJ, Lupski JR (1994) Genomic fingerprinting of bacteria using repetitive sequence based polymerase chain reaction. Methods Mol Cell Biol 5:25–40
Wu SL, Jiang LM (2012) Recent advances in mangrove actinomycetes. Curr Biotechnol 2:335–340
Yu H, Zhang L, Li L, Zheng C, Guo L, Li W, Sun P, Qin L (2010) Recent developments and future prospects of antimicrobial metabolites produced by endophytes. Microbiol Res 165:437–449
Yuan M, Yu Y, Li HR, Dong N, Zhang XH (2014) Phylogenetic diversity and biological activity of actinobacteria isolated from the Chukchi shelf marine sediments in the Arctic Ocean. Mar Drugs 12:1281–1297
Zhao K, Penttinen P, Guan T, Xiao J, Chen Q, Xu J, Lindström K, Zhang L, Zhang X, Strobel GA (2011) The diversity and antimicrobial activity of endophytic actinomycetes isolated from medicinal plants in Panxi plateau, China. Curr Microbiol 62:182–190
Zinniel DK, Lambrecht P, Harris NB, Feng Z, Kuczmarski D, Higley P, Ishimaru CA, Arunakumari A, Barletta RG, Vidaver AK (2002) Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl Environ Microbiol 68:2198–2208
Acknowledgements
Authors are thankful to the DBT-Bioinformatics Infrastructure Facility (BIF) Centre, CSIR-NEIST for providing computational facility and also to the Director, CSIR-NEIST for the necessary laboratory resources.
Funding
The work was supported by grants from the DBT sponsored twinning project (No. BT/209/NE/TBP/2011 dated 30/05/2012), the Government of India, New Delhi.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 3871 kb)
Rights and permissions
About this article
Cite this article
Gohain, A., Sarma, R.K., Debnath, R. et al. Phylogenetic affiliation and antimicrobial effects of endophytic actinobacteria associated with medicinal plants: prevalence of polyketide synthase type II in antimicrobial strains. Folia Microbiol 64, 481–496 (2019). https://doi.org/10.1007/s12223-018-00673-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-018-00673-0