Abstract
Objective
The aim of the this study was to analyze the status of sex-determining region Y-related high-mobility group box 4 (SOX4) expression in varied human cancers and its correlation with overall survival in patients with human cancers.
Methods
To observe initially the expression status of SOX4 in twenty kinds of human cancers at protein database (The Human Protein Atlas). We systematically and carefully searched the studies from electronic databases and seriously identified according to eligibility criteria. The correlation between SOX4 expression and overall survival in human cancers was evaluated through Review Manager.
Results
We found that SOX4 expression was significantly positive in most types of human cancer tissues, and the positive rate of SOX4 expression was about 78 % in overall cancer tissues. Furthermore, a total of 10 studies which included 1348 cancer patients were included in the final analysis. Meta-analysis showed that SOX4 overexpression was correlated with a poor overall survival and the pooled hazard ratio (HR), and corresponding 95 % confidence interval (CI) was 1.67 (95 % CI 1.01–2.78). From subgroup analyses, we present evidence that SOX4 overexpression was an unfavorable prognostic factor for colorectal cancer patients’ recurrence-free survival and gastric cancer patients’ overall survival, and the pooled HRs (95 % CI) were 1.73 (95 % CI 1.04–2.88) and 3.74 (95 % CI 1.04–13.45), respectively.
Conclusions
In summary, SOX4 is a potential prognostic biomarker in human cancers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
SOX4 is a major member of the SOX family [1]. SOX4 gene is located on 6p22.3 and encodes a protein of 474 amino acids with three distinguishable domains: an HMG box, a glycine-rich region, and a serine-rich region [2]. The HMG box acts as a DNA-binding region, whereas the serine-rich domain acts as a transactivation domain. The central domain containing the glycine-rich region located between the HMG box and serine-rich domains acts as a novel functional region for promoting apoptotic cell death [3]. SOX4 involves in many developmental processes such as the T cell differentiation and the development of thymocyte, nervous system, and embryonic cardiovascular system [4–6].
In recent years, the clinical significance of SOX4 has gained more and more attention, with many reports suggesting that SOX4 may involve in tumor development and progression. Present studies have suggested that SOX4 as one of the most frequently overexpressed protein in several types of human cancers mediated regulation of tumorigenesis and tumor progression such as hepatocellular carcinoma [7, 8], prostate cancer [9, 10], breast cancer [11, 12], colorectal cancer [13, 14], and bladder cancer [15, 16]. Moreover, SOX4 overexpression in tumor cells has been shown to be an independent prognostic factor in several types of tumors, which has a favorable or unfavorable prognostic significance according to tumor types [8, 14–26]. Some studies indicated that positive SOX4 expression was significantly associated with a poor prognosis in colorectal cancer [14, 17], gastric cancer [18, 21], acute lymphoblastic leukemia [20], and prostate cancer [24], while others studies suggested patients with higher levels of SOX4 expression had better survival than those with lower levels of SOX4 expression in bladder carcinoma [15, 23], melanoma [19], and hepatocellular carcinoma [8].
In order to identify the pathological roles of SOX4 in human cancers, we evaluated the status of SOX4 expression in twenty kinds of human cancers through a protein database and performed a meta-analysis aiming to evaluate the relationship between SOX4 expression and prognosis in patients with cancers.
Materials and methods
Analysis of protein database
The data of SOX4 expression in twenty kinds of human cancers were retrieved from the protein database (The Human Protein Atlas, www.proteinatlas.org). The status of SOX4 expression was evaluated in twenty kinds of human cancer samples including breast cancer, carcinoid, cervical cancer, colorectal cancer, endometrial cancer, glioma, head and neck cancer, liver cancer, lung cancer, lymphoma, melanoma, ovarian cancer, pancreatic cancer, prostate cancer, renal cancer, skin cancer, stomach cancer, testis cancer, thyroid cancer, and urothelial cancer.
