Abstract
This study aims to evaluate the potential of filamentous fungi consortiums to design a bioprocess for producing hydrolytic enzymatic cocktails on lignocellulosic biomass. Four different microorganisms were analyzed in solid and submerged monoculture fermentation systems: Aspergillus niger, Trichoderma harzianum, Rhizopus oryzae, and Thermomyces lanuginosus. A. niger and R. oryzae were selected as the most suitable to form a consortium. Two different inoculation strategies were assessed, one was simultaneous and the other was a 12-h post-phase inoculation. Xylanase, cellulase, amylase, pectinase, and protease activities were evaluated in mono and mixed cultures. Furthermore, the secretomes of the fermentation systems were analyzed by mass spectrometry to compare and determine the difference in the protein profiles excreted. The mixed culture of A. niger and R. oryzae, which were inoculated simultaneously in solid-state fermentation, showed the highest levels of enzyme at 96 h, 30°C, 1 × 107 conidia, and 65% relative humidity. The proportion of xylanase (98 IU/gds), cellulase (27 IU/gds), amylase (30 IU/gds), pectinase (21 IU/gds) and protease (108,000 IU/gds) was enhanced by 25% regarding the monoculture, demonstrating that the use of microbial consortiums can be a promising alternative to obtain enzymatic cocktails with high synergism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the last years, one of the most important challenges of enzyme market has been the isolation and development of enzyme-producing microorganisms to meet the demands of a constantly growing market. This continued growth introduced enzymes into a wide variety of industrial processes and products like textiles, detergent, paper, and second-generation biofuel production industries [1]. This market is estimated to reach $ 8.5 billion by 2022 [2].
Most of the enzymes produced by this industry have hydrolytic activity and are the most demanded by the second-generation biofuel industry. The synergistic effect of multiple different functional hydrolytic enzymes is required for the hydrolysis of lignocellulosic material in order to obtain fermentable sugars to be converted into biofuels [3, 4]. The formulation of these hydrolytic enzymes with a defined concentration is considered an enzymatic cocktail [5].
The enzymes that conform the cocktails are generally produced through monocultures of only one microorganism in submerged (SmF) or solid fermentation (SSF) and in many cases using lignocellulosic waste as substrates [6, 7]. In contrast, in a natural system, lignocellulose degradation is carried out by multiple lignocellulolytic microorganisms, being fungi the most capable of producing a variety of lignocellulolytic enzymes. The production of enzymes by filamentous fungi presents many advantages when compared to those produced by yeast and bacteria, such as higher yields and the secretion of these enzymes to the extracellular medium [8]. These microorganisms establish synergistic relationships among each other to enhance the substrate degradation rates [9]. For this reason, fungi mixed cultures are a novel strategy to produce an enzymatic cocktail enriched in a unique fermentation step avoiding having to carry out successive fungal monocultures.
Cellulose, pectin, and hemicellulose are the main components of lignocellulosic biomass forming complex networks on which filamentous fungi grow, producing different combinations of xylanase, cellulase, and pectinase to degrade these structures [10]. Filamentous fungi such as Aspergillus ssp., Trichoderma ssp., and Thermomyces ssp. are some of the most studied cellulolytic and hemicellulolytic microorganisms. Mixed fermentation of fungi with different catalytic potentials is a novel strategy to improve the yield of enzyme production and obtain a blend of enzymes. This strategy can be adopted by biorefineries or lignocellulose-based bioproduction industries [11]. Yadav et al. [12] propose that a fungal consortium conformed by filamentous fungi is a tool to achieve the global demand for biofuels.
The aim of this work was to design a strategy to improve hydrolytic enzyme production by filamentous fungi. After evaluating the compatibility between different fungi strains and their enzymatic production profile, we selected A. niger and R. oryzae for being the most prominent to construct an efficient fungal consortium for the expression of protease and lignocellulolytic enzymes (xylanolytic, cellulolytic, amylolytic, and pectinolytic), by the utilization of lignocellulolytic substrates in SmF and SSF.
A combination of different lignocellulosic substrates orange peel (OP), Brewer’s spent grain (BSG), soybean hulls (SH), and by-products obtained from the processing of wheat flour (WF) were selected as their rich compositions for fungi growth and enzyme production [10, 13]. Furthermore, the different enzyme profiles produced by mono and mixed cultures were studied by SDS-page and mass spectrometry.
Materials and Methods
Chemicals and Substrates
Beechwood xylan, carboxymethyl cellulose, pectin and starch, 3,5-dinitrosalicylic acid (DNS), D-xylose, D-glucose, bicinchoninic acid, and bovine serum albumin were purchased from Sigma (St. Louis, MO, USA). BSG and SH were kindly provided by Nomada microbrewer and Molinos Río de la Plata S. A. respectively (Rosario, Argentina). The WF was kindly provided by Moliser S. A. (Venado Tuerto, Argentina) and the OP was obtained from a local producer. All lignocellulosic substrates were dry to continuous weight, ground in a laboratory grinder (Mod. TDMC, Tecno Dalvo, Argentina), and sieved through a 0.841 mm sieve. Finally, they were stored at − 20 °C.
Microorganisms
The strain of Aspergillus niger NRRL3 was provided by the culture collection of Agricultural Research Service, USDA. The strain of Thermomyces lanuginosus CBS 288.54 was provided by CBS-KNAW Fungal Biodiversity Center (an Institute of the Royal Netherlands Academy of Arts and Sciences). The strains of Rhizopus oryzae MUCL 28,168 and Trichoderma harzianum T104 were provided by the Department of Food Research of the Faculty of Chemistry of the Autonomous University of Coahuila, Saltillo, Mexico. The inoculums were prepared by culturing the microorganisms on potato-glucose agar medium for 5 days at 30 °C in an Erlenmeyer flask. Then, asexual propagules (spores and conidia) were suspended on 20% w/v glycerol and stored at − 20 °C. Propagule concentration was determined using a Neubauer chamber as described by Grigoryev [10].
