Abstract
The COVID-19 outbreak began in December 2019 and has affected people worldwide. It was declared a pandemic in 2020 by the World Health Organization. Developing rapid and reliable diagnostic techniques is crucial for identifying COVID-19 early and preventing the disease from becoming severe. In addition to conventional diagnostic techniques such as RT-PCR, computed tomography, serological assays, and sequencing methods, biosensors have become widely accepted for identifying and screening COVID-19 infection with high accuracy and sensitivity. Their low cost, high sensitivity, specificity, and portability make them ideal for diagnostics. The use of nanomaterials improves the performance of biosensors by increasing their sensitivities and limiting detection by several orders of magnitude. This manuscript briefly reviews the COVID-19 outbreak and its pathogenesis. Furthermore, it comprehensively discusses the currently available biosensors for SARS-CoV-2 detection, with a special emphasis on nanomaterials-based biosensors developed to detect this emerging virus and its variants efficiently.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coronavirus disease-19 (COVID-19) was detected first in China in December 2019 and declared as a global pandemic by WHO in March 2020 (https://www.who.int/health-topics/coronavirus). The virus affects not only the respiratory system but also the heart, liver, GI tract, kidney, and central nervous system, eventually leading to multiple organ failure. Due to its similarity with previous severe acute respiratory syndrome coronavirus (SARS-CoV), it was named SARS-CoV-2 by the International Committee on Taxonomy of Viruses (ICTV), and the disease was named COVID-19. As of now, there have been 700 million reported cases of COVID-19, resulting in 6.97 million deaths and 671 million recoveries (https://www.worldometers.info/coronavirus). The number of cases is still rising, posing a major health concern, and strong strategies are needed to combat this life-threatening disease. People infected with SARS-CoV-2 can be divided into three categories based on the severity of the disease: (1) asymptomatic cases with or without detectable virus, (2) mild symptomatic or non-severe cases that usually recover after infection, and (3) severe respiratory symptomatic patients [1] with greater viral load that requires hospitalization, suffer respiratory failure, and in some cases, multiorgan failure (5–10%) [1,2,3,4].
To date, various strategies such as vaccines, drug repurposing, new antivirals, and therapeutic antibodies have been effectively used to treat SARS-CoV-2 [5]. Alongside these treatments, early clinical diagnosis, prompt isolation of affected individuals, effective treatment planning, and implementing preventive measures are crucial in preventing disease severity, community transmission, and potential new waves of COVID-19 or any future pandemics.
Several successful approaches in COVID-19 diagnostics have been employed to achieve public health goals and manage the pandemic. These techniques include real-time reverse transcription polymerase chain reaction (RT-PCR), serological assays, and computed tomography (CT). While these methods have well-established regulatory pathways, they often require skilled personnel, expensive instruments, and laboratory infrastructure [5].
Currently, there is a preference for sensitive, reliable, and "Naked-eye" methods of SARS-CoV-2 detection to provide immediate test outcomes at the sample collection site. Researchers are now exploring various biosensor types to improve detection and make it more suitable for at-home testing. Biosensors are expected to address several unmet needs in the diagnostics sector due to their portability and quick turnaround times.
Biosensors
A biosensor is a device that measures chemical or biological reactions by generating signals proportional to the concentration of the substance of interest, known as the analyte. A typical biosensor consists of the following components: (a) Analyte: the substance that needs to be detected. (b) Bioreceptor: a molecule, such as an enzyme, cell, DNA, antibody, or aptamer, that interacts with the analyte with high specificity through a biorecognition event. (c) Transducer: converts the biorecognition event into a measurable optical or electrical signal. (d) Electronics/Amplifier: processes the transduced signal, amplifies it, and converts it from analog to digital form. (e) Display: a computer, printer, or liquid crystal display that generates data or graphics understandable by users [6].
Various types of biosensors are currently available in the market, including enzyme biosensors, DNA biosensors, immunosensors (based on the bioreceptor), and electrochemical, optical, piezoelectric, and thermometric biosensors (based on the transducer) [6,7,8]. Biosensors have been proven to detect different types of viruses including HIV [9, 10], hepatitis virus [11], Zika virus [12], human coronavirus [13], dengue virus [14], and influenza virus [15]; plants pathogens [16], and bacteria [17,18,19,20]. The first biosensor was created by Clark and Lyons [21], to detect glucose concentration in biological samples by fabricating the electrochemical biosensor by immobilizing glucose oxidase (GOD) enzyme molecules on the surface of an oxygen electrode. The potentiometric urea-specific enzyme electrode developed in 1969 is a significant advancement in the development of biosensors [22]. These biosensors rely on the use of oxidase and dehydrogenase enzyme kinds of electrodes and are categorized as biosensors of the first generation. In the second generation, biosensors, auxiliary enzymes, and co-reactants are co-immobilized with the analyte-converting enzyme to enhance analytical quality operation [23]. In the third-generation biosensors, including SPR (Surface Plasmon Resonance) biosensors, biomolecules are used as biosensing material. The third-generation biosensor consists of only a direct electron transfer (DET)-type enzyme and an electrode. Since the system does not require a mediator, it has the advantages of low cost, reduced risk of side reactions, and superior biocompatibility [24,25,26]. Finally, advancements in MEMS/NEMS/BioNEMS (Micro, Nano, or BioNano Electro-Mechanical Systems, nanotechnology, and biotechnology, have led to development of the fourth generation of biosensors. The fourth generation of biosensors is anticipated to have features such as diagnostic efficiency and cost-effectiveness. These sensors utilize nanomaterials and do not require any enzymes,and hence exhibit better shelf life, improved sensitivity, enhanced reproducibility, excellent stability, and less detection time [27].
Nanomaterials in the Development of Biosensors
Nanomaterials and nanostructures play a pivotal role in the development of biosensors, especially in the context of real-world applications. Nanomaterials are mainly used as transducer materials and are essential for constructing biosensors [28]. Compared to their bulk counterparts, nanomaterials possess unique optical, electrical, chemical, and physical characteristics. Their nanoscale dimensions provide a high surface-to-volume ratio, allowing for more biomolecular interactions. They also exhibit excellent electrocatalytic behaviors, which are vital for detecting various biomolecules. Nanomaterials offer high stability, sensitivity, and selectivity, which are crucial for obtaining reliable biosensor readings over time, detecting analytes at very low concentrations, and binding to specific targets, respectively. Additionally, nanoparticles are easily functionalized with target substrates and operate at the same scale as biological processes [29,30,31,32]. Furthermore, these are devices that the average person can use without much complexity, even in the absence of access to high-tech laboratories [28].
Nanomaterials have remarkable properties and can be utilized in various types of biosensors, including those used for environmental monitoring, food processing, biomedicine, and health care [33,34,35,36,37,38,39,40]. Examples of nanomaterials include carbon nanomaterials, quantum dots, metal nanoclusters, polymer nanocomposites, plasma nanoparticles, and other nanomaterials. These materials enhance the affinity, selectivity, and sensitivity of viral detection [25, 41,42,43,44,45,46,47,48]. Additionally, nanostructures can be designed with multiplexing capability to detect multiple targets simultaneously, which is beneficial for diagnosing diseases like COVID-19 that require the identification of several biomarkers [49].
Some of the nanomaterials extensively used in developing biosensors for COVID-19 diagnostics include:
Gold nanostructures Gold nanoparticles (GNPs) are widely used in point-of-care biosensors due to their excellent electrical, and surface plasmon resonance properties. They are capable of detecting amino acids, enzymes, DNA, and RNA in samples, making them suitable for identifying various mutated forms of coronaviruses [50,51,52,53,54,55].
Metal oxides These nanostructures are used for their semiconducting properties in biosensing applications. They can interact with viral proteins related to SARS-COV-2 infection [56,57,58].
Carbon-based nanomaterials This category includes graphene and carbon nanotubes, known for their high surface area and electrical conductivity, which are beneficial for biosensor platforms [59,60,61,62,63].
Quantum dots These semiconductor nanostructures are used for their optical properties and can be functionalized to detect specific biomolecules related to COVID-19 [64, 67].
Polymer-based nanocomposites These are used for creating flexible biosensors for wearable diagnostics, providing continuous monitoring capabilities [68,69,70]
Silica nanoparticles Due to their biocompatibility and ease of functionalization, silica nanoparticles can be used in biosensors to detect viral particles or antibodies against SARS-COV-2 [71, 72].
These nanostructures are incorporated into various biosensor designs such as paper-based, film-based, and sample-to-answer chip-based biosensors, as well as graphene-based, thread-based, and carbon-based biosensors [73]. Integration of these nanostructures into biosensors represents a significant step forward in the fight against the pandemic. The review will briefly discuss the COVID-19 pathogenesis and the various diagnostic approaches in the next section.
SARS-CoV-2 Diagnostic Targets and Diagnostic Devices
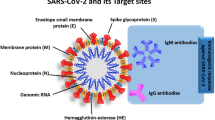
The SARS-COV-2 virus shares a similar cell receptor, angiotensin-converting enzyme II (ACE2), with SARS-CoV and has a genomic similarity of 75–80%. It also exhibits 50% similarity with the Middle East Respiratory Syndrome coronavirus (MERS-CoV) and 96% similarity with a bat coronavirus [74, 75]. Structurally, the SARS-Cov-2 genome contains a large positive-sense strand RNA with a length of about 30kb. It encodes 25 non-structural and accessory proteins as well as four structural proteins. These proteins play various roles ranging from viral assembly to infection, replication, survival, immune evasion, and transmission in host cells [76, 77].
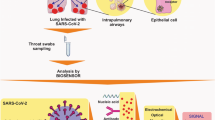
Pathologically, the priming of the virus occurs via viral spike protein (S) through transmembrane protease serine-2 (TMPRSS2) protein on the host membrane which then binds with the angiotensin-converting enzyme 2 (ACE2) receptor as shown in Fig. 1. The ACE2 receptor is widely expressed on the surface of lung alveolar epithelium and enterocytes of the small intestine, contributing to the viral entry of SARS-CoV-2. The SARS-CoV-2 spike protein (S protein) binds to ACE2 with higher affinity compared to SARS-CoV, which might be a factor for increased viral transmissibility and severity of COVID-19 [78].
Pathogenesis of SARS-CoV-2 infection: The coronavirus life cycle begins when (1) the virion attaches itself to the host cell receptor ACE2 using its spike protein. After attaching to the receptor, the virus enters the cytosol through the acid-dependent proteolytic cleavage of the S protein via TMPRSS2. (2) This is followed by the viral and cellular membranes fusing with the help of the S2 subunit and release of viral genome. (3) The replicase is then translated from the genomic RNA. (4) Viral RNA synthesis occurs, and (5) viral replication-transcription complexes are formed. (6) Viral structure proteins (S, E, and M) are translated from the RNA (7) These proteins are inserted into the endoplasmic reticulum, and then move to the endoplasmic reticulum-Golgi intermediate compartment (ERGIC). (8) Genomic RNA is packaged into helical structures (ribonucleoprotein complexes) in the cytoplasm by multiple copies of the nucleocapsid (N protein), which then interact with hydrophobic M proteins (envelope protein) in the ERGIC to control virion assembly. (9) As virions bud from the ERGIC membranes, they are subsequently taken out of the cell by exocytosis
Patients with obesity and diabetes exhibit upregulated expression of ACE2, increasing the susceptibility to SARS-CoV-2 infection and leading to more severe disease in comorbid situations [79]. The virus-host interaction stimulates the production of various proinflammatory cytokines [e.g. Interleukin-1 (IL-1), Interleukin-6 (IL-6), and tumor necrosis factor-α (TNF-α)] and chemokines [e.g. chemokine ligand 5 (CCL5), chemokine (C-X-C motif) ligand 8 (CXCL8)], leading to immunopathogenesis [38, 80,81,82,83]. Dysfunction or exhaustion of CD4 + and CD8 + T cells has been observed [84,85,86,87] and might be responsible for severe immune injury and acute respiratory distress syndrome (ARDS) in patients [87,88,89].