Inclusion and exclusion criteria
Studies eligible for inclusion in this meta-analysis met the following criteria: (1) Expression of SOX4 was evaluated in all cancers by cDNA microarray, tissue microarray, or immunohistochemistry; (2) hazard ratios (HRs) for overall survival related to SOX4 expression were provided or were extractable from the published data; (3) all patients diagnosed with malignant tumors or cancer must be confirmed by histopathologic examinations; and (4) there was sufficient information about study population, origin of country, and cancer type. The following criteria were used to exclude published studies: (1) Any studies that did not meet all inclusion criteria and (2) studies of case reports, letters, and reviews without original data. When a study reporting the same patient cohort was included in several publications, the most complete study was selected.
Search strategy
The MEDLINE, EMBASE, SCOPUS, WEB OF SCIENCE and Cochrane Library databases were systematically searched till October 2014. Publications with the following search words in the title, abstract, or keywords were included: SOX4, SRY (sex-determining region Y)-box 4, Carcinoma, Cancer, Tumor and Prognosis. Keywords were combined and Boolean operators (and, or, not) were used in the search strategies. The studies identified through the search were independently screened by two authors (JC and HJ) for inclusion. Any disagreements were arbitrated by a third author (BL). We did not limit our search by country, race, or date.
Data extraction
Data were independently extracted by two authors (JC and XY) using the same standardized table. Data were extracted from the included studies, and the major information included the following: first name, publication year, country, cancer type, total cases, survival analysis method (multivariate or univariate), end point results, and hazard ratio with 95 % confidence interval (CI). For articles with the same population resources or overlapping datasets, data were extracted and reported as a single trial. Overall survival in relation to survivin expression was estimated by the HR. If authors reported HR and 95 % CI, these data were extracted from the included articles. Otherwise, HR and 95 % CI were calculated as described previously [27, 28]. The number of patients at risk in each group, number of events, and the log-rank statistic or its p value were used to calculate the HR estimate and its variance. If the study did not provide a HR but reported the survival curve, we need to obtain data from survival curves. Survival curve could be read by Engauge Digitizer.
Statistical analysis
The meta-analysis was performed through using Cochrane Collaboration Review Manager 5.1 software. The details of statistical analysis were shown as described previously [29]. Heterogeneity of hazard ratio was assessed by use of the Chi-square and I 2 tests. When heterogeneity was significant (I 2 > 50 %and p < 0.05 for χ2), we used a random effects model with the DerSimonian and Laird method for the meta-analysis. Otherwise, we used a fixed effects model with the inverse variance method.
Overall survival (OS) of gastric cancer and recurrence-free survival (RFS) of colorectal cancer were determined in subgroup analyses. The sensitivity was analyzed by changing the effect model to estimate confidence. Funnel plot was executed for evaluating the potential publication bias. An asymmetric plot indicates there was potential publication bias; otherwise, the plot should be shaped like a funnel.
Results
SOX4 expression is positive in most types of human cancer tissues
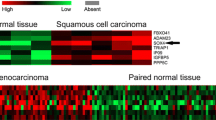
From protein database, SOX4 expression was significantly positive in most types of human cancer tissues such as breast cancer, carcinoid, cervical cancer, colorectal cancer, endometrial cancer, glioma, head and neck cancer, liver cancer, lung cancer, lymphoma, melanoma, ovarian cancer, pancreatic cancer, prostate cancer, renal cancer, skin cancer, stomach cancer, testis cancer, thyroid cancer, and urothelial cancer (Fig. 1A; SOX4 expression in normal tissues was showed in http://www.proteinatlas.org/ENSG00000124766-SOX4/tissue). The high and medium staining rate was about 78 % in overall cancer tissues (Fig. 1B). Specific SOX4 protein staining was found in nucleus at twenty kinds of human cancers (Fig. 1C).
Status of SOX4 in human cancer tissues through protein database (The Human Protein Atlas). A The expression of SOX4 in twenty kinds of human cancers. B Overall cancer tissues staining statistics of SOX4 expression. C Immunohistochemical staining of SOX4 expression in twenty kinds of human cancers: a breast cancer, b carcinoid, c cervical cancer, d colorectal cancer, e endometrial cancer, f glioma, g head and neck cancer, h liver cancer, i lung cancer, j lymphoma, k melanoma, l ovarian cancer, m pancreatic cancer, n prostate cancer, o renal cancer, p skin cancer, q stomach cancer, r testis cancer, s thyroid cancer, and t urothelial cancer
Eligible studies
Electronic database search identified a total of 684 articles by using our search criteria (Fig. 2). After carefully reading the title and abstract, 668 articles were excluded, because they did not present any data about the association of SOX4 expression with patients’ outcome. After reviewing the full text of the remaining 15 studies, we ultimately included 10 studies in the final analysis. Five studies were excluded from the final review because of insufficient survival data (n = 5) [8, 15, 16, 25, 26].
Study characteristics
A total of 1348 cases from the 10 studies that had pathological results and clinical data were included in this meta-analysis. Publication year of selected studies ranged from 2006 to 2014. These cases of 11 studies were from different populations (six studies in China and other four studies in Canada, USA, Denmark, and France, respectively) and eight kinds of cancers (lung cancer, gastric cancer, colorectal cancer, pancreatic cancer, cutaneous melanoma, acute lymphoblastic leukemia, gallbladder carcinoma, and prostate cancer). SOX4 expression was evaluated by immunohistochemistry, cDNA microarray, or tissue microarray. The positive expression rate of SOX4 varied from 32.8 to 80.0 %. Overall, five studies calculated HR and 95 % CI by multivariate analysis; the other five studies used univariate analysis. Table 1 shows the main characteristics of all of the studies.
Meta-analysis
There was obvious heterogeneity among those eight studies (I 2 = 82.0 %). Thus, the random effects model was used to calculate the pooled HR with corresponding 95 % CI. Meta-analysis of those eight studies showed that SOX4 expression was obviously associated with poor OS outcome in various cancers, with the pooled HR of 1.67 (95 % CI 1.01–2.78) (Fig. 3).
Subgroup analysis was performed to explore the between-study heterogeneity. No significant heterogeneity was observed among the studies on colorectal cancer for RFS (I 2 = 0.0 %, P = 0.93) and on gastric cancer for OS (I2 = 75.0 %, P = 0.05). The random effects model was used to calculate the pooled HR (95 % CI) in subgroups of colorectal cancer and gastric cancer. The result indicated that pooled HRs (95 % CI) were 1.73 (95 % CI 1.04–2.88) for colorectal cancer patients’ RFS and 3.74 (95 % CI 1.04–13.45) for gastric cancer patients’ OS (Fig. 4).
Sensitivity analysis suggested that changing the effect model had little effect on pooled HR and did not change the strength of the association between SOX4 expression and OS for patients with cancers. The HRs (95 % CI) were 1.75 (95 % CI 1.42–2.15) for changing the effect model. Publication bias of this meta-analysis was evaluated by funnel plot. No evidence of asymmetry was observed in the funnel plots (Fig. 5).
Discussion
SOX4, which is a member of the SOX family, has been implicated in multiple biological processes for the T cell differentiation and the development of thymocyte, nervous system, and embryonic cardiovascular system [4–6]. In recent years, more and more evidences have been presented suggesting that SOX plays an important role in tumor development and progression through involving in cell proliferation, migration, invasion, apoptosis, and epithelial-to-mesenchymal transition [3].
In our study, we found that SOX4 expression was significantly positive in twenty kinds of human cancer tissues including breast cancer, carcinoid, cervical cancer, colorectal cancer, endometrial cancer, glioma, head and neck cancer, liver cancer, lung cancer, lymphoma, melanoma, ovarian cancer, pancreatic cancer, prostate cancer, renal cancer, skin cancer, stomach cancer, testis cancer, thyroid cancer, and urothelial cancer through protein database, and the total positive rate was about 78 % in overall cancer tissues. Our result is consistent with reviews about the relationship between SOX4 and cancer performed by Jafarnejad et al. [2] and Vervoort et al. [3]. These studies consistently imply that overexpressed SOX4 may serve as unfavorable role in human cancer pathogenesis. Furthermore, the experiment in vitro showed that downregulated SOX4 expression decreased cell viability and induced apoptosis, and promoted malignant phenotype of adenoid cystic carcinoma cells [30]. Vervoort et al. [31] reported that TGFB1 induced the mesenchymal phenotype of mammary epithelial cells during epithelial-to-mesenchymal transition depending on upregulation of SOX4 expression. Similar to Zhang et al. [11] study in breast cancer, they found that SOX4 expression induced epithelial-to-mesenchymal transition and accelerated metastasis in vitro and in vivo.
In the past 10 years, SOX4 overexpression has been suggested to be an independent predictor for OS in various human cancers, which has a favorable or unfavorable prognostic significance according to the type of cancer. In primary gallbladder carcinoma, SOX4 overexpression was significantly correlated with better OS and disease-free survival, and was an independent favorable prognostic factor for OS and disease-free survival through multivariate analyses [23]. Moreover, Jafarnejad et al. reported that the SOX4 expression in metastatic melanoma was remarkably decreased in comparison with primary melanoma, and increased SOX4 expression associated with a better disease-specific survival for melanoma patients and was an independent favorable prognostic factor [19]. Conversely, there was more evidence suggesting that overexpression of SOX4 was unfavorable prognosis factor in gastric cancer [18, 21], acute lymphoblastic leukemia [20], colorectal cancer [14, 17], etc. In gastric cancer patients, higher level of SOX4 expression was markedly associated with distant metastasis, low differentiation, and unfavorable prognosis [21]. Similarly, Ramezani-Rad et al. [20] study showed that acute lymphoblastic leukemia (ALL) patients at the time of diagnosis with SOX4 overexpression in ALL cells had a poor OS outcome, and SOX4 was identified as a key regulator of MAPK and PI3K/AKT signaling in ALL cells. Meanwhile, SOX4 overexpression also has been shown to be an unfavorable prognostic factor in prostate cancer [24].
In order to evaluate the prognostic value of SOX4 in human cancer, we are to carry out a meta-analysis to analyze the association between SOX4 and human cancer. In the meta-analysis of 10 studies containing 1348 cases, we found that SOX4 expression was obviously associated with unfavorable OS in varied cancers, with the pooled HR of 1.67 (95 % CI 1.01–2.78). Furthermore, the subgroup analysis was performed to identify the heterogeneity of tumor types. The subgroup analysis showed that there was no significant subgroup difference between gastric cancer and colorectal cancer (both I 2 = 17.2 %). The SOX4 overexpression was significantly correlated with unfavorable OS in gastric cancer (HR 3.74, 95 %CI 1.04–13.45) and unfavorable RFS in colorectal cancer (HR 1.73, 95 %CI 1.04–2.88). Among those 10 studies, SOX4 overexpression served as a favorable prognostic factor only in gallbladder carcinoma (HR 1.73, 95 %CI 0.10–0.91) [23]. A similar report which could not be included in this meta-analysis because of insufficient data shows that SOX4 overexpression had a favorable OS outcome for bladder carcinoma patients through tissue microarray analysis [15]. The discrepancy of SOX4 expression in different types of human cancers may be attributed to tumor heterogeneity. We thought that SOX4 overexpression was a credible favorable prognostic factor for bladder carcinoma patients through systematically reviewing two relevant studies.
The meta-analysis may have some potential limitations. On the one hand, although 1348 cases of 10 studies were included in this meta-analysis, well-designed and large clinical studies for each cancer were insufficient. On the other hand, there was statistical heterogeneity in this meta-analysis which may be attributed to the differences in evaluation criterions of SOX4 expression, population, disease stages, and tumor types.
In summary, SOX4 overexpression was significantly correlated with unfavorable OS in most types of cancers except bladder carcinoma. SOX4 may be considered to serve as a credible prognostic factor in human cancers. In the future, more studies are necessary to verify and strengthen the role of SOX4 in human cancers.
Abbreviations
- SOX4:
-
Sex-determining region Y-related high-mobility group box 4
- OS:
-
Overall survival
- RFS:
-
Recurrence-free survival
- HR:
-
Hazard ratio
- CI:
-
Confidence interval
References
Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2010;42(3):425–8. doi:10.1016/j.biocel.2009.07.018.
Jafarnejad SM, Ardekani GS, Ghaffari M, Li G. Pleiotropic function of SRY-related HMG box transcription factor 4 in regulation of tumorigenesis. Cell Mol Life Sci CMLS. 2013;70(15):2677–96. doi:10.1007/s00018-012-1187-y.
Vervoort SJ, van Boxtel R, Coffer PJ. The role of SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? Oncogene. 2013;32(29):3397–409. doi:10.1038/onc.2012.506.
Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, et al. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380(6576):711–4. doi:10.1038/380711a0.
Ya J, Schilham MW, de Boer PA, Moorman AF, Clevers H, Lamers WH. Sox4-deficiency syndrome in mice is an animal model for common trunk. Circ Res. 1998;83(10):986–94.
Cheung M, Abu-Elmagd M, Clevers H, Scotting PJ. Roles of Sox4 in central nervous system development. Brain Res Mol Brain Res. 2000;79(1–2):180–91.
Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene. 2008;27(42):5578–89. doi:10.1038/onc.2008.168.
Hur W, Rhim H, Jung CK, Kim JD, Bae SH, Jang JW, et al. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: clinical implication and functional analysis in vitro. Carcinogenesis. 2010;31(7):1298–307. doi:10.1093/carcin/bgq072.
Liu P, Ramachandran S, Ali Seyed M, Scharer CD, Laycock N, Dalton WB, et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res. 2006;66(8):4011–9. doi:10.1158/0008-5472.can-05-3055.
Moreno CS. The Sex-determining region Y-box 4 and homeobox C6 transcriptional networks in prostate cancer progression: crosstalk with the Wnt, Notch, and PI3K pathways. Am J Pathol. 2010;176(2):518–27. doi:10.2353/ajpath.2010.090657.
Zhang J, Liang Q, Lei Y, Yao M, Li L, Gao X, et al. SOX4 induces epithelial-mesenchymal transition and contributes to breast cancer progression. Cancer Res. 2012;72(17):4597–608. doi:10.1158/0008-5472.can-12-1045.
Graham JD, Hunt SM, Tran N, Clarke CL. Regulation of the expression and activity by progestins of a member of the SOX gene family of transcriptional modulators. J Mol Endocrinol. 1999;22(3):295–304.
Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, et al. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol Cell Biol. 2007;27(22):7802–15. doi:10.1128/mcb.02179-06.
Andersen CL, Christensen LL, Thorsen K, Schepeler T, Sorensen FB, Verspaget HW, et al. Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer. Br J Cancer. 2009;100(3):511–23. doi:10.1038/sj.bjc.6604884.
Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, et al. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66(7):3434–42. doi:10.1158/0008-5472.can-05-3456.
Gunes S, Yegin Z, Sullu Y, Buyukalpelli R, Bagci H. SOX4 expression levels in urothelial bladder carcinoma. Pathol Res Pract. 2011;207(7):423–7. doi:10.1016/j.prp.2011.05.005.
Barrier A, Boelle PY, Roser F, Gregg J, Tse C, Brault D, et al. Stage II colon cancer prognosis prediction by tumor gene expression profiling. J Clin Oncol. 2006;24(29):4685–91. doi:10.1200/jco.2005.05.0229.
Shen R, Pan S, Qi S, Lin X, Cheng S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun. 2010;394(4):1047–52. doi:10.1016/j.bbrc.2010.03.121.
Jafarnejad SM, Wani AA, Martinka M, Li G. Prognostic significance of Sox4 expression in human cutaneous melanoma and its role in cell migration and invasion. Am J Pathol. 2010;177(6):2741–52. doi:10.2353/ajpath.2010.100377.
Ramezani-Rad P, Geng H, Hurtz C, Chan LN, Chen Z, Jumaa H, et al. SOX4 enables oncogenic survival signals in acute lymphoblastic leukemia. Blood. 2013;121(1):148–55. doi:10.1182/blood-2012-05-428938.
Fang CL, Hseu YC, Lin YF, Hung ST, Tai C, Uen YH, et al. Clinical and prognostic association of transcription factor SOX4 in gastric cancer. PLoS One. 2012;7(12):e52804. doi:10.1371/journal.pone.0052804.
Huang HY, Cheng YY, Liao WC, Tien YW, Yang CH, Hsu SM, et al. SOX4 transcriptionally regulates multiple SEMA3/plexin family members and promotes tumor growth in pancreatic cancer. PLoS One. 2012;7(12):e48637. doi:10.1371/journal.pone.0048637.
Wang C, Zhao H, Lu J, Yin J, Zang L, Song N, et al. Clinicopathological significance of SOX4 expression in primary gallbladder carcinoma. Diagn pathology. 2012;7:41. doi:10.1186/1746-1596-7-41.
Wang L, Zhang J, Yang X, Chang YW, Qi M, Zhou Z, et al. SOX4 is associated with poor prognosis in prostate cancer and promotes epithelial-mesenchymal transition in vitro. Prostate Cancer Prostatic Dis. 2013;16(4):301–7. doi:10.1038/pcan.2013.25.
de Bont JM, Kros JM, Passier MM, Reddingius RE, Sillevis Smitt PA, Luider TM, et al. Differential expression and prognostic significance of SOX genes in pediatric medulloblastoma and ependymoma identified by microarray analysis. Neuro Oncology. 2008;10(5):648–60. doi:10.1215/15228517-2008-032.
Wang L, Li Y, Yang X, Yuan H, Li X, Qi M, et al. ERG-SOX4 interaction promotes epithelial-mesenchymal transition in prostate cancer cells. Prostate. 2014;74(6):647–58. doi:10.1002/pros.22783.
Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–34.
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi:10.1186/1745-6215-8-16.
Wu KP, Li Q, Lin FX, Li J, Wu LM, Li W, et al. MT1-MMP is not a good prognosticator of cancer survival: evidence from 11 studies. Tumour Bio J Int Soc Oncodev Biol Med. 2014;. doi:10.1007/s13277-014-2567-8.
Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25(41):5626–39. doi:10.1038/sj.onc.1209566.
Vervoort SJ, Lourenco AR, van Boxtel R, Coffer PJ. SOX4 mediates TGF-beta-induced expression of mesenchymal markers during mammary cell epithelial to mesenchymal transition. PLoS One. 2013;8(1):e53238. doi:10.1371/journal.pone.0053238.
Acknowledgments
This study was supported by the plan project of science and technology (2014).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical approval
No ethics approval was required.
Additional information
J. Chen and H. L. Ju are co-first authors.
Rights and permissions
About this article
Cite this article
Chen, J., Ju, H.L., Yuan, X.Y. et al. SOX4 is a potential prognostic factor in human cancers: a systematic review and meta-analysis. Clin Transl Oncol 18, 65–72 (2016). https://doi.org/10.1007/s12094-015-1337-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-015-1337-4