Enzyme Production
Monoculture Fermentation
SmF and SSF were carried out for enzyme production. In both cases, the fermentation was performed in 250-mL Erlenmeyer flasks considered as bioreactors. The bioreactors were sterilized with 4 g of dry substrate (gds) of a homogeneous mixture containing 1 gds of each lignocellulosic substrate (BSG, SH, WF, and OP) and an appropriate volume of distilled water. The fermentable mass, containing 50 mL of distilled water in SmF and 10 mL in SSF, was inoculated with an appropriate volume of inoculums to reach a final concentration of 1 × 106 conidia/mL in SmF and SSF systems, and incubated at 30 °C for 5 days. The SmF systems were incubated under continuous stirring (125 rpm). After the incubation, the fermented broth of SSF was obtained by adding 30 mL of distilled water to each bioreactor. Both systems were shaken for 30–35 min using a shaker at 140 rpm agitation speed, centrifuged at 3200 rpm for 5 min in order to remove fungal mycelia, and then filtered through a 0.45 µm membrane [14]. Samples from both systems were taken daily and the obtained culture supernatant was directly employed for enzyme assays to determine the fermentation day (24, 48, 72, 96, or 120 h) that shows the highest enzymatic activity. The systems were performed in triplicate and in a randomized distribution.
Mixed Culture Fermentation
The enzyme production of the mixed fermentation was performed as described before for both SmF and SSF. The substrate was inoculated with 1 × 106 conidia/mL in a 1:1 ratio by both fungi. According to the growth rates of each species constituting the fungal consortium, two models of inoculation were tested: (A) simultaneous and (B) out of phase for 12 h. This was done to make a growth comparison at maximum speed with their monocultures.
The evaluation of the growth and production of enzymes by fungal consortia was performed in the same conditions as in monoculture fermentations. The enzymatic activity and total proteins were determined on the enzymatic extract. In order to compare the enzymatic activities obtained in SmF and SSF, the total IU was calculated as follows:
Then, the IU value was divided by the grams of dry substrates used in each fermentation and the activity was informed as IU/gds.
The systems were performed in triplicate and in a randomized distribution.
Enzymatic Activity and Total Protein Assays
The enzymatic activities of xylanolytic enzymes (XE), cellulolytic enzymes (CE), pectinolytic enzymes (PE), and amylolytic enzymes (AE) in the enzymatic extracts from all fermentations were analyzed.
Beechwood xylan (1% w/v), carboxymethyl cellulose (1% w/v), pectin (1% w/v), and starch (1% w/v) solutions were incubated with the required volume of enzymatic extract in a 50 mM citrate buffer (pH 5.30) at 50 °C for 10 min to determine XE [15], CE [16], PE [17], and AE [18] activity, respectively. The reducing groups released after the incubation period were established by the DNS method [19]. The absorbance of each sample was measured at 560 nm [20]. Solutions of the substrate alone and enzyme alone were used as controls. Calibration curves with D-xylose, D-glucose, and glucuronic acid were carried out under the same experimental conditions as the samples. A unit of enzymatic activity is defined as the amount of enzyme required to release 1 µmol of the equivalent reducing sugar per minute (μmol sugar/min).
Proteolytic activity was assayed according to the method described by Castro and Sato [21]. The reaction mixture, containing azocasein (1% w/v), 20 mM Tris–HCl buffer (pH 8.00), and the enzymatic extract was incubated at 37 °C for 30 min. The incubation was stopped by adding TCA (10% w/v) and then centrifuged at 5500 rpm for 15 min. The supernatant was neutralized with 1.8 N NaOH and the absorbance was measured at 420 nm.
The total extracellular proteins excreted were quantified using the bicinchoninic acid method described by Smith, where bovine serum albumin was used as the standard protein [22].
Stability Determination
The enzymatic stability of the extract produced by A. niger against the protease activity of the enzymatic extracts produced by R. oryzae and T. lanuginosus was evaluated. The enzymatic extract collected on the last day of fermentation was centrifuged at 8000 rpm for 20 min at 4 °C and filtered aseptically through a sterile membrane with a 0.45-μm pore size. The fungal extracts of A. niger were mixed with those of R. oryzae and T. lanuginosus in equal parts in Falcon tubes at a final volume of 10 mL for 21 days at 20 °C. The stability of the enzymatic extract was monitored by measuring XE, CE, and PE activity in all systems daily.
Growth Rate of Individual Fungal Species
For the individual growth studies, the fungus was inoculated with 1 × 106 conidia/mL onto the middle of a Petri plate containing 4 gds and incubated at 30 °C. The mycelial growth ratio of the fungal colony—growth rate per day (mm/day)—was calculated according to Mead and coauthors [23] as follows:
where Gr is the growth rate, DGf is the final diametral growth expressed in mm, DGi is the initial diametral growth expressed in mm, Tf is the final time in which fungal growth ends or final day, and Ti is the initial time.
The inhibition behavior shown by the different microorganisms was grouped [23] as follows:
-
Negative: absence of a zone of inhibition and normal growth of the fungal colony, similar to the control.
-
Positive: presence of a defined zone of inhibition.
Interaction Studies
To study the interactions between A. niger and R. oryzae, equal conidia concentration of both fungi were inoculated 40 mm apart from each other in the same Petri plate containing the substrate mixture described in “Enzyme Production.” Controls of both species were done by the cultivation of each one in different Petri plates. The plates were incubated at a controlled temperature of 30 °C for 10 days. The interactions were visually analyzed using the postulates enunciated by Skidmore and Dickinson in 1976 [24] and Stahl and Christensen in 1992 [25], both based on the observations made by Porter in 1924 [26].
Antagonism Studies
The evaluation of antagonism was carried out according to the M27-Ed4 document, Wayne, PA: Clinical and Laboratory Standards Institute [27]. The enzymatic extracts of A. niger and R. oryzae, collected on the last day of both fermentations, were centrifuged at 8000 rpm for 20 min at 4 °C. The supernatants were supplemented with a culture medium consisting of 2% w/v glucose as a carbon source and 1% w/v peptone as a nitrogen source. Both supernatant and culture medium was filtered aseptically through a sterile membrane with 0.22-μm pore size and stored at 4 °C. Finally, combinations of serial dilutions of sterile fungal extracts supplemented with culture medium and inoculated with A. niger and R. oryzae were tested to determine the minimum inhibitory concentration (MIC) in 96-well microplates. Table 1 shows the protein concentration of serial dilutions of the sterile fungal extracts. Three controls were included: an extract sterility control (culture medium and extract); a culture medium sterility control (culture medium); and a growth control (culture medium and inoculum of the fungus to be evaluated). The microplates were homogenized at 150 rpm for 5 min on an orbital shaker platform Innova 4000 (New Brunswick Scientific, NJ, USA) and incubated at 30 °C for 4 days or until evidencing growth of the fungus was evaluated. The MIC was taken at the lowest protein concentration of enzymatic extract that triggered complete fungal growth inhibition and was demonstrated visually.
Electrophoresis
SDS-page was performed on 12.5% w/v polyacrylamide gel according to the Laemmli method [28]. For the visualization of the proteins, Coomassie brilliant blue G250 (Cat. Num. 42,655, Biopack) staining was performed. A mixture of six highly purified proteins was used as a molecular weight marker (Amersham Low Molecular Weight Calibration Kit, code 17–0446-01, GE Healthcare).
Nano-LC–MS/MS Analysis
Proteins of the enzymatic extract monoculture and a mixed culture of A. niger and R. oryzae in SSF were analyzed using mass spectrometry. In order to obtain a single electrophoretic band containing all the proteins, the enzymatic solutions were run on SDS-page electrophoresis gel up to 1 cm after entering the resolution gel, revealing it with Coomassie brilliant blue. The bands were extracted and digested with trypsin. The resulting peptides were extracted and dried in a Speed-Vac. The samples were analyzed by nano-LC–MS/MS using an EASY-nLC 1000 system (Thermo Scientific) coupled to a LTQ Orbitrap XL mass spectrometer (Thermo Scientific, Bremen, Germany). Peptides were analyzed using the Xcalibur 3.0.63 (Thermo Scientific) software. The acquired raw MS/MS data files were searched by Proteome Discoverer software (version 1.4, Thermo Scientific) and Homo sapiens Uniprot database. The combination of equal volumes of enzyme extracts from monocultures of A. niger and R. oryzae was defined as a theoretical mixed culture. The relative abundance percentage of the theoretical mixed culture was calculated as half the sum of the relative abundances of each enzyme extract from the monocultures of A. niger and R. oryzae.
Mass spectrometry analysis was carried out at the Proteomics Core Facility CEQUIBIEM, at the University of Buenos Aires/CONICET.
Statistical Analysis
All experiments were performed in triplicate. Data, considered as the xylanolytic, cellulolytic, pectinolytic, amilolytic, and proteolytic activities in fermentations extracts of the different fermentation systems, were tested using the analysis of variance (ANOVA), and Tukey’s test was used to compare the means. The significance of the means was measured at p < 0.05. The statistical analysis of the data was performed with the statistical software package “Minitab” (Version 17, PA, USA), and all graphics were carried out using the software “Sigma Plot 10.0.”
Results and Discussion
Analysis of the Enzyme Production Profile of Monocultures
The production of XE, CE, PE, AE, and proteases was evaluated under SSF and SmF conditions for 5 days. In all systems, A. niger, T. harzianum, R. oryzae, and T. lanuginosus were able to colonize, penetrate, and grow on the combination of the different lignocellulosic biomass.
However, not all of them presented the same enzyme-producing capacity. A. niger showed the highest activity values for XE, CE, and PE in both SSF and SmF conditions, probably because A. niger has a stronger potential for degrading biomass compared to the rest of the microorganisms [29]. T. harzianum and T. lanuginosus were also capable of complete degradation of plant cell wall polysaccharides, although they showed lower activity values than A. niger for XE, CE, and PE activities. Regarding the other evaluated fungus, R. oryzae presented the best production of proteases between all the species in both SSF and SmF conditions. Table 2 shows the highest enzymatic activity of XE, CE, PE, AE, and proteases and the time when these activities were registered.
Maximum XE (73.7 and 56.6I IU/gds) activities for A. niger were observed at 96 and 120 h in SSF and SmF conditions (Table 2). Moran-Aguilar et al. [30] evaluated three A. niger strains in SSF for the production of xylanase. A higher xylanase activity (1400.80 IU/gds) was reached than the higher activity registered in this work using brewery-spent grains pretreated in an autoclave as a carbon source.
In this work, the XE activity of T. harzianum (23.0 IU/gds) reached only 31% of the maximum activity of A. niger. However, Marques and coauthors [31] observed a higher level of XE activity (20.71 IU/mL) in a culture of T. harzianum after 48 h of fermentation at 28 °C using a complex medium that contains minerals, trace elements, and 0.5% of corn-soybean-based starter feeds for broiler chicks [31].
Solid-state fermentation is considered a method with great potential for the production of CE by filamentous fungi. In addition, the production of CE by SSF is beneficial over submerged cultivation due to the use of low-cost substrates, a lower requirement of water for the reaction, and easy management procedures [32]. However, the optimal conditions for CE production evaluated in this work were 12.3 IU/gds at 96 h and 23.5 IU/gds 48 h, in SSF and SmF conditions for A. niger fermentations (Table 2). The use of a suitable substrate is essential to enhance the production of CE by filamentous fungi. Kumar et al. [33] observed that the maximum endoglucanase activity (26.33 IU/gds) was detected with wheat bran as substrate after evaluating eight solid substrates in SSF. Sharma et al. [34] observed maximum endoglucanase activity (21.5 IU/gds) in SSF using pea pod waste by A. niger. Taddia and coauthors [10] analyzed the time profiling for the production of xylanolytic and cellulolytic enzymes and obtained the highest levels of these enzymes on the fourth day of fermentation.
The highest PE activities were observed in SSF conditions for all species. The maximum PE activity (10.5 IU/gds) was at 72 h of SSF fermentation for A. niger (Table 2). The other fungal species showed significantly lower activities than the mentioned for A. niger (p˃0.005). These findings were in concordance with Abdulla et al. [35] who reported that A. niger was the best producer of PE among twenty-five different fungal strains isolated from different sources. Furthermore, Ortiz et al. [36] reported that SSF was the best condition for the production of these enzymes, reaching, after 2 days of fermentation with A. giganteus at 28 °C, a PE activity of 55 IU/gds using a mixture of wheat bran and orange peel in a 70:30 proportion as substrates. Even though the activity values obtained in this work were lower than those previously reported, this system was not optimized.
Conversely, it was observed that the highest AE production was recorded for T. lanuginosus (29.0 IU/gds) and R. oryzae (23.0 IU/gds) in SmF at 72 h of fermentation (Table 2). Carbon source influences the metabolic activities of the microorganisms and thus the production of enzymes largely depends on it. Therefore, the rich composition of the combined substrates employed induced maximum production in contrast with other substrates used in previous works [10, 13].
Maximum protease activities (41,880 and 33,660 IU/gds) for R. oryzae were observed at 120 and 96 h in SSF and SmF conditions respectively (Table 2). Also, T. lanuginosus presented a higher protease activity (9000 IU/gds) in SSF. Negi et al. [37] optimized the parameters temperature, pH, and carbon source for the production of protease from R. oryzae. The 412.8 IU/gds after 72 h of fermentation at 28 °C and pH 6 was the maximum protease activity reached, with wheat bran and soybean as substrates in a 4:1 ratio [37]. Benabda and coauthors [38] optimized the production of protease by R. oryzae in SSF. The highest levels of protease production (2400 IU/gds) were obtained at 120 h, 30 °C, and pH 5.5 using bread wastes as substrate [38]. Ahmed and Abood [39] studied the effect of some growth factors on protease production by R. oryzae in SSF. The enzyme production reached a maximum of 17,410 IU/gds after 144 h of fermentation at pH 5.0 and 30 °C using sunflower residue as substrate [39]. The protease activities reported in this work for monocultures were superior to those of the bibliography. Despite this, we did not study the effect of temperature and pH in the enzyme production by fungi; these factors were contemplated according to bibliography. The aforementioned authors found that 30 °C and a range of 5–6 pH were the suitable conditions to produce these enzymes. Both SmF and SSF presented a range of 5.2–5.5 pH due to the lignocellulosic biomass dissolved in distilled water.
In conclusion, A. niger was the best producer of EX, CE, and PE, and a poor producer of proteases, while R. oryzae was the best producer of proteases and a poor producer of the other enzymes. Furthermore, R. oryzae presented the best production of AE. T. lanuginosus was a good producer of proteases and amylases. This might be because low levels of free sugars induce the production of proteases as a reflection of physiological stress [40]. Therefore, A. niger, T. lanuginosus, and R. oryzae were selected to be evaluated in a second stage to study their compatibility to integrate a mixed culture.
Enzymatic Stability Experiment
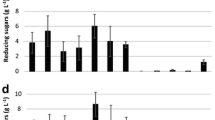
The stability of XE, CE, PE, and AE in presence of fungal protease was relevant to broaden the industrial applications of enzymes, as well as to increase the efficiency of industrial processes when the enzymatic cocktail is applied. Figure 1 shows the residual activity of XE of an enzymatic extract produced in SSF by A. niger in the presence of an enzymatic extract of T. lanuginosus and R. oryzae. The enzymatic extract of A. niger was produced under the conditions mentioned before for SSF after 120 h of fermentation and adjusted to pH 6.00.
Effect of protease activity of R. oryzae (RO) and T. lanuginosus (TL) on the xylanases stability of enzymatic extracts of A. niger (AN) at 22 °C and pH 6.00. All cultures were obtained on the fifth day by solid-state fermentation at 30 °C using 4 gds of a homogeneous mixture of lignocellulosic substrates
The XE activity of an extract produced by A. niger in the absence of other enzymatic extracts (control) decreased by 15% of its initial value during the period tested. The same behavior was observed for the enzymatic extract of A. niger in the presence of the enzymatic extract produced by R. oryzae. The CE and PE of A. niger presented the same tendencies in presence of R. oryzae extract (data not shown). However, when the enzymatic extract of A. niger was in the presence of the enzymatic extract produced by T. lanuginosus, it showed a constant decrease of the XE activity beginning in the second day of incubation until the decrease of 84% of the initial activity. The same behavior was observed for CE and PE activities of the enzymatic extract of A. niger (data not shown).
The protease activity in the enzymatic extract of R. oryzae was 41,880 IU/gds, four times higher than the protease activity from T. lanuginosus extract (9000 IU/gds). Therefore, the mixture of A. niger enzyme extract and the extract of R. oryzae and T. lanuginosus, where the stability of the enzyme was analyzed, was prepared considering that each one had the same protease concentration. These theoretical combinations simulated an enzyme extract obtained by mixed fermentation.
Proteases are a complex group of enzymes that vary greatly in their physicochemical and catalytic properties. They are generally classified by optimum pH and catalytic mechanisms. On the optimum pH basis, they are classified into acid, neutral, and alkaline proteases [41]. Regarding their catalytic mechanism, they are classified into serine proteases, cysteine proteases, aspartic proteases, and metallo proteases, determined indirectly through reactivity towards inhibitors of particular amino acid residues in the active site region [42].
Negi and coauthors [37] found that R. oryzae produces alkaline proteases. While the proteases of T. lanuginosus have a very broad optimum pH with no difference in their properties between pH 5.00–9.00 [42]. Therefore, T. lanuginosus proteases cannot be classified into acid, neutral, or alkaline proteases. Although the proteases of T. lanuginosus and R. oryzae were at the same concentration in this work, the stability tests were performed at pH 6.00, where the proteases of T. lanuginosus were active but not those of R. oryzae. In the enzymatic extracts produced by A niger where the pH was adjusted over 6.50, the XE, CE, and PE activities decreased progressively in presence of the enzymatic extract of T. lanuginosus and R. oryzae (data not shown).
This would explain how the XE of A. niger was stable at pH 6.00 in presence of the R. oryzae extract and not in presence of the T. lanuginosus extract. The results obtained suggest that the hydrolytic enzymes of A. niger are more stable in the presence of R. oryzae proteases than those of T. lanuginosus under these assayed conditions.
Growth Rate of Individual Fungal Species
The results of the growth rate of the individual strains evaluated in the PDA medium were 4.2 mm/day for A. niger and 15 mm/day for R. oryzae; the results are shown in Table S1. This behavior can be explained based on the colonization strategies of the microorganisms in a particular environment. R. oryzae appears to have adopted a strategy in which much of its metabolic energy is used to occupy the environment as quickly as possible, regardless of its high nutrient content. Fungi that follow this behavior are characterized for their easy adaptation to environmental stress with low availability of nutrients. The maximum growth rate for A. niger was 24 h and 12 h for R. oryzae after they were inoculated in the medium. In these systems, A. niger presented a slower metabolism and a slower colonization than R. oryzae.
Interaction Studies
The fungal consortium used in mixed fermentation can improve the processes of substrate degradation and enzyme production instead of the fungal fermentation in monocultures of microorganisms. For this reason, the compatibility between the filamentous fungi involved must be established.
The interaction study of A. niger against R. oryzae shown in Figure SF1 was observed that no mycelium could conquer the territory occupied by the other microorganism which was in concord with the postulate of deadlock by Stahl and Christensen [25]. They also found that this is the most common interaction between fungi when growing in a nutrient-rich environment. In some cases, A. niger produces a yellow halo when it meets the mycelium of another species.
Antagonism Studies
The antifungal potential of extracellular metabolites from A. niger and R. oryzae against some fungi was previously reported by different authors. Liu et al. [43] found that A. niger produced a chitinase which could degrade mycelial waste with high efficiency. Abdel-Rahman et al. [44] evaluated the relation between antimicrobial activities of terpenoids extracted from different isolated endophytic fungi like A. niger. Jin et al. [45] conducted a study of bioprospect for saponin-producing endophytic fungi and evaluated the antimicrobial activity of saponins, where they isolated an Aspergillus ssp. capable of producing a saponin extract which exhibited antimicrobial activity. Christ-Ribeiro et al. [46] studied the antifungal potential of a phenolic extract obtained from rice bran fermented with Rhizopus oryzae and found eighteen compounds (organic acids and gallates) with antifungal potential activity against strains of Fusarium, Aspergillus, and Penicillium.
In this work, it was visually established that the microbial growth in the broth dilution assay with cellular fungal extracts from A. niger and R. oryzae did not exhibit an inhibitory effect between the fungi. Finally, A. niger and R. oryzae species did not mutually inhibit each other by the production of antagonistic metabolites.
Analysis of the Enzyme Production Profile of Mixed Culture Fermentation
Kull [47] defined a consortium or mixed culture as a group of microorganisms interlinked through relations, or groups of interspecific semiosic links in biocoenosis.
Enhanced production of hydrolytic enzymes can be obtained with cocultures compared to monocultures. Moreover, the coculture technique can be viewed as a feasible bioprocess to produce enzymes for the complete hydrolysis of lignocellulosic materials.
Some authors have reported the mixed culture method they used; Dhilon et al. [48] produced cellulases and xylanases with a mixed culture of A. niger and Trichoderma reseei; and Kshirsagar et al. [49] generated a mixed culure of A. niger and Trichoderma viride to produce hydrolytic enzymes. The use of Trichoderma spp. and Aspergillus spp. is common and it is generally associated with the production of hydrolytic enzymes for biotechnology processes, and there are no studies about a consortium combining A. niger with R. oryzae. Hence, an association of A. niger and R. oryzae was evaluated for an efficient and eco-friendly enzyme cocktail production.
Due to the time differences in which A. niger (24 h) and R. oryzae (12 h) reach their maximum growth rate since they were inoculated, two inoculation strategies were proposed as fermentation strategies. One was the simultaneous phase inoculation of A. niger and R. oryzae (A + R) and the second which was out of phase was R. oryzae was inoculated 12 h later than A. niger (A + 12R). Both species were inoculated to reach the same final concentration in all systems. The mixed culture conditions were the same as those described for monocultures; fermentation systems were incubated at 30 °C using 4 gds of a mixture of lignocellulosic substrates.
The highest XE, CE, PE, and AE activities were observed when both species were inoculated simultaneously in solid and submerged fermentation systems (Table 3), and these activities values were higher compared to fermentations of A. niger or R. oryzae in monocultures.
The XE activity reached a maximum value of 98.0 IU/gds at 96 h of SSF and 69.0 IU/gds at 120 h of SmF. However, the highest CE activity was observed in SmF (39.0 IU/gds at 120 h), as well as in the monoculture fermentation of the mentioned species (Table 2).
The highest values of PE activities were observed at 96 h of SSF (14.0 IU/gds) and at 120 h of SmF (21.5 IU/gds). The last activity was two times higher than those observed in monocultures of A. niger. The same behavior was observed for AE with the highest AE activity (30 IU/gds) at 72 h of SSF.
Regarding proteases, the highest activity was recorded at 72 h of SSF (108,000 IU/gds). The significantly higher values of activities obtained can be attributed to the immobilized substrate of the support and the microorganism, which generates a mechanism of adaptation to promote growth, resulting in the extracellular production of needed enzymes [40]. In addition to this, SSF favors the possibility of controlling the water activity as a selection parameter during the cocultivation of fungi, as different fungi have different water demands.
Moreover, it was possible to explain the results obtained in the SSF mixed culture by the mechanisms of metabolic regulation. The simultaneous production of enzymes of the hemicellulolytic and cellulolytic complex by filamentous fungi contributes to synergistically degrading the cell wall of the substrate used. In Aspergillus ssp., the XlnR protein intervenes in the regulation of the enzymes that constitute the xylanolytic and cellulolytic complexes [19, 50], activating the transcription of the genes xlnA, xlnB, and xlnC, of the xylanolytic system, and xlnD encoding the endoxylanases A, B, and C and β-xylosidase proteins and other accessory enzymes involved in the degradation of hemicellulose, as well as the transcription of genes that code for endoglucanases A B (eglA and eglB) and others involved in cellulose degradation. In the coinoculated mixed culture, the higher growth rate of R. oryzae leads to the production of xylanolytic enzymes and an increase in D-xylose units (inducer of XlnR transcription); this favors the synthesis of enzymes of the xylanolytic and cellulolytic complex by A. niger, and a consequent increase in proteins of the system.
Furthermore, in Aspergillus ssp., the production of amylases is also regulated by CreA. This protein binds specifically in the alpha-amylase transcription promoter, indicating that CreA could be involved in the repression by glucose of amylolytic complex enzymes; consequently, it limits the expression of the enzymes [51].
Secretomes Obtained by Monocultures and Mix Cultures
The enzymatic extract obtained by SSF of A. niger and R. oryzae monocultures and a mixed culture of both strains inoculated simultaneously were analyzed by SDS-page. A higher concentration of enzymes in the mixed culture compared to the monoculture was observed (figure not shown).
The enzymes produced by A. niger and R. oryzae were identified by mass spectrometry. The enzymes produced by A. niger were CE (108.8 and 93.2 kDa, respectively); XE (87.2 kDa); PE (98.7 and 42.1 kDa, respectively); AE (68.3 and 55.2 kDa, respectively); and proteases (41.3 kDa). The enzymes produced by R. oryzae were proteases (92.8, 41.8, 41.5, 41.4, and 41.2 kDa, respectively); and AE (65 and 62.1 kDa, respectively). To analyze similarities and differences in enzyme production by mono- and mixed culture of SSF, the identified proteins by mass spectrometry were grouped into two: I, enzymes selected in this work (EX, EG, EP, EA, and proteases), and II, other extracellular proteins. Figure 2 shows that the proportion of the selected enzymes (group I) was enhanced in the mixed culture fermentation with respect to A. niger and R. oryzae monoculture fermentation.
In the extract from A. niger monoculture, 49.77% of the total identified proteins belong to group I and the rest to group II. Regarding the R. oryzae monoculture extract, 49.74% of the proteins belonged to group I, while 50.26% corresponded to group II.
Finally, in the extract from the mixed culture fermentation of both fungi, 85.12% of the found proteins belonged to group I. The contribution of the selected enzymes produced by A. niger in the mixed culture was enhanced by 23.37% regarding the total of proteins secreted by A. niger in the monoculture, while the enzymes produced by R. oryzae in the mixed culture were enhanced by 48.45%. 14.88% of the proteins found in the extract of the mixed culture fermentation corresponded to other unselected proteins. Table 4 summarizes the relative abundance of the proteins of group I in the secretomes of the monoculture of A. niger and R. oryzae, and in a mixed culture of both species inoculated simultaneously. Culture conditions were the same as described before. Detailed information on the A. niger, R. oryzae, and mixed culture secretomes are shown in Tables S2, S3, and S4, respectively.
The relative abundance percentages of CE, AE, and proteolytic enzymes increased in the enzymatic extract of the mixed culture compared with the theoretical mixed culture. However, the relative abundance percentages of the XE and PE remained constant in the enzymatic extract of the fungal consortium compared to the theoretical mixed culture.
Both species contribute to the extract with the production of hydrolytic enzymes as expected for each species. A. niger contributes XE, CE, PE, and AE, while R. oryzae contributes with proteases, AE, and PE. Therefore, mixed culture may offer an efficient strategy to produce a cocktail enriched by enzymes from various species at one fermentation step. No similar data were found on bibliography to do a comparison.
Conclusions
Mixed cultures of A. niger and R. oryzae were generated both in SmF and SSF using lignocellulosic waste as substrates and the compatibility studies indicated good compatibility between the two fungi. These mixed cultures enhanced protein secretion—in abundance and complexity—compared to monocultures, with an increase in the production of XE, AE, CE, PE, and proteases. It was concluded that mixed culture is a suitable fermentation strategy to produce an enzymatic cocktail enriched by several species and with a higher production rate in a single fermentation step rather than successive monoculture fermentations.
References
Kirk O, Borchert TV, Fuglsang CC (2002) Industrial enzyme applications. Curr Opin Biotechnol 13:345–351. https://doi.org/10.1016/S0958-1669(02)00328-2
Zhang Y, He S, Simpson BK (2018) Enzymes in food bioprocessing — novel food enzymes, applications and related techniques. Curr Opin Food Sci 19:30–35. https://doi.org/10.1016/j.cofs.2017.12.007
Gao W, Li Z, Liu T, Wang Y (2021) Production of high-concentration fermentable sugars from lignocellulosic biomass by using high solids fed-batch enzymatic hydrolysis. Biochem Eng J 176:108186. https://doi.org/10.1016/j.bej.2021.108186
Dood D, Cann IKO (2009) Enzymatic deconstruction of xylan for biofuel production. GCB Bioenergy 1:2–17. https://doi.org/10.1111/j.1757-1707.2009.01004.x
Jørgensen H (2016) Enzyme recycling in lignocellulosic biorefineries. Biofuels Bioprod Bioref. https://doi.org/10.1002/bbb.1724
Silva DF, Hergesel LM, Campioni TS et al (2018) Evaluation of different biological and chemical treatments in agroindustrial residues for the production of fungal glucanases and xylanases. Process Biochem 67:29–37. https://doi.org/10.1016/j.procbio.2018.02.008
Londoño-Hernandez L, Ruiz-Leza H, Ramírez Toro C, et al (2020) Advantages and progress innovations of solid-state fermentation to produce industrial enzymes. In: Springer (ed) Microbial enzyme: roles and application in Industries. Springer Netherlands, p 20
Polizeli MLTM, Rizzatti ACS, Monti R, et al (2005) Xylanases from fungi: properties and industrial applications. 577–591. https://doi.org/10.1007/s00253-005-1904-7
Cortes-Tolalpa L, Jiménez DJ, de Lima Brossi MJ et al (2016) Different inocula produce distinctive microbial consortia with similar lignocellulose degradation capacity. Appl Microbiol Biotechnol 100:7713–7725. https://doi.org/10.1007/s00253-016-7516-6
Taddia A, Boggione J, Tubio G (2019) Screening of different agroindustrial by-products for industrial enzymes production by fermentation processes. Int J Food Sci Technol 54:1027–1035. https://doi.org/10.1111/ijfs.13915
Intasit R, Cheirsilp B, Suyotha W, Boonsawang P (2021) Synergistic production of highly active enzymatic cocktails from lignocellulosic palm wastes by sequential solid state-submerged fermentation and co-cultivation of different filamentous fungi. Biochem Eng J 173:108086. https://doi.org/10.1016/j.bej.2021.108086
Yadav AN, Singh S, Mishra S, Gupta A (2019) Recent advancement in white biotechnology through fungi. Galway, Ireland
Podestá MV, Morilla EA, Allasia MB, et al (2019) An eco-friendly method of purification for xylanase from Aspergillus niger by polyelectrolyte precipitation. J Polym Environ 27: https://doi.org/10.1007/s10924-019-01571-3
Taddia A, Brandaleze GN, Boggione MJ et al (2020) An integrated approach to the sustainable production of xylanolytic enzymes from Aspergillus niger using agro-industrial by-products. Prep Biochem Biotechnol 50:979–991. https://doi.org/10.1080/10826068.2020.1777425
Bailey MJ, Biely P, Poutanen K (1992) Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol 23:257–270. https://doi.org/10.1016/0168-1656(92)90074-J
Ghose TK (1987) Measurement of cellulase activities. Pure Appl Chem 59:257–268. https://doi.org/10.1351/pac198759020257
Biz A, Farias FC, Motter FA, et al (2014) Pectinase activity determination: an early deceleration in the release of reducing sugars throws a spanner in the works! PLoS One 9: https://doi.org/10.1371/journal.pone.0109529
Hernández MS, Rodríguez MR, Guerra NP, Rosés RP (2006) Amylase production by Aspergillus niger in submerged cultivation on two wastes from food industries. J Food Eng 73:93–100. https://doi.org/10.1016/j.jfoodeng.2005.01.009
Solís-Pereira S, Favela-Torres E, Viniegra-González G, Gutiérrez-Rojas M (1993) Effects of different carbon sources on the synthesis of pectinase by Aspergillus niger in submerged and solid state fermentations. Appl Microbiol Biotechnol 39:36–41. https://doi.org/10.1007/BF00166845
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. https://doi.org/10.1007/BF00166845
Janser R, De CS, Sato HH (2013) Synergistic effects of agroindustrial wastes on simultaneous production of protease and alpha-amylase under solid state fermentation using a simplex centroid mixture design. Ind Crop Prod 49:813–821. https://doi.org/10.1016/j.indcrop.2013.07.002
Mallia AK, Frovenzano MD, Fujimoto EK et al (1985) Measurement of protein using bicinchoninic acid. Anal Biochem 85:76–85
Mead GC, Hudson WR, Hinton MH (1993) Microbiological survey of five poultry processing plants in the UK. Br Poult Sci 34:497–503. https://doi.org/10.1080/00071669308417605
Skidmore AM, Dickinson CH (1976) Colony interactions and hyphal interference between Septoria nodorum and phylloplane fungi. Trans Br Mycol Soc 66:57–64. https://doi.org/10.1016/S0007-1536(76)80092-7
Stahl PD, Christensen M (1992) In vitro mycelial interactions among members of a soil microfungal community. 24:309–316. https://doi.org/10.1016/0038-0717(92)90190-9
Porter CL (1924) Concerning the characters of certain fungi as exhibited by their growth in the presence of other fungi. Am J Bot 11:168–188
Giacone L, Cordisco E, Garrido MC, et al (2019) Photodynamic activity of Tagetes minuta extracts against superficial fungal infections. Med Mycol 1–13. https://doi.org/10.1093/mmy/myz114
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680. https://doi.org/10.1038/227680a0
Culleton H, Mckie V, De Vries RP (2013) Physiological and molecular aspects of degradation of plant polysaccharides by fungi: what have we learned from Aspergillus? Biotechnol J 8:884–894. https://doi.org/10.1002/biot.201200382
Moran-Aguilar MG, Costa-Trigo I, Calderón-Santoyo M, et al (2021) Production of cellulases and xylanases in solid-state fermentation by different strains of Aspergillus niger using sugarcane bagasse and brewery spent grain. Biochem Eng J 172: https://doi.org/10.1016/j.bej.2021.108060
Marques SFF, Minafra CS, Cafe MB et al (2016) Production and characterization of a Trichoderma harzianum multienzyme complex and its application in broiler chicks’ diets. Curr Biotechnol 7:26–33. https://doi.org/10.2174/2211550104666150930204632
Srivastava N, Elgorban AM, Mishra PK et al (2020) Enhance production of fungal cellulase cocktail using cellulosic waste. Environ Technol Innov 19:100949. https://doi.org/10.1016/j.eti.2020.100949
Kumar BA, Amit K, Alok K, Dharm D (2018) Wheat bran fermentation for the production of cellulase and xylanase by Aspergillus niger NFCCI 4113. Res J Biotechnol 13:11–18
Sharma R, Rawat R, Bhogal RS, Oberoi HS (2015) Multi-component thermostable cellulolytic enzyme production by Aspergillus niger HN-1 using pea pod waste: appraisal of hydrolytic potential with lignocellulosic biomass. Process Biochem 50:696–704. https://doi.org/10.1016/j.procbio.2015.01.025
Abdullah R, Jafer A, Nisar K et al (2018) Process optimization for pectinase production by locally isolated fungal strain using submerged fermentation. Biosci J 34:1025–1032. https://doi.org/10.14393/BJ-v34n1a2018-39947
Ortiz GE, Ponce MC, Diego M et al (2017) Pectinase production by Aspergillus giganteus in solid-state fermentation: optimization, scale-up, biochemical characterization and its application in olive-oil extraction. J Ind Microbiol Biotechnol 44:197–211. https://doi.org/10.1007/s10295-016-1873-0
Negi S, Jain S, Raj A (2020) Combined ANN/EVOP factorial design approach for media screening for cost-effective production of alkaline proteases from Rhizopus oryzae (SN5)/NCIM-1447 under SSF. AMB Express 10: https://doi.org/10.1186/s13568-020-00996-7
Benabda O, M’Hir S, Kasmi M, et al (2019) Optimization of protease and amylase production by Rhizopus oryzae cultivated on bread waste using solid-state fermentation. J Chem 2019: https://doi.org/10.1155/2019/3738181
Ahmed SA, Abood NH (2017) Effect of some growth factors on protease production by Rhizopus oryzae. J Al-Nahrain Univ 20:90–95. https://doi.org/10.22401/jnus.20.2.12
Aguilar CN, Contreras-Esquivel JC, Rodriguez R, Prado LA, Loera O (2004) Differences in fungal enzyme productivity in submerged and solid state cultures. In: Food Science and Biotechnology. pp 109–113
Guo Y, Tu T, Zheng J et al (2020) A novel thermostable aspartic protease from Talaromyces leycettanus and its specific autocatalytic activation through an intermediate transition state. Appl Microbiol Biotechnol 104:4915–4926. https://doi.org/10.1007/s00253-020-10569-0
Li D-C, Yang Y-J, Shen C-Y (1997) Protease production by the thermophilic fungus Thermomyces lanuginosus. Mycol Res 101:18–22. https://doi.org/10.1017/S0953756296002109
Liu T, Han H, Wang D et al (2020) Potent fungal chitinase for the bioconversion of mycelial waste. J Agric Food Chem 68:5384–5390. https://doi.org/10.1021/acs.jafc.0c01342
Abdel-Rahman T, Hussein A-S, Beshir S, et al (2019) Antimicrobial activity of terpenoids extracted from Annona muricata seeds and its endophytic Aspergillus niger strain SH3 either singly or in combination. Open Access Maced J Med Sci 3127–3131. https://doi.org/10.3889/oamjms.2019.793
Jin Z, Gao L, Zhang L et al (2017) Antimicrobial activity of saponins produced by two novel endophytic fungi from Panax notoginseng. Nat Prod Res 31:2700–2703. https://doi.org/10.1080/14786419.2017.1292265
Christ-Ribeiro A, Graça CS, Kupski L et al (2019) Cytotoxicity, antifungal and anti mycotoxins effects of phenolic compounds from fermented rice bran and Spirulina sp. Process Biochem 80:190–196. https://doi.org/10.1016/j.procbio.2019.02.007
Kull K (2010) Ecosystems are made of semiosic bonds: consortia, umwelten, biophony and ecological codes. Biosemiotics 3:347–357. https://doi.org/10.1007/s12304-010-9081-1
Dhillon GS, Oberoi HS, Kaur S et al (2011) Value-addition of agricultural wastes for augmented cellulase and xylanase production through solid-state tray fermentation employing mixed-culture of fungi. Ind Crops Prod 34:1160–1167. https://doi.org/10.1016/j.indcrop.2011.04.001
Kshirsagar S, Waghmare P, Saratale G et al (2020) Composition of synthesized cellulolytic enzymes varied with the usage of agricultural substrates and microorganisms. Appl Biochem Biotechnol 191:1695–1710. https://doi.org/10.1007/s12010-020-03297-8
Pedraza-Zapata DC, Sánchez-Garibello AM, Quevedo-Hidalgo B et al (2017) Promising cellulolytic fungi isolates for rice straw degradation. J Microbiol 55:711–719. https://doi.org/10.1007/s12275-017-6282-1
Kato M, Sekine K, Tsukagoshi N (1996) Sequence-specific binding sites in the taka-amylase a G2 promoter for the CreA repressor mediating carbon catabolite repression. Biosci Biotechnol Biochem 60:1776–1779. https://doi.org/10.1271/bbb.60.1776
Acknowledgements
We would like to thank Tec. Marina Scrocchi Kretzschmar and the staff from the Statistical Area for the data processing and the English Department (Faculty of Biochemical and Pharmaceutical Sciences, UNR) for the language correction of the manuscript. We thank Dra. Silvia Moreno from CEQUIBIEM for performing the analysis of protein samples by LC-MS.
Funding
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica under Grant PICT-II-B-2018 2054 and the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) under Grant PUE BD20160041CO and Secretary of Science and Technology of National University of Rosario under Grant 80020180100033UR and 80020180100036UR.
Author information
Authors and Affiliations
Contributions
Esteban Amador Morilla: conceptualization; methodology; investigation; validation; writing—original draft. Antonela Taddia: investigation; methodology; validation; writing—original draft. Maximiliano Sortino: conceptualization, investigation, review and editing. Gisela Tubio: conceptualization, investigation, resources, funding acquisition, supervision, original draft, writing, review and editing.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Morilla, E.A., Taddia, A., Sortino, M. et al. Mixed Cultures of Aspergillus niger and Rhizopus oryzae Using Lignocellulosic Substrates to Improve Hydrolytic Enzyme Production. Bioenerg. Res. 16, 2285–2296 (2023). https://doi.org/10.1007/s12155-023-10567-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-023-10567-w