Evidence suggests that COVID-19 affects not only the elderly and adults, but also children of all ages, especially newborns, preterm infants, and children with co-morbidities [90,91,92,93,94].
Conventional Diagnostic Techniques for COVID-19 Détection
Early clinical diagnostics is crucial for disease detection, treatment planning, and disease prognosis. CT scans, RT-PCR, and serological tests are the routinely used diagnostic techniques to detect COVID-19 infection in affected individuals as briefly discussed here:
Real-time PCR Real-time PCR (RT-PCR) tests are considered the gold standard for detecting COVID-19. They detect the genetic material (RNA) of the SARS-CoV-2 virus with high sensitivity and specificity, but can sometimes produce false results. However, RT-PCR tests take time (usually 2–3 days) as the test is done in specialized laboratories, away from the site of sample collection. This technique requires specialized laboratory equipment and trained personnel for sample processing and analysis, which can strain healthcare resources. Additionally, RT-PCR tests only identify the presence of an active virus and do not reveal whether a patient had previously been infected and has since recovered [95,96,97].
Serological assay Many serology-based commercial assays, such as an enzyme-linked immunosorbent assay (ELISA), a chemiluminescence immunoassay (CLIA), and lateral flow immunoassay (LFIA) detect antibodies against viral proteins or the SARS-CoV-2 virus. These detection methods are cost-effective, less time-consuming (about 30 min), and require trained operators. However, they are less sensitive than RT-PCR tests due to the absence of target amplification. RT-PCR tests along with serological immunoassays provide high detection rates of the virus (98.6%). Serological assays are suitable for detecting past infections, but may not be reliable for detecting active infections as antibodies take some time to develop. There is also a risk of cross-reactivity with other viruses leading to false positive results [98,99,100].
CT scans CT scans efficiently image lung variations in symptomatic patients, but can be inaccurate in asymptomatic individuals. They are less time-consuming (30–60 min) and offer higher sensitivity than RT-PCR [101]. However, CT scans are costly, less specific, require expertise, and can have adverse effects on long-term health. Additionally, differentiating COVID-19 infection symptoms from those of other cases of pneumonia is challenging for radiologists [102,103,104,105,106,107,108]. Therefore, CT could be used in conjunction with RT-PCR for a more accurate diagnosis.
Therefore, it is essential to develop cost-effective, time-efficient, and reliable methods to detect SARS-CoV-2 that do not rely on expensive lab equipment or highly trained staff. This is important for making testing more accessible, especially in remote areas where the overall cost of testing can be a burden.
Biosensors offer several advantages over conventional diagnostic techniques, particularly in the rapid and sensitive detection of diseases like COVID-19. They provide real-time data, require minimal reagents, and offer portability, automation, and multi-target analysis, making them suitable for use as point-of-care (POC) devices [109,110,111,112,113]. In the long term, biosensors can streamline the diagnostic process and reduce the need for multiple tests, thus reducing costs. This article provides a comprehensive review of promising biosensor technologies and current biosensors for the diagnosis of COVID-19 (Table 1), with a special emphasis on nanomaterial biosensors.
Nanomaterial-Based Biosensors for COVID-19 Diagnosis
Electrochemical Biosensor
The electrochemical sensing platform provides a sensitive, low-cost, compact, and portable method for assessing metabolic components. Electrochemical biosensors have become valuable tools for detecting pathogens, viral infections, drug levels, and monitoring health indicators like blood glucose, lactate, cholesterol, cancer markers, and cardiovascular disease indicators [124,125,126,127,128,129,130,131,132].
Electrochemical biosensors convert biological events into electrical impulses. They utilize electrodes to immobilize biological molecules, and the use of nanoparticles with large surface areas increases loading capacity and reactant mass transfer, resulting in high analytical sensitivity performance [127, 133,134,135,136]. These biosensors have emerged as a crucial development in the diagnostics of COVID-19, offering several advantages and facing some limitations.
An electrochemical paper-based peptide nucleic acid biosensor has been developed for COVID-19 diagnostics, capable of detecting the SARS-CoV-2 nucleocapsid gene in nasopharyngeal swabs. This sensor applies to real clinical samples with an amplification-free system and provides a high sensitivity and specificity against standard RT-PCR assay. Under optimal conditions, this biosensor has a linear range from 0.1 to 200 nM and a detection limit (LOD) of 1.0 pM [121]. In another approach, where an effective paper-based electrochemical platform was shown to be capable of COVID-19 diagnosis, the sensing strategy was based on the immunocomplex formation between the immobilized spike protein of SARS-CoV-2 and the captured immunoglobulins produced in response to SARS-CoV-2 in humans, which disrupts the redox conversion ([Fe (CN)6]3−/4−). With this approach, SARS-CoV-2 antibodies were detected in 30 min with a sensitivity of 1 ng/ml which was three times higher than the colorimetric lateral flow assay [137].
Another electrochemical biosensor for rapid, sensitive, and nucleic-acid-amplification-free detection of SARS-CoV-2 was based on the trans-cleavage activity of CRISPR/Cas13a. In this biosensor, a nanocomposite and gold nanoflower (AuNF) were progressively coated on the electrode surface to improve the conductivity and increase the sensing area of the surface. With a limit of detection (LOD) of 4.4 × 10−2 fg/ml for the open reading frame (ORF) gene and 8.1 × 10−2 fg/ml for the S gene, the CRISPR/Cas1 3a-assisted electrochemical sensor could detect the ORF and S genes in a wide linear dynamic range from 1.0 × 10−1 to 1.0 × 105 fg/ml. This biosensor has the potential for pre-amplification-free detection of ultra-low concentrations of SARS-CoV-2 RNA and on-site and rapid diagnostic testing for COVID-19 [122].
An electrochemical biosensor has also been developed to identify SARS-CoV-2 RdRp RNA in pharyngeal swabs using an accession approach with a detection limit of 21.69 aM. This detection method showed excellent sensitivity in buffer solution and pharyngeal swabs but is unable to identify target RNA in a homogenous system, which is a drawback when compared to more straightforward PCR techniques. However, it is useful in the clinical detection of SARS-CoV-2 RNA because of its benefits of high sensitivity and high specificity due to facile separation based on the solid phase system detection [138]. Au@Ti3C2@PEI-Ru(dcbpy)32+ nanocomposite-based electrochemiluminescence (ECL) biosensor has also been rationally designed for sensitive detection of the RNA-dependent RNA polymerase (RdRp) gene of SARS-CoV-2. This ECL biosensor sensitively detects the SARS-CoV-2 RdRp gene with a detection range of 1 fM to 100 pM and a limit of detection of 0.21 fM [123].
The dual function of graphene as a transducing platform and an electroactive label for the detection of 2019-nCoV target sequences has been exploited to utilize graphene oxide nano colloids (GONCs) as the electroactive nanomaterial. Briefly, the original electrochemical signal was reduced upon immobilization of the DNA probe (the biorecognition element) onto the GONC platform. This signal was subsequently partially recovered upon the introduction of the 2019-nCoV target. Over a broad linear range from 10−10 to 10−5 M, a direct link was found between the electrochemical signal resulting from the intrinsic electroactivity of GONC and the 2019-nCoV concentration. This method upon incorporation into a DNA amplifier provides a low-cost and portable means of on-site identification of the 2019-CoV virus in infected patients, eliminating the need to transport the sample to centralized lab facilities [139].
Graphene oxide–gold (GO–Au) nanocomposites-based electrochemical immunosensors have also been utilized for simultaneous detection of SARS-CoV-2 antigen and SARS-CoV-2 antibody. The SARS-CoV-2 antigen immunosensor has a linear detection range of 10.0 ag/ml to 50.0 ng/ml and a predicted LOD of 3.99 ag/ml, while the antibody immunosensor has a range of 1.0 fg/ml- 1.0 ng/ml and a high sensitivity of 1.0 fg/ml. The low LOD can be used for the detection of the virus in the serum and patient swab samples from COVID-19-affected individuals [140].
Thus, nanomaterial-based electrochemical biosensors show promise for COVID-19 diagnostics due to their high sensitivity and specificity, even at low concentrations. These biosensors produce results much faster than traditional laboratory methods, which is crucial for early virus detection and containment. Their small size makes them suitable for point-of-care testing, enabling on-site diagnosis without the need for complex lab equipment. They are generally less expensive to produce than other diagnostic tests, making them more accessible for widespread use. Additionally, they are user-friendly and require minimal training to use. However, challenges related to their fabrication, stability, and potential interference leading to false positive results, must be addressed to fully realize their potential. A study suggests that a method called shuffle shepherd optimization-based deep convolution fuzzy network (SSO-GDCFN) diagnoses COVID-19 with 99.99% accuracy. This system uses a variety of biosensors, including electrochemical sensors, and is integrated with AI for improved performance [141].
Surface Plasmon Resonance (SPR)
Surface plasmon resonance (SPR) is a widely used optical sensing technology for real-time, label-free analysis of protein–protein interactions. It helps in determining the kinetic and equilibrium properties of these interactions [142]. SPR biosensors offer a cost-effective and less time-consuming alternative to complex instrumentation and target-labeling procedures. In clinical settings, they are used for various applications such as detecting cancer biomarkers, food allergens, and pathogens [143,144,145]. The SPR-based binding technique involves immobilization of a ligand on the surface of a sensor chip connected to a gold surface. Optical biosensors, including SPR and LSPR, have been commercially available since the early 1990s and have been widely utilized in lab settings to identify viral strains, such as those linked to SARS and MERS, as extensively reviewed by [146]. SPR biosensors have emerged as a powerful tool in COVID-19 diagnostics.
SPR-based sensors can detect specific nucleocapsid antibodies against SARS-CoV-2 in undiluted human serum as opposed to oropharynx swabs. SPR sensor coated with a peptide monolayer and functionalized with the recombinant protein of the SARS-CoV-2 nucleocapsid detects anti-SARS-CoV-2 antibodies in the nM range. It is a fast, label-free bioassay that diagnoses samples within 15 min of sample/sensor contact [147]. Utilizing photonic crystal fiber (PCF) in conjunction with a multilayered surface plasmon resonance (SPR) biosensor based on PCF (Au/BaTiO3/graphene) is helpful in terms of speedy detection of the novel coronavirus and is recommended. The biosensor quickly detects COVID-19 in scenarios where the virus spike receptor-binding domain (RBD) acts as an analyte and monoclonal antibodies (mAbs) as a probe ligand or vice versa [148]. Through the interface, a coupling medium is required for the excitation of photon energy [149]. The fiber-optic absorbance platform (P-FAB) biosensor-based SPR with a gold nanoparticle coating detects COVID-19 [150] without requiring pre-processing of patients’ saliva samples.
Severe Acute Respiratory Syndrome (SARS) kinetics was measured and characterized using SPR. Following its presence in 2002–2003, a study assessed the kinetics of SARS binding to RNA during the phosphorylation and nonphosphorylation of the SARS N-protein. The SPR analysis suggested that nonphosphorylated and phosphorylated N protein bound with the same affinity to viral RNA [151]. SARS N- protein can be detected using an optical QDs-based RNA aptamer chip at as low as 0.1 pg/ml. In this method, the immobilized SARS-CoV nucleoprotein (N-protein) can be selectively hybridized with an RNA aptamer coupled with quantum dots (QDs) on the surface of a glass chip. The optical signal fluctuation of an RNA aptamer supported by QDs serves as the basis for the detection [152].
A five-layer SPR biosensor coated in graphene has been proposed as a quick way to identify the spike protein (S-protein) of the SARS-CoV-2 virus in the early stages without producing false positives. The suggested sensor not only identifies the virus spike protein, but also the spike protein blocking COVID-19, and the virus single-stranded RNA in a fast, less expensive, and more accurate manner [153]. Recently, a novel surface plasmon resonance (SPR) biosensor utilizing sandwiched structures has been described to identify the spike S1 protein of SARS-CoV-2. The sensor exhibits a low detection limit of 12 fg/ml (S/N = 3) with a wide linear range of 0.0001 to 1000 ng/ml and combines a Ti3C2-MXene nanosheets modified with polydopamine (PDA)-Ag nanoparticle (AgNP)/anti-SARS-CoV-2 spike S1 protein nanoconjugate signal enhancers. The MXene-based SPR biochip for SARS-CoV-2 antigen recognition offers a quick and easy method for diagnosing COVID-19 and encourages the use of 2D nanomaterial-based sensing chips in clinical diagnosis and disease screening [154]. Similarly, a surface plasmon resonance (SPR) biosensor based on the dual signal-amplification strategy of Au@Ag@Au nanoparticles (NPs) and graphene oxide (GO) was developed and a sandwich immunoassay was utilized to sensitively and efficiently detect SARS-CoV-2 N protein in artificial saliva simulated samples. In this method, Au@Ag@Au nanoparticles amplify the SPR response signal which was further augmented by GO due to its larger surface area and abundant oxygen-containing functional group. The biosensor efficiently detects SARS-CoV-2 N protein with a detection limit of 0.083 ng/ml, and a linear range of 0.1 ng/ml ~ 1000 ng/ml [155].
SPR in microscale and LSPR in nanoscale viral sensing systems are believed to be beneficial as next-generation detection techniques. In comparison to SPR on a single substrate such as a thin metal film, localized SPR (LSPR) is the plasmon phenomenon produced by irradiating light on metal nanoparticles. In SPR, the optical alterations generated upon binding of the virus with a prism connected to a metal film are used as a signal whereas in LSPR the strong plasmons produced locally on the nanoparticles are utilized as a signal or as an enhancer to increase the intensity of fluorescent compound. Thus, SPR and LSPR reduce false-positive and false-negative results that are common in current viral detection methods [156] and can effectively detect low viral load at an early stage.
A facile wavelength-based SPR sensor built using 3D printing technology with air-stable Near Infrared (NIR)-emitting perovskite nanocomposites as a light source provides a suitable biosensor that is lightweight, compact, plug-less, portable, and appropriate for onsite sample detection. Experimentally, the detection limit of the NIR SPR biosensor is comparable with that of state-of-the-art portable SPR sensors for refractive index change i.e. 10–6 RIU (Refractive index unit). The biosensor was capable of effectively diagnosing COVID-19 patients from health subjects within 15 min due to the high specificity of the used antibody against SARS-CoV-2 [157].
Because of high sensitivity, quick reaction, and no labeling required, surface plasmon resonance (SPR) detection technology has developed quickly and has become a common quantitative analysis method in the domains of biosensing, biomedicine, biochemistry, and biopharmaceuticals. However, SPR faces some limitations such as the design and operation of SPR biosensors can be complex, requiring specialized knowledge and equipment. These biosensors are susceptible to surface fouling, negatively affecting their accuracy and reproducibility, and have limited multiplexing capabilities allowing few targets to be detected at a time. The initial setup and operation costs can also be high compared to other diagnostics making them less suitable in resource-limited settings.
Localized Surface Plasmon Coupled Fluorescence (LSPCF)
Localized surface plasmon stimulates the LSPCF fiber-optic biosensor, which is made up of a complex of immobilized biomolecules stacked in a sandwich pattern on the surface of an optical fiber. Numerous benefits come with this biosensor, including early disease identification, simplicity, and convenience of use [158]. The LSPCF system is a chip-based assay that accurately measures viral proteins in serum, both quantitatively and qualitatively, and has been used in COVID-19 diagnostics. LSPCF fiber-optic biosensor combining a sandwich immunoassay with the LSP technique employing gold nanoparticles (AuNPs) can detect very low concentrations (approx. 1 pg/ml) of recombinant SARS-CoV N-protein (GST-N) in serum. A linear range was observed from 0.1 pg/ml to 1 ng/ml in diluted serum/buffer solution. Using the same monoclonal antibodies, the detection limit of the LSPCF fiber-optic biosensor for the GST-N protein (was improved at least 104 fold in comparison with conventional ELISA. Hence, it is suitable for the diagnosis of SARS infection even before the symptoms appear [116, 159].
The N-protein can also be detected by plasmonic fiber-optic absorbance biosensor (P-FAB), which is a portable device based on a U-bent optical fiber probe. The P-FAB platform detects the N-protein using the label-free and labeled assay [150]. The label-free probe can be used for diagnosis of COVID-19 by introducing the patient’s saliva sample. The binding of SARS-CoV-2 or free N-protein results in a decrease in the light intensity within 15 min. In labeled bioassay, detector antibodies were labeled with gold nanoparticles. The sample from the infected individual was combined with conjugated detector antibody for 5 min, and this complex was added to the bent probe's detecting area formed by immobilization of capture antibody on the U-bent fiber optic probe. The interaction leads to an absorption shift generating signal within 5 min and can identify viral loads as low as 106 particles/ml, which is sufficient to induce infection [150].
Hence, LSPCF significantly enhances the fluorescence signals, leading to improved sensitivity in detecting virus particles. It allows for label-free, rapid detection of the virus, simplifying the diagnostic process and reducing the time and cost associated with labeling. However, the technique requires precise control over the experimental condition and can be complex to set up and interpret. Similar to other plasmonic techniques, LSPCF also suffers from surface fouling producing false positive results in some cases. The high initial equipment set-up cost may also limit its accessibility in resource-limited settings.
Biosensor Field-Effect Transistor (Bio-FET or BioFET)
Field effector biosensors (FETs) are ideal for undertaking a quick, real-time, and label-free detection of many biological analytes, including viruses, cancer biomarkers, and environmental toxins, since they are sensitive to surface charge [114, 160]. They also play a role in personalized medicine by assessing individual response to drugs. To detect a biological element and to enhance the generated electrical signal, the surfaces of the biosensors are replaced with biocompatible and conductive materials.
A unique type of FET utilized for sensing biomolecules in a fluid system is the biofield effect transistor. Because the BioFETs follow electro-analysis of charged biomolecules such as virus-related biomarkers, they are among the best options for coronavirus detection [161]. The source, gate, and drain electrodes; make up the three terminals of a traditional field effect transistor. A gate controls the current flow between the source and the drain in a similar way that a valve controls fluid flow because of the pressure difference between two locations in a pipe. Utilizing silicon nanowires in BioFETs with the integration of microfluidics (channels, valves, etc.) on the same chip forms a fully functional system for at-home diagnostics of viruses and infectious diseases or routine health checkup functions [162].
The term "BioFET sensors" refers to any family of FETs, including gene-, enzyme-, and cell-FETs. They are specifically designed to assess charge-induced field effects in various bio-interface contexts. BioFETs need to have their surface functionalized with specific biorecognition elements (BREs) to detect antigen, antibody, nucleic acid, etc. with high accuracy and specificity. Certain BREs can be immobilized in their ideal configuration to form complementary complexes, which alter the conductance of the channel area. Chem/BioFETs can be broadly divided into three groups: apta-, geno-, and immunosensors. Chem/BioFET are rapid detectors with a LOD down to fM, incredible sensitivity in comparison to other biosensors. Thy are of lower cost when integrated with complementary metal oxide semiconductor (CMOS) technology, and hence have the potential to be used for at-home point of care (POC) diagnostics for COVID-19 detection.
A FET-based biosensor for detecting SARS-CoV-2 in bodily fluids was demonstrated by Seo and associates in which graphene sheets were modified as sensing materials for the device using a monoclonal antibody that was specific to the SARS-CoV-2 spike protein. This method successfully detected SARS-CoV-2 in clinical samples and culture medium, with LODs of 1.6 × 101 pfu/ml and 2.42 × 102 copies/ml, respectively. As a result, this COVID-19 immunosensing method that uses an off-chip readout system appears promising and doesn't require sample pre-processing [163].
In a similar approach, electrical field effect sensors utilized crumpled graphene for probing the charge in the biological entities. Three kind of proteins were captured with specific antibodies including two COVID-19-related proteins, nucleocapsid (N-) and spike (S-) proteins and IL-6. The biomarkers were detectable with the highest sensitivity and extremely low LODs (Hwang et al. 2021). A saliva-based portable COVID-19 antigen test using the electrical double layer (EDL)-gated field-effect transistor-based biosensor (BioFET) has been developed to be used by medical providers and frontline health workers. It detects SARS-CoV-2 nucleocapsid (N) with LODs of 0.34 ng/ml (7.44 pM) and 0.14 ng/ml (2.96 pM) in phosphate buffer saline (PBS, 1x) and artificial saliva, respectively [164]. This is in line with another study where the EDL-BioFET detects the SARS-CoV-2 N-gene cDNAs and viral RNAs in diluted human saliva, with a detection limit of ≈1 fm within 20 min. [165]. In addition, another study by Torel Lopez et al. provides evidences that graphene BioFET (GBioFET) can detect SARS-CoV-2 with high sensitivity and accuracy [166].
Based on the carbon nanotube field-effect transistor (CNT-FET), Zamzami and colleagues have created a rapid (2–3 min), user-friendly, affordable, and quantitative electrochemical biosensor that enables digital detection of the SARS-CoV-2 S1 in fortified saliva samples for prompt and precise detection of SARS-CoV-2 S1 antigens. The biosensor was created via CNT printing on a Si/SiO2 surface. Through non-covalent contact, the SARS-CoV-2 S1 antibody was immobilized on the CNT surface and the commercial SARS-CoV-2 S1 antigen was employed to assess the CNT-FET biosensor's electrical output. The CNT-FET biosensor successfully identified the SARS-CoV-2 S1 antigen at concentrations ranging from 0.1 fg/mL to 5.0 pg/mL in the 10 mM AA buffer pH 6.0. The developed CNT-FET biosensor had a limit of detection (LOD) of 4.12 fg/mL and higher selectivity (no reaction to SARS-CoV-1 S1 or MERS-CoV S1 antigen). The biosensor is time-saving, incredibly sensitive, and may provide a useful platform for quickly detecting SARS-CoV-2 S1 antigen in patient saliva. [167]
Thus, Bio-FETs provide highly sensitive, and label-free real-time detection of COVID-19 and is suitable for portable, point-of-care devices. Incorporation of graphene, metal nanoparticles, and nanotubes further enhances their performance. However, complex fabrication, stability issues, and interference issues limit its use. In addition, current CNT -FET based biosensors are typically capable of detecting only one analyte at a time, limiting their use in complex diagnostic situations. Detection of viruses in complex media such as saliva or blood also poses challenges in the detection of specific signals leading to false positives or negatives.
Colorimetric Biosensor
Colorimetric biosensors are frequently incorporated with biomarker detectors and have the following characteristics: easy to use, less expensive, simple interpretation, visibility, etc. Colorimetric paper-based biosensors are in high demand and most appealing as the presence of a specific pathogen can be easily detected by a simple change in color, which can be differentiated with the naked eye without the use of expensive and complex apparatus [168]. They have been used as point-of-care devices for the detection of pathogenic bacteria in clinical and environmental settings [169, 170]. Low sensitivity is the main drawback of colorimetric assays since it might be challenging to convert observable signals into a color readout [171]. Utilizing nanomaterial in calorimeter biosensors enhances the detection capability of biosensors [157, 172]. Noble metal nanoparticles (gold/silver) incorporated into colorimetric biosensors have increased sensitivity for identifying target biological molecules, including pathogenic bacteria and viruses, DNA, poisons, proteins, and others [172, 173]. Due to their ease of synthesis, chemical and physical stability, superior biocompatibility, distinctive optoelectronic behavior, and ease of modification with organic and bioactive substances, gold nanoparticles (AuNPs) are widely utilized in colorimetric biomedical assays [170, 174,175,176]. AuNPs have a very important property called localized surface plasmon resonance (LSPR). The change of monodispersed AuNPs to an aggregated state is observed by a change in color. Thus, size-induced LSPR effects of AuNPs have been exploited for colorimetric detection [174]. The antibody-functionalized AuNP are even more useful for rapid colorimetric screening of pathogenic viruses [174]. In the case of detection of the COVID-19 virus in human saliva, intense color changes were noted. The observations were made with the naked eye owing to plasmon coupling when f-AuNPs form clusters on the virus, with high sensitivity and a detection limit of 0.28 PFU/ml (PFU stands for plaque-forming units). Plasmon coupling was corroborated with computer simulations using the finite-difference time-domain (FDTD) method and the diagnosis was carried out with a smartphone camera. It provides COVID-19 diagnosis with 100% accuracy for small quantities of saliva samples [177].
For the simultaneous detection of DNA linked to bacterial and viral infectious illnesses, including human papillomavirus (HPV), Mycobacterium tuberculosis (MTB), and Middle East respiratory syndrome coronavirus (MERS-CoV), a multiplex paper-based colorimetric DNA sensor (PAD) was created. AgNPs were employed in DNA detection based on acpcPNA-induced nanoparticle aggregation as a colorimetric reagent. Regarding single-base mismatch, two-base mismatch, and noncomplementary target DNA, this colorimetric DNA sensor demonstrated excellent selectivity. The LOD for MERS-CoV, MTB, and HPV under optimal conditions was determined to be 1.53, 1.27, and 1.03 nM, respectively. Thus, this multiplex colorimetric PAD could be a disposable, low-cost substitute instrument for quick screening and detection in infectious disease diagnostics [178] and can be utilized for COVID-19 diagnostics.
The RNA-extraction-free nano-amplified colorimetric test is another approach that utilizes plasmonic gold nanoparticles capped with an antisense oligonucleotide (ASoS) to provide rapid and naked-eye molecular diagnosis of COVID-19. A unique dual-prong approach that integrates nucleic acid (NA) amplification and plasmonic sensing for point-of-care detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was employed in this test, with a detection time of less than 1 h. The accuracy, specificity, and sensitivity of the test were found to be very high and was > 98.4%, > 96.6%, and 100%, respectively when tested for clinical samples, with a detection limit of 10 copies/μl [120].
Thus, colorimetric biosensors provide a simple, low-cost, portable, and rapid quantitative screening tool with minimal instrumentation for the diagnosis of COVID-19. These devices can be used in point-of-care settings in places with low resources. The results can often be visualized with the naked eye; without the need for complex instruments, which is beneficial in resource-limited settings. However, compared with other diagnostic methods, colorimetric biosensors are less sensitive and have potential for interference which could result in a high rate of false negatives. Also, while they are good for qualitative results, they may not provide precise quantitative data in various testing environments.
CRISPR-Cas Based Biosensors
Clustered regularly interspaced short palindromic repeats (CRISPR)-based detection techniques have emerged as a novel rapid diagnostic approach for accurately detecting pathogens (such as viruses or bacteria) by targeting their genetic material [179]. They have been explored for detecting the RNA genome of SARS-CoV-2 in patient samples in less than an hour. Different research groups around the world have developed methodologies to detect COVID-19 more quickly as reviewed by [180, 181]. Recently, in the United States, an easy-to-implement and accurate CRISPR-Cas12-based lateral flow assay has been developed for the detection of SARS-CoV-2 from RNA extracted from the respiratory swab in less than 40 min. The method has been validated using contrived reference samples and clinical samples from patients affected with COVID-19 infection (n = 36 patients) and with other viral respiratory infections (n = 42 patients). The CRISPR-Cas-based assays are emerging as a promising alternative to RT-PCR assay with 95% and 100% positive and negative predictive agreement, respectively [182].
Mammoth Biosciences in 2018, introduced a technique named DETECTR (DNA Endonuclease-Targeted CRISPR Trans Reporter). It detects sensitive DNA using LbCas12a isolated from the Lachnospiraceae bacterium ND2006. DETECTR accompanied by RT-LAMP (Reverse-transcription-Loop Mediated Isothermal Amplification) is used to detect SARS-CoV-2 with high precision within 30 min [164]. Based on this method, a research group from Saudi Arabia developed a robust molecular diagnostic device “iSCAN”, to facilitate the effective management and control of COVID-19. The group exploited the specific detection ability of SARS-CoV-2 by CRISPR-Cas12 to develop a one-step nucleic acid amplification method known as Reverse transcription-loop-mediated isothermal amplification (RT-LAMP). RT-LAMP is conducted at a single temperature and is utilized for colorimetric virus detection along with CRISPR-Cas12 and CRISPR-Cas13 systems which perform indiscriminate RNA and single-stranded deoxyribonucleic acid (ssDNA) cleavage, respectively. The module is highly efficient, fast, sensitive, specific, and user-friendly for SARS-CoV-2 detection and when combined with lateral flow cells enables highly efficient point-of-care SARS-CoV-2 detection [183]. iSCAN-V2, a one-pot reverse transcription-recombinase polymerase amplification (RT-RPA)-coupled CRISPR/Cas12b-based assay is another point-of-care assay developed for SARS-CoV-2 detection. The assay is coupled with a low-cost, commercially available fluorescence visualizer to enable on-site detection of the virus at a single temperature in less than an hour. Cas12b was more efficient than Cas12a in the iSCAN-V2 detection platform. iSCAN-V2 detection exhibited 93.75% sensitivity and 100% specificity when clinically validated with RT-qPCR on patient samples [184].
Another rapid, sensitive, one-pot point-of-care assay for visual SARS-CoV-2 detection is the All-In-One Dual CRISPR-Cas12a (AIOD-CRISPR) assay. This assay targets SARS-CoV-2’s nucleoprotein gene and two CRISPR RNAs without protospacer adjacent motif (PAM) site limitation and detects the nucleic acids with a high sensitivity. Using a low-cost hand warmer (approximately $0.3) as an incubator for the AIOD-CRISPR assay enables the detection of clinical samples within 20 min. This allows for instrument-free, visual SARS-CoV-2 detection at the point of care [185].
Kumar et al. Developed a lateral flow test capable of detecting a Cas9 protein that binds to a nucleic acid sequence present in a specific mutant strain of SARS-CoV-2. If used only for diagnosis, it can detect mutations in the SARS-CoV-2 virus at a fraction of the price of next-generation sequencing (NGS) approaches. It not only offers a quick and cheap method to monitor the spread of mutant SARS-CoV-2 strains; but also provides a way to determine vaccine efficacy against new viral strains [186]. FnCas9 Editor Linked Uniform Detection Assay (FELUDA) utilizes a direct Cas9-based enzymatic readout for detecting nucleobase and nucleotide sequences without trans cleavage of reporter molecules. FELUDA is 100% accurate in detecting single nucleotide variants (SNVs), including heterozygous carriers, and presents a simple web tool JATAYU to aid end-users. FELUDA uses a lateral flow readout and demonstrates 100% sensitivity and 97% specificity for detecting the virus in clinical samples within 1 h, across all ranges of viral loads. When combined with RT-RPA and a smartphone app called True Outcome Predicted via Strip Evaluation (TOPSE), FELUDA provides a prototype for detecting CoV-2 closer to home [187, 188].
Thus, CRISPR assays are rapid, customizable, and scalable, with high specificity and sensitivity to detect SARS-CoV-2 and its variant strains at home or any point of care with limited resources. Although, simpler than RT-PCR, CRISPR-Cas based biosensors, still require some level of equipment and technical expertise to perform the assay accurately. Therefore, further improvements and adaptations are needed to overcome the current limitations of its widespread application.
Conclusion
The global COVID-19 vaccination effort has demonstrated efficacy in combatting the debilitating SARS-CoV-2 virus, which posed a significant threat to humanity in recent years. Despite this progress, the presence of various virus mutations and increased genetic diversity continue to pose challenges to effective treatment. In addition to vaccination, it is crucial to manage critical COVID-19 patients to reduce disease severity and mortality caused by different COVID-19 variants. Rapid detection of pathogenic DNA/RNA sequences or variants through point-of-care diagnostics has proven the be valuable in expediting clinical prognosis during the recent COVID-19 outbreak. Traditional diagnostic techniques such as qRT-PCR, CT scans, and serological assays are labor-intensive, time-consuming, and may not be readily applicable in remote or resource-constrained settings due to the requirement for laboratory infrastructure.
Biosensors have made significant advancements as an effective diagnostic tool, although they come with associated advantages and limitations. Developing biosensors in point-of-care settings, such as clinics or pharmacies, can facilitate swift screening and alleviate the burden on centralized laboratories. Various commercially available point-of-care devices; including chip-based and paper-based biosensors, offer low-cost and user-friendly solutions. Additionally, nanomaterial-based biosensors provide biocompatibility, selectivity, sensitivity, wearability, and a limit of detection in home settings, making them suitable for large-scale screening techniques.
Approaches such as; nanomaterial-assisted paper-based microfluidics hold great potential in managing the pandemic by providing rapid and accessible results. Further research and innovation to improve the sensitivity, specificity, and scalability of paper-based microfluidic tests will create exciting possibilities for point-of-care testing and clinical diagnostics. Furthermore, the current nano biosensors for COVID-19 diagnostics are multiplexed. Several strategies can be considered to simplify the practical implementation of multiplexed nano sensors. These include integrating nano sensors into portable devices, employing AI and machine-learning algorithms for automated data analysis, and utilizing smartphone components for reading diagnostic assays, thereby reducing the need for specialized technical expertise.
Moreover, enhancing production scalability, collaborating with regulatory bodies, and increasing public awareness and training can strengthen the acceptance and effective utilization of nanomaterial-based biosensors. The early and accurate diagnosis, superior performance, and mass production of nanomaterial-based biosensors could yield long-term cost savings and benefit public health initiatives.
References:
Hernandez Acosta RA, Esquer Garrigos Z, Marcelin JR, Vijayvargiya P (2022) COVID-19 pathogenesis and clinical manifestations. Infect Dis Clin North Am. https://doi.org/10.1016/j.idc.2022.01.003
Boyton RJ, Altmann DM (2021) The immunology of asymptomatic SARS-CoV-2 infection: What are the key questions? Nat Rev Immunol. https://doi.org/10.1038/s41577-021-00631-x
Gibson PG, Qin L, Puah SH (2020) COVID-19 acute respiratory distress syndrome (ARDS): clinical features and differences from typical pre-COVID-19 ARDS. Med J Aust. https://doi.org/10.5694/mja2.50674
Saba SK, Khan F, Lamia S, Shahid MI, Anika T, Akhter S et al (2022) Updates on COVID-19: virology, etiology, epidemiology, pathogenesis, diagnosis, transmission and prevention. Bangladesh Pharm J. https://doi.org/10.3329/bpj.v25i2.60966
Majumder J, Minko T (2021) Recent developments on therapeutic and diagnostic approaches for COVID-19. AAPS J. https://doi.org/10.1208/s12248-020-00532-2
Bhalla N, Jolly P, Formisano N, Estrela P (2016) Introduction to biosensors. Essays Biochem. https://doi.org/10.1042/EBC20150001
Alhadrami HA (2018) Biosensors: classifications, medical applications, and future prospective. Biotechnol Appl Biochem. https://doi.org/10.1002/bab.1621
Mehrotra P (2016) Biosensors and their applications - a review. J Oral Biol Craniofac Res. https://doi.org/10.1016/j.jobcr.2015.12.002
Guo Y, Chen J, Chen G (2013) A label-free electrochemical biosensor for detection of HIV related gene based on interaction between DNA and protein. Sens Actuators B Chem. https://doi.org/10.1016/j.snb.2013.04.046
Farzin L, Shamsipur M, Samandari L, Sheibani S (2020) HIV biosensors for early diagnosis of infection: the intertwine of nanotechnology with sensing strategies. Talanta. https://doi.org/10.1016/j.talanta.2019.120201
Yao CY, Fu WL (2014) Biosensors for hepatitis B virus detection. World J Gastroenterol. https://doi.org/10.3748/wjg.v20.i35.12485
Khristunova E, Dorozhko E, Korotkova E, Kratochvil B, Vyskocil V, Barek J (2020) Label-free electrochemical biosensors for the determination of flaviviruses: dengue, zika, and japanese encephalitis. Sensors (Switzerland). https://doi.org/10.3390/s20164600
Drobysh M, Ramanaviciene A, Viter R, Chen CF, Samukaite-Bubniene U, Ratautaite V, Ramanavicius A (2022) Biosensors for the determination of SARS-CoV-2 virus and diagnosis of COVID-19 infection. Int J Mol Sci. https://doi.org/10.3390/ijms23020666
Dutta R, Rajendran K, Jana SK, Saleena LM, Ghorai S (2023) Use of graphene and its derivatives for the detection of dengue virus. Biosensors. https://doi.org/10.3390/bios13030349
Wei-Wen Hsiao W, Fadhilah G, Lee CC, Endo R, Lin YJ, Angela S, Ku CC, Chang HC, Chiang WH (2023) Nanomaterial-based biosensors for avian influenza virus: a new way forward. Talanta. https://doi.org/10.1016/j.talanta.2023.124892
Khater M, de la Escosura-Muñiz A, Merkoçi A (2017) Biosensors for Plant Pathogen Detection. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2016.09.091
Castillo-Henríquez L, Brenes-Acuña M, Castro-Rojas A, Cordero-Salmerón R, Lopretti-Correa M, Vega-Baudrit JR (2020) Biosensors for the detection of bacterial and viral clinical pathogens. Sensors (Switzerland). https://doi.org/10.3390/s20236926
Li D, Liu L, Huang Q, Tong T, Zhou Y, Li Z, Bai Q, Liang H, Chen L (2021) Recent advances on aptamer-based biosensors for detection of pathogenic bacteria. World J Microbiol Biotechnol. https://doi.org/10.1007/s11274-021-03002-9
Yu T, Xianyu Y (2021) Array-based biosensors for bacteria detection: from the perspective of recognition. Small. https://doi.org/10.1002/smll.202006230
Mobed A, Malehmir S, Ahmad alipour A, Azizimoghaddam Y, Sarabi HS, Ghazi F (2022) Biosensors, modern technology for the detection of cancer-associated bacteria. Biotechnol Lett. https://doi.org/10.1007/s10529-022-03257-8
Clark LC, Lyons C (1962) Electrode systems for continuous monitoring in cardiovascular surgery. Ann N Y Acad Sci. https://doi.org/10.1111/j.1749-6632.1962.tb13623.x
Guilbault GG, Montalvo JG (1969) A urea-specific enzyme electrode. J Am Chem Soc. https://doi.org/10.1021/ja01036a083
Scheller FW, Schubert F, Neumann B, Pfeiffer D, Hintsche R, Dransfeld I et al (1991) Second generation biosensors. Biosens Bioelectron. https://doi.org/10.1016/0956-5663(91)80010-U
Adachi T, Sowa K (2023) Research and development of third-generation biosensors. Bunseki Kagaku. https://doi.org/10.2116/bunsekikagaku.72.483
Malik S, Singh J, Goyat R, Saharan Y, Chaudhry V, Umar A et al (2023) Nanomaterials-based biosensor and their applications: a review. Heliyon. https://doi.org/10.1016/j.heliyon.2023.e19929
Zhang W, Li G (2004) Third-generation biosensors based on the direct electron transfer of proteins. Anal Sci. https://doi.org/10.2116/analsci.20.603
Naikoo GA, Awan T, Salim H, Arshad F, Hassan IU, Pedram MZ et al (2022) Fourth-generation glucose sensors composed of copper nanostructures for diabetes management: a critical review. Bioeng Transl Med. https://doi.org/10.1002/btm2.10248
Malhotra BD, Ali Md. A (2018) Chapter 1 - Nanomaterials in biosensors: fundamentals and applications. Micro and Nano Technologies
Kumar A, Purohit B, Maurya PK, Pandey LM, Chandra P (2019) Engineered nanomaterial assisted signal-amplification strategies for enhancing analytical performance of electrochemical biosensors. Electroanalysis. https://doi.org/10.1002/elan.201900216
Lee SH, Sung JH, Park TH (2012) Nanomaterial-based biosensor as an emerging tool for biomedical applications. Ann Biomed Eng. https://doi.org/10.1007/s10439-011-0457-4
Su H, Li S, Jin Y, Xian Z, Yang D, Zhou W et al (2017) Nanomaterial-based biosensors for biological detections. Adv Health Care Technol. https://doi.org/10.2147/ahct.s94025
Yun YH, Eteshola E, Bhattacharya A, Dong Z, Shim JS, Conforti L et al (2009) Tiny medicine: nanomaterial-based biosensors. Sensors. https://doi.org/10.3390/s91109275
Al-Douri Y, Mansoob Khan M, Robert Jennings J, Abd El-Rehim AF (2022) Nanomaterial-based biosensors for COVID-19 detection. Critical Rev Solid State Mater Sci. https://doi.org/10.1080/10408436.2021.1989665
Britto JSJ, Guan X, Tran TKA, Lei Z, Bahadur R, Patel V et al (2024) Emerging multifunctional carbon-nanomaterial-based biosensors for cancer diagnosis. Small Sci. https://doi.org/10.1002/smsc.202300221
Das S, Saha B, Tiwari M, Tiwari DK (2023) Diagnosis of cancer using carbon nanomaterialbased biosensors. Sensors Diagn. https://doi.org/10.1039/d2sd00182a
Lan L, Yao Y, Ping J, Ying Y (2017) Recent advances in nanomaterial-based biosensors for antibiotics detection. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2017.01.007
Taguchi M, Ptitsyn A, McLamore ES, Claussen JC (2014) Nanomaterial-mediated biosensors for monitoring glucose. J Diabetes Sci Technol. https://doi.org/10.1177/1932296814522799
Zhang C, Jiang C, Lan L, Ping J, Ye Z, Ying Y (2021) Nanomaterial-based biosensors for agro-product safety. TrAC - Trends Anal Chem. https://doi.org/10.1016/j.trac.2021.116369
Zhang W, Asiri AM, Liu D, Du D, Lin Y (2014) Nanomaterial-based biosensors for environmental and biological monitoring of organophosphorus pesticides and nerve agents. TrAC - Trends Anal Chem. https://doi.org/10.1016/j.trac.2013.10.007
Zheng L, Jin W, Xiong K, Zhen H, Li M, Hu Y (2023) Nanomaterial-based biosensors for the detection of foodborne bacteria: a review. Analyst. https://doi.org/10.1039/d3an01554h
Karabulut G, Beköz Üllen N, Karakuş S (2022) Nanostructures in biosensors: development and applications. https://doi.org/10.5772/intechopen.108508
Khan R, Radoi A, Rashid S, Hayat A, Vasilescu A, Andreescu S (2021) Two-dimensional nanostructures for electrochemical biosensor. Sensors. https://doi.org/10.3390/s21103369
Kumar A, Sharma S, Pandey LM, Chandra P (2018) Nanoengineered material based biosensing electrodes for enzymatic biofuel cells applications. Mater Sci Energy Technol. https://doi.org/10.1016/j.mset.2018.04.001
Napi MLM, Sultan SM, Ismail R, How KW, Ahmad MK (2019) Electrochemical-based biosensors on different zinc oxide nanostructures: a review. Materials. https://doi.org/10.3390/ma12182985
Naresh V, Lee N (2021) A review on biosensors and recent development of nanostructured materials-enabled biosensors. Sensors (Switzerland). https://doi.org/10.3390/s21041109
Wang H, Wang T, Yuan X, Wang Y, Yue X, Wang L et al (2023) Plasmonic nanostructure biosensors: a review. Sensors. https://doi.org/10.3390/s23198156
Welch EC, Powell JM, Clevinger TB, Fairman AE, Shukla A (2021) Advances in biosensors and diagnostic technologies using nanostructures and nanomaterials. Adv Funct Mater. https://doi.org/10.1002/adfm.202104126
Yeom SH, Kang BH, Kim KJ, Kang SW (2011) Nanostructures in biosensor-a review. Front Biosci. https://doi.org/10.2741/3731
Xi H, Jiang H, Juhas M, Zhang Y (2021) Multiplex biosensing for simultaneous detection of mutations in SARS-CoV-2. ACS Omega. https://doi.org/10.1021/acsomega.1c04024
Beduk T, Beduk D, de Oliveira Filho JI, Zihnioglu F, Cicek C, Sertoz R et al (2021) Rapid point-of-care COVID-19 diagnosis with a gold-nanoarchitecture-assisted laser-scribed graphene biosensor. Anal Chem. https://doi.org/10.1021/acs.analchem.1c01444
El-Said WA, Al-Bogami AS, Alshitari W (2022) Synthesis of gold nanoparticles@reduced porous graphene-modified ITO electrode for spectroelectrochemical detection of SARS-CoV-2 spike protein. Spectrochim Acta A Mol Biomol Spectrosc. https://doi.org/10.1016/j.saa.2021.120237
Hatamluyi B, Rezayi M, Amel Jamehdar S, Rizi KS, Mojarrad M, Meshkat Z et al (2022) Sensitive and specific clinically diagnosis of SARS-CoV-2 employing a novel biosensor based on boron nitride quantum dots/flower-like gold nanostructures signal amplification. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2022.114209
Hryniewicz BM, Volpe J, Bach-Toledo L, Kurpel KC, Deller AE, Soares AL et al (2022) Development of polypyrrole (nano)structures decorated with gold nanoparticles toward immunosensing for COVID-19 serological diagnosis. Mater Today Chem. https://doi.org/10.1016/j.mtchem.2022.100817
Khan J, Rasmi Y, Kırboğa KK, Ali A, Rudrapal M, Patekar RR (2022) Development of gold nanoparticle-based biosensors for COVID-19 diagnosis. Beni Suef Univ J Basic Appl Sci. https://doi.org/10.1186/s43088-022-00293-1
Vidotti M, Carvalhal RF, Mendes RK, Ferreira DCM, Kubota LT (2011) Biosensors based on gold nanostructures. J Braz Chem Soc. https://doi.org/10.1590/S0103-50532011000100002
Hou S, Wu M, Li H, Gong HR, Gao Z, Shi R et al (2023) Ultrasensitive detection of SARS-CoV-2 by flexible metal oxide field-effect transistors. Adv Funct Mater. https://doi.org/10.1002/adfm.202301268
Jouyandeh M, Sajadi SM, Seidi F, Habibzadeh S, Munir MT, Abida O et al (2022) Metal nanoparticles-assisted early diagnosis of diseases. OpenNano. https://doi.org/10.1016/j.onano.2022.100104
Mondal J, An JM, Surwase SS, Chakraborty K, Sutradhar SC, Hwang J et al (2022) Carbon nanotube and its derived nanomaterials based high performance biosensing platform. Biosensors (Basel). https://doi.org/10.3390/bios12090731
Balkourani G, Brouzgou A, Archonti M, Papandrianos N, Song S, Tsiakaras P (2021) Emerging materials for the electrochemical detection of COVID-19. J Electroanal Chem. https://doi.org/10.1016/j.jelechem.2021.115289
Fahmy HM, Abu Serea ES, Salah-Eldin RE, Al-Hafiry SA, Ali MK, Shalan AE et al (2022) Recent progress in graphene- and related carbon-nanomaterial-based electrochemical biosensors for early disease detection. ACS Biomater Sci Eng. https://doi.org/10.1021/acsbiomaterials.1c00710
Haghayegh F, Salahandish R, Hassani M, Sanati-Nezhad A (2022) Highly stable buffer-based zinc oxide/reduced graphene oxide nanosurface chemistry for rapid immunosensing of SARS-CoV-2 antigens. ACS Appl Mater Interfaces. https://doi.org/10.1021/acsami.1c24475
Kim SY, Lee JC, Seo G, Woo JH, Lee M, Nam J et al (2022) Computational method-based optimization of carbon nanotube thin-film immunosensor for rapid detection of SARS-CoV-2 Virus. Small Sci. https://doi.org/10.1002/smsc.202100111
Mishra S, Aamna B, Parida S, Dan AK (2023) Carbon-based biosensors: next-generation diagnostic tool for target-specific detection of SARS-CoV-2 (COVID-19). Talanta Open. https://doi.org/10.1016/j.talo.2023.100218
Byakodi M, Shrikrishna NS, Sharma R, Bhansali S, Mishra Y, Kaushik A et al (2022) Emerging 0D, 1D, 2D, and 3D nanostructures for efficient point-of-care biosensing. Biosens Bioelectron X. https://doi.org/10.1016/j.biosx.2022.100284
Manivannan S, Ponnuchamy K (2020) Quantum dots as a promising agent to combat COVID-19. Appl Organomet Chem. https://doi.org/10.1002/aoc.5887
Zhao Y, Chen J, Hu Z, Chen Y, Tao Y, Wang L et al (2022) All-solid-state SARS-CoV-2 protein biosensor employing colloidal quantum dots-modified electrode. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2022.113974
Zheng Y, Song K, Cai K, Liu L, Tang D, Long W et al (2022) B-cell-epitope-based fluorescent quantum dot biosensors for SARS-CoV-2 enable highly sensitive COVID-19 antibody detection. Viruses. https://doi.org/10.3390/v14051031
Tran VV (2022) Conjugated polymers-based biosensors for virus detection: lessons from COVID-19. Biosensors (Basel). https://doi.org/10.3390/bios12090748
Wang S, Wang C, Xin Y, Li Q, Liu W (2022) Core–shell nanocomposite of flower-like molybdenum disulfide nanospheres and molecularly imprinted polymers for electrochemical detection of anti COVID-19 drug favipiravir in biological samples. Microchim Acta. https://doi.org/10.1007/s00604-022-05213-9
Yaiwong P, Wiratchan S, Semakul N, Bamrungsap S, Jakmunee J, Ounnunkad K (2024) Label-free electrochemical immunosensor employing new redox probes/porous organic polymers/graphene oxide nanocomposite towards multiplex detection of three SARS-COV2-induced storming proteins for severe COVID-19 diagnosis. Mater Today Chem. https://doi.org/10.1016/j.mtchem.2024.101906
Mudiganti M, Magna G, di Zazzo L, Stefanelli M, Capuano R, Catini A et al (2022) Porphyrinoids coated silica nanoparticles capacitive sensors for COVID-19 detection from the analysis of blood serum volatolome. Sens Actuators B Chem. https://doi.org/10.1016/j.snb.2022.132329
Theobald N (2020) Emerging vaccine delivery systems for COVID-19: Functionalised silica nanoparticles offer a potentially safe and effective alternative delivery system for DNA/RNA vaccines and may be useful in the hunt for a COVID-19 vaccine. Drug Discov Today. https://doi.org/10.1016/j.drudis.2020.06.020
Ramesh M, Janani R, Deepa C, Rajeshkumar L (2023) Nanotechnology-enabled biosensors: a review of fundamentals, design principles, materials, and applications. Biosensors (Basel). https://doi.org/10.3390/bios13010040
Arabi YM, Murthy S, Webb S (2020) COVID-19: a novel coronavirus and a novel challenge for critical care. Intensive Care Med. https://doi.org/10.1007/s00134-020-05955-1
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H et al (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet. https://doi.org/10.1016/S0140-6736(20)30251-8
Brian DA, Baric RS (2005) Coronavirus genome structure and replication. Curr Top Microbiol Immunol. https://doi.org/10.1007/3-540-26765-4_1
Chen Y, Liu Q, Guo D (2020) Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. https://doi.org/10.1002/jmv.25681
Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O et al (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 1979:367. https://doi.org/10.1126/science.aax0902
Herman-Edelstein M, Guetta T, Barnea A, Waldman M, Ben-Dor N, Barak Y et al (2021) Expression of the SARS-CoV-2 receptorACE2 in human heart is associated with uncontrolled diabetes, obesity, and activation of the renin angiotensin system. Cardiovasc Diabetol. https://doi.org/10.1186/s12933-021-01275-w
Cevik M, Kuppalli K, Kindrachuk J, Peiris M (2020) Virology, transmission, and pathogenesis of SARS-CoV-2. The BMJ. https://doi.org/10.1136/bmj.m3862
Xu G, Liu Y, Liao M, Gou J, Wang X, Yuan J et al (2020) Persistent viral activity, cytokine storm, and lung fibrosis in a case of severe COVID-19. Clin Transl Med. https://doi.org/10.1002/ctm2.224
Zhang L, Han C, Zhang S, Duan C, Shang H, Bai T et al (2021) Diarrhea and altered inflammatory cytokine pattern in severe coronavirus disease 2019: Impact on disease course and in-hospital mortality. J Gastroenterol Hepatol (Australia). https://doi.org/10.1111/jgh.15166
Zhu C, He G, Yin Q, Zeng L, Ye X, Shi Y et al (2021) Molecular biology of the SARs-CoV-2 spike protein: a review of current knowledge. J Med Virol. https://doi.org/10.1002/jmv.27132
Huang B, Cai Y, Li N, Li K, Wang Z, Li L et al (2021) Sex-based clinical and immunological differences in COVID-19. BMC Infect Dis. https://doi.org/10.1186/s12879-021-06313-2
Martonik D, Parfieniuk-Kowerda A, Starosz A, Grubczak K, Moniuszko M, Flisiak R (2023) Effect of antiviral and immunomodulatory treatment on a cytokine profile in patients with COVID-19. Front Immunol. https://doi.org/10.3389/fimmu.2023.1222170
Rezaei M, Marjani M, Mahmoudi S, Mortaz E, Mansouri D (2021) Dynamic changes of lymphocyte subsets in the course of COVID-19. Int Arch Allergy Immunol. https://doi.org/10.1159/000514202
Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C et al (2020) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. https://doi.org/10.1016/S2213-2600(20)30076-X
Gozzi-Silva SC, Oliveira LM, Alberca RW, Pereira NZ, Yoshikawa FS, Pietrobon AJ et al (2022) Generation of cytotoxic T cells and dysfunctional CD8 T cells in severe COVID-19 patients. Cells. https://doi.org/10.3390/cells11213359
Rha MS, Shin EC (2021) Activation or exhaustion of CD8+ T cells in patients with COVID-19. Cell Mol Immunol. https://doi.org/10.1038/s41423-021-00750-4
Cheng DR, Schrader S, McMinn A, Crawford NW, Tosif S, McNab S et al (2023) Paediatric admissions with SARS-CoV-2 during the Delta and Omicron waves: an Australian single-centre retrospective study. BMJ Paediatr Open. https://doi.org/10.1136/bmjpo-2023-001874
Choi JH, Choi SH, Yun KW (2022) Risk factors for severe COVID-19 in children: a systematic review and meta-analysis. J Korean Med Sci. https://doi.org/10.3346/JKMS.2022.37.E35
Chun JY, Jeong H, Kim Y (2022) Age-varying susceptibility to the delta variant (B.1.617.2) of SARS-CoV-2. JAMA Netw Open. https://doi.org/10.1001/jamanetworkopen.2022.3064
Cui X, Zhao Z, Zhang T, Guo W, Guo W, Zheng J et al (2021) A systematic review and meta-analysis of children with coronavirus disease 2019 (COVID-19). J Med Virol. https://doi.org/10.1002/jmv.26398
Han MS, Kim KM, Oh KJ, Chang JY, Lee SY, Choi JE et al (2023) Distinct clinical and laboratory features of COVID-19 in children during the pre-delta, delta and omicron wave. Pediatric Infect Dis J. https://doi.org/10.1097/INF.0000000000003872
Rahbari R, Moradi N, Abdi M (2021) rRT-PCR for SARS-CoV-2: analytical considerations. Clin Chim Acta. https://doi.org/10.1016/j.cca.2021.01.011
Tombuloglu H, Sabit H, Al-Khallaf H, Kabanja JH, Alsaeed M, Al-Saleh N et al (2022) Multiplex real-time RT-PCR method for the diagnosis of SARS-CoV-2 by targeting viral N, RdRP and human RP genes. Sci Rep. https://doi.org/10.1038/s41598-022-06977-z
Torretta S, Zuccotti G, Cristofaro V, Ettori J, Solimeno L, Battilocchi L et al (2021) Diagnosis of SARS-CoV-2 by RT-PCR using different sample sources: review of the literature. Ear Nose Throat J. https://doi.org/10.1177/0145561320953231
Ismail AAA (2020) Serological tests for COVID-19 antibodies: limitations must be recognized. Ann Clin Biochem. https://doi.org/10.1177/0004563220927053
Lisboa Bastos M, Tavaziva G, Abidi SK, Campbell JR, Haraoui LP, Johnston JC et al (2020) Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. The BMJ. https://doi.org/10.1136/bmj.m2516
Tantuoyir MM, Rezaei N (2021) Serological tests for COVID-19: Potential opportunities. Cell Biol Int. https://doi.org/10.1002/cbin.11516
Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W et al (2020) Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. https://doi.org/10.1148/radiol.2020200642
de Smet K, de Smet D, Ryckaert T, Laridon E, Heremans B, Vandenbulcke R et al (2020) Diagnostic performance of chest CT for SARS-CoV-2 infection in individuals with or without COVID-19 symptoms. Radiology. https://doi.org/10.1148/RADIOL.2020202708
Jamal Z, Bozdar N, Sohu DM, Ullah I, Phul AH, Hafeez R (2022) Role of computed tomography scan chest for management. Pak J Med Health Sci. https://doi.org/10.53350/pjmhs2216761
Kwee TC, Kwee RM (2020) Chest ct in covid-19: What the radiologist needs to know. Radiographics. https://doi.org/10.1148/rg.2020200159
Pontone G, Scafuri S, Mancini ME, Agalbato C, Guglielmo M, Baggiano A et al (2021) Role of computed tomography in COVID-19. J Cardiovasc Comput Tomogr. https://doi.org/10.1016/j.jcct.2020.08.013
Shao JM, Ayuso SA, Deerenberg EB, Elhage SA, Augenstein VA, Heniford BT (2020) A systematic review of CT chest in COVID-19 diagnosis and its potential application in a surgical setting. Colorectal Dis. https://doi.org/10.1111/codi.15252
Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J (2020) Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. J Clin Microbiol. https://doi.org/10.1148/radiol.2020200343
Yoon SH, Lee JH, Kim BN (2023) Chest CT findings in hospitalized patients with SARS-CoV-2: delta versus omicron variants. Radiology. https://doi.org/10.1148/radiol.220676
Choi JR (2020) Development of point-of-care biosensors for COVID-19. Front Chem. https://doi.org/10.3389/fchem.2020.00517
Gowri A, Ashwin Kumar N, Suresh Anand BS (2021) Recent advances in nanomaterials based biosensors for point of care (PoC) diagnosis of Covid-19—a minireview. TrAC - Trends Anal Chem. https://doi.org/10.1016/j.trac.2021.116205
Malekjahani A, Sindhwani S, Syed AM, Chan WCW (2019) Engineering steps for mobile point-of-care diagnostic devices. Acc Chem Res. https://doi.org/10.1021/acs.accounts.9b00200
Strong EB, Schultz SA, Martinez AW, Martinez NW (2019) Fabrication of miniaturized paper-based microfluidic devices (MicroPADs). Sci Rep. https://doi.org/10.1038/s41598-018-37029-0
Yadav SK, Verma D, Yadav U, Kalkal A, Priyadarshini N, Kumar A et al (2023) Point-of-care devices for viral detection: COVID-19 pandemic and beyond. Micromachines (Basel). https://doi.org/10.3390/mi14091744
Panahi A, Sadighbayan D, Forouhi S, Ghafar-Zadeh E (2021) Recent advances of field-effect transistor technology for infectious diseases. Biosensors (Basel). https://doi.org/10.3390/bios11040103
Sadighbayan D, Hasanzadeh M, Ghafar-Zadeh E (2020) Biosensing based on field-effect transistors (FET): recent progress and challenges. TrAC - Trends Anal Chem. https://doi.org/10.1016/j.trac.2020.116067
Huang JC, Chang YF, Chen KH, Su LC, Lee CW, Chen CC et al (2009) Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2009.07.012
Yang Y, Murray J, Haverstick J, Tripp RA, Zhao Y (2022) Silver nanotriangle array based LSPR sensor for rapid coronavirus detection. Sens Actuators B Chem. https://doi.org/10.1016/j.snb.2022.131604
Naikoo GA, Arshad F, Hassan IU, Awan T, Salim H, Pedram MZ, Ahmed W, Patel V, Karakoti AS, Vinu A (2022) Nanomaterials-based sensors for the detection of COVID-19: a review. Bioeng Transl Med. https://doi.org/10.1002/btm2.10305
Pandey PS, Raghuwanshi SK, Shadab A, Ansari MTI, Tiwari UK, Kumar S (2022) SPR based biosensing chip for COVID-19 diagnosis - a review. IEEE Sens J. https://doi.org/10.1109/JSEN.2022.3181423
Alafeef M, Moitra P, Dighe K, Pan D (2021) RNA-extraction-free nano-amplified colorimetric test for point-of-care clinical diagnosis of COVID-19. Nat Protoc. https://doi.org/10.1038/s41596-021-00546-w
Lomae A, Preechakasedkit P, Hanpanich O, Ozer T, Henry CS, Maruyama A et al (2023) Label free electrochemical DNA biosensor for COVID-19 diagnosis. Talanta. https://doi.org/10.1016/j.talanta.2022.123992
Heo W, Lee K, Park S, Hyun KA, Jung HI (2022) Electrochemical biosensor for nucleic acid amplification-free and sensitive detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA via CRISPR/Cas13a trans-cleavage reaction. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2021.113960
Yao B, Zhang J, Fan Z, Ding Y, Zhou B, Yang R et al (2021) Rational engineering of the DNA walker amplification strategy by using a Au@Ti3C2@PEI-Ru(dcbpy)32+nanocomposite biosensor for detection of the SARS-CoV-2 RdRp gene. ACS Appl Mater Interfaces. https://doi.org/10.1021/acsami.1c04453
Aamri ME, Yammouri G, Mohammadi H, Amine A, Korri-Youssoufi H (2020) Electrochemical biosensors for detection of microRNA as a cancer biomarker: pros and cons. Biosensors (Basel). https://doi.org/10.3390/BIOS10110186
Bakirhan NK, Ozcelikay G, Ozkan SA (2018) Recent progress on the sensitive detection of cardiovascular disease markers by electrochemical-based biosensors. J Pharm Biomed Anal. https://doi.org/10.1016/j.jpba.2018.07.021
Caldevilla R, Morais SL, Cruz A, Delerue-Matos C, Moreira F, Pacheco JG et al (2023) Electrochemical chemically based sensors and emerging enzymatic biosensors for antidepressant drug detection: a review. Int J Mol Sci. https://doi.org/10.3390/ijms24108480
Curulli A (2023) Functional nanomaterials enhancing electrochemical biosensors as smart tools for detecting infectious viral diseases. Molecules. https://doi.org/10.3390/molecules28093777
Liu M, Yang M, Wang M, Wang H, Cheng J (2022) A flexible dual-analyte electrochemical biosensor for salivary glucose and lactate detection. Biosensors (Basel). https://doi.org/10.3390/bios12040210
Ma R, Liu R, Xia F (2023) “Electrochemical biosensors for drugs detection. Electrochem Biosens Whole Blood Anal. https://doi.org/10.1007/978-981-99-5644-9_6
Pan M, Zhao Y, Qiao J, Meng X (2024) Electrochemical biosensors for pathogenic microorganisms detection based on recognition elements. Folia Microbiol (Praha). https://doi.org/10.1007/s12223-024-01144-5
Rudewicz-Kowalczyk D, Grabowska I (2022) electrochemical biosensors for cardiovascular diseases’ (CVDs) biomarkers detection. Engineering. https://doi.org/10.3390/engproc2022021011
Sadighbayan D, Sadighbayan K, Tohid-kia MR, Khosroushahi AY, Hasanzadeh M (2019) Development of electrochemical biosensors for tumor marker determination towards cancer diagnosis: recent progress. TrAC - Trends Anal Chem. https://doi.org/10.1016/j.trac.2019.05.014
Bi R, Ma X, Miao K, Ma P, Wang Q (2023) Enzymatic biosensor based on dendritic gold nanostructure and enzyme precipitation coating for glucose sensing and detection. Enzyme Microb Technol. https://doi.org/10.1016/j.enzmictec.2022.110132
Farzin L, Shamsipur M (2018) Recent advances in design of electrochemical affinity biosensors for low level detection of cancer protein biomarkers using nanomaterial-assisted signal enhancement strategies. J Pharm Biomed Anal. https://doi.org/10.1016/j.jpba.2017.07.042
Kalambate PK, Noiphung J, Rodthongkum N, Larpant N, Thirabowonkitphithan P, Rojanarata T et al (2021) Nanomaterials-based electrochemical sensors and biosensors for the detection of non-steroidal anti-inflammatory drugs. TrAC - Trends Anal Chem. https://doi.org/10.1016/j.trac.2021.116403
Naikoo GA, Awan T, Hassan IU, Salim H, Arshad F, Ahmed W et al (2021) Nanomaterials-based sensors for respiratory viral detection: a review. IEEE Sens J. https://doi.org/10.1109/JSEN.2021.3085084
Yakoh A, Pimpitak U, Rengpipat S, Hirankarn N, Chailapakul O, Chaiyo S (2021) Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2020.112912
Zhou K, Dai J (2023) A duplex-specific nuclease based electrochemical biosensor for the assay of SARS-CoV-2 RdRp RNA. Anal Biochem. https://doi.org/10.1016/j.ab.2022.114983
Ang WL, Lim RRX, Ambrosi A, Bonanni A (2022) Rapid electrochemical detection of COVID-19 genomic sequence with dual-function graphene nanocolloids based biosensor. FlatChem. https://doi.org/10.1016/j.flatc.2022.100336
Sadique MA, Yadav S, Ranjan P, Khan R, Khan F, Kumar A et al (2022) Highly sensitive electrochemical immunosensor platforms for Dual detection of SARS-CoV-2 antigen and antibody based on gold nanoparticle functionalized graphene oxide nanocomposites. ACS Appl Bio Mater. https://doi.org/10.1021/acsabm.2c00301
Alam MM, Alam MM, Mirza H, Sultana N, Sultana N, Pasha AA et al (2023) A Novel COVID-19 diagnostic system using biosensor incorporated artificial intelligence technique. Diagnostics. https://doi.org/10.3390/diagnostics13111886
Wang S, Shan X, Patel U, Huang X, Lu J, Li J et al (2010) Label-free imaging, detection, and mass measurement of single viruses by surface plasmon resonance. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.1005264107
Bellassai N, D’Agata R, Jungbluth V, Spoto G (2019) Surface plasmon resonance for biomarker detection: advances in non-invasive cancer diagnosis. Front Chem. https://doi.org/10.3389/fchem.2019.00570
Chiu NF, Tai MJ, Nurrohman DT, Lin TL, Wang YH, Chen CY (2021) Immunoassay-amplified responses using a functionalized MoS2-based SPR biosensor to detect PAPP-A2 in maternal serum samples to screen for fetal down’s syndrome. Int J Nanomed. https://doi.org/10.2147/IJN.S296406
Souto DEP, Volpe J, Gonçalves CC, Ramos CHI, Kubota LT (2019) A brief review on the strategy of developing SPR-based biosensors for application to the diagnosis of neglected tropical diseases. Talanta. https://doi.org/10.1016/j.talanta.2019.120122
Antiochia R (2020) Nanobiosensors as new diagnostic tools for SARS, MERS and COVID-19: from past to perspectives. Microchim Acta. https://doi.org/10.1007/s00604-020-04615-x
Djaileb A, Charron B, Jodaylami MH, Thibault V, Coutu J, Stevenson K et al (2020) A rapid and quantitative serum test for SARS-CoV-2 antibodies with portable surface plasmon resonance sensing. ChemRxiv
Gupta A, Singh T, Singh RK, Tiwari A (2023) Numerical analysis of coronavirus detection using photonic crystal fibre-based SPR sensor. Plasmonics. https://doi.org/10.1007/s11468-022-01761-1
Taha BA, Al Mashhadany Y, Bachok NN, Ashrif A, Bakar A, Hafiz Mokhtar MH, Dzulkefly Bin Zan MS et al (2021) Detection of covid-19 virus on surfaces using photonics: challenges and perspectives. Diagnostics. https://doi.org/10.3390/diagnostics11061119
Murugan D, Bhatia H, Sai VVR, Satija J (2020) P-FAB: a fiber-optic biosensor device for rapid detection of COVID-19. Trans Indian Natl Acad Eng. https://doi.org/10.1007/s41403-020-00122-w
Chen H, Gill A, Dove BK, Emmett SR, Kemp CF, Ritchie MA et al (2005) Mass spectroscopic characterization of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding by using surface plasmon resonance. J Virol. https://doi.org/10.1128/jvi.79.2.1164-1179.2005
Roh C, Jo SK (2011) Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip. J Chem Technol Biotechnol. https://doi.org/10.1002/jctb.2721
Akib TBA, Mou SF, Rahman MM, Rana MM, Islam MR, Mehedi IM et al (2021) Design and numerical analysis of a graphene-coated SPR biosensor for rapid detection of the novel coronavirus. Sensors. https://doi.org/10.3390/s21103491
Wu Q, Wu W, Chen F, Ren P (2022) Highly sensitive and selective surface plasmon resonance biosensor for the detection of SARS-CoV-2 spike S1 protein. Analyst. https://doi.org/10.1039/d2an00426g
Fan L, Du B, Pei F, Hu W, Feng S, Liu B et al (2023) A novel SPR immunosensor based on dual signal amplification strategy for detection of SARS-CoV-2 nucleocapsid protein. Biosensors (Basel). https://doi.org/10.3390/bios13050549
Takemura K (2021) Surface plasmon resonance (Spr)-and localized spr (lspr)-based virus sensing systems: optical vibration of nano-and micro-metallic materials for the development of next-generation virus detection technology. Biosensors (Basel). https://doi.org/10.3390/bios11080250
Chen LC, Li MC, Chen KR, Cheng YJ, Wu XY, Chen SA et al (2023) Facile and unplugged surface plasmon resonance biosensor with NIR-emitting perovskite nanocomposites for fast detection of SARS-CoV-2. Anal Chem. https://doi.org/10.1021/acs.analchem.2c05661
Zhang H, Zhou X, Li X, Gong P, Zhang Y, Zhao Y (2023) Recent advancements of LSPR fiber-optic biosensing: combination methods, structure, and prospects. Biosensors (Basel). https://doi.org/10.3390/bios13030405
Demeke Teklemariam A, Samaddar M, Alharbi MG, Al-Hindi RR, Bhunia AK (2020) Biosensor and molecular-based methods for the detection of human coronaviruses: a review. Mol Cell Probes. https://doi.org/10.1016/j.mcp.2020.101662
Vu CA, Chen WY (2019) Field-effect transistor biosensors for biomedical applications: recent advances and future prospects. Sensors (Switzerland). https://doi.org/10.3390/s19194214
Poghossian A, Jablonski M, Molinnus D, Wege C, Schöning MJ (2020) Field-effect sensors for virus detection: from ebola to SARS-CoV-2 and plant viral enhancers. Front Plant Sci. https://doi.org/10.3389/fpls.2020.598103
Meyyappan M, Lee JS (2013) Nanowire BioFETs: an overview. Nanowire Field Effect Transistors: Principles Appl. https://doi.org/10.1007/978-1-4614-8124-9_9
Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, Lee CS, Jun S, Park D, Kim HG, Kim SJ, Lee JO, Kim BT, Park EC, Kim SIl (2020) Correction: rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor (ACS Nano (2020) 14:4 (5135-5142) DOI: 10.1021/Acsnano.0c02823). ACS Nano. 2020. https://doi.org/10.1021/acsnano.0c06726
Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA (1979) CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360(6387).
Paulose AK, Huang CC, Chen PH, Tripathi A, Chen PH, Huang YS, Wang YL (2022) A rapid detection of COVID-19 viral RNA in human saliva using electrical double layer-gated field-effect transistor-based biosensors. Adv Mater Technol 7(1). https://doi.org/10.1002/admt.202100842
Toral-Lopez A, Kokh DB, Marin EG, Wade RC, Godoy A (2022) Graphene BioFET sensors for SARS-CoV-2 detection: a multiscale simulation approach. Nanoscale Adv. https://doi.org/10.1039/d2na00357k
Zamzami MA, Rabbani G, Ahmad A, Basalah AA, Al-Sabban WH, Nate Ahn S et al (2022) Carbon nanotube field-effect transistor (CNT-FET)-based biosensor for rapid detection of SARS-CoV-2 (COVID-19) surface spike protein S1. Bioelectrochemistry. https://doi.org/10.1016/j.bioelechem.2021.107982
Fang Y, Ramasamy RP (2015) Current and prospective methods for plant disease detection. Biosensors. https://doi.org/10.3390/bios5030537
Pires ACDS, Soares NDFF, Da Silva LHM, Da Silva MDCH, De Almeida MV, Le Hyaric M et al (2011) A colorimetric biosensor for the detection of foodborne bacteria. Sens Actuators B Chem. https://doi.org/10.1016/j.snb.2010.09.069
Song X, Wang H, Xu X (2022) Amikacin- and AuNP-mediated colorimetric biosensor for the rapid and sensitive detection of bacteria. LWT. https://doi.org/10.1016/j.lwt.2022.114189
Choi Y, Hwang JH, Lee SY (2018) Recent trends in nanomaterials-based colorimetric detection of pathogenic bacteria and viruses. Small Methods. 2018. https://doi.org/10.1002/SMTD.201700351
Malekzad H, Sahandi Zangabad P, Mirshekari H, Karimi M, Hamblin MR (2017) Noble metal nanoparticles in biosensors: recent studies and applications. Nanotechnol Rev. https://doi.org/10.1515/ntrev-2016-0014
Yu L, Li N (2019) Noble metal nanoparticles-based colorimetric biosensor for visual quantification: a mini review. Chemosensors. https://doi.org/10.3390/chemosensors7040053
Cordeiro M, Carlos FF, Pedrosa P, Lopez A, Baptista PV (2016) Gold nanoparticles for diagnostics: advances towards points of care. Diagnostics. https://doi.org/10.3390/diagnostics6040043
Liu L, Yang D, Liu G (2019) Signal amplification strategies for paper-based analytical devices. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2019.04.043
Nguyen QH, Kim MIl (2020) Nanomaterial-mediated paper-based biosensors for colorimetric pathogen detection. TrAC - Trends Anal Chem. https://doi.org/10.1016/j.trac.2020.116038
Materón EM, Gómez FR, Almeida MB, Shimizu FM, Wong A, Teodoro KBR, Silva FSR, Lima MJA, Angelim MKSC, Melendez ME, Porras N, Vieira PM, Correa DS, Carrilho E, Oliveira ON, Azevedo RB, Goncalves D (2022) Colorimetric detection of SARS-CoV-2 using plasmonic biosensors and smartphones. ACS Appl Mater Interfaces. https://doi.org/10.1021/acsami.2c15407
Teengam P, Siangproh W, Tuantranont A, Vilaivan T, Chailapakul O, Henry CS (2017) Multiplex paper-based colorimetric DNA sensor using pyrrolidinyl peptide nucleic acid-induced AgNPs aggregation for detecting MERS-CoV, MTB, and HPV oligonucleotides. Anal Chem. https://doi.org/10.1021/acs.analchem.7b00255
Zhang X, Shi Y, Chen G, Wu D, Wu Y, Li G (2022) CRISPR/Cas systems-inspired nano/biosensors for detecting infectious viruses and pathogenic bacteria. Small Methods. https://doi.org/10.1002/smtd.202200794
Gupta R, Kazi TA, Dey D, Ghosh A, Ravichandiran V, Swarnakar S et al (2021) CRISPR detectives against SARS-CoV-2: a major setback against COVID-19 blowout. Appl Microbiol Biotechnol. https://doi.org/10.1007/s00253-021-11583-6
Selvam K, Najib MA, Khalid MF, Mohamad S, Palaz F, Ozsoz M et al (2021) Rt-lamp crispr-cas12/13-based sars-cov-2 detection methods. Diagnostics. https://doi.org/10.3390/DIAGNOSTICS11091646
Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J et al (2020) CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol. https://doi.org/10.1038/s41587-020-0513-4
Ali Z, Aman R, Mahas A, Rao GS, Tehseen M, Marsic T et al (2020) iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res. https://doi.org/10.1016/j.virusres.2020.198129
Aman R, Marsic T, Sivakrishna Rao G, Mahas A, Ali Z, Alsanea M et al (2022) iSCAN-V2: A one-Pot RT-RPA–CRISPR/Cas12b assay for point-of-care SARS-CoV-2 detection. Front Bioeng Biotechnol. https://doi.org/10.3389/fbioe.2021.800104
Ding X, Yin K, Li Z, Lalla RV, Ballesteros E, Sfeir MM et al (2020) Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun. https://doi.org/10.1038/s41467-020-18575-6
Kumar M, Gulati S, Ansari AH, Phutela R, Acharya S, Azhar M et al (2021) FnCas9 based CRISPR diagnostic for rapid and accurate detection of major SARS-CoV2 variants on a paper strip. Elife. https://doi.org/10.7554/eLife.67130
Azhar M, Phutela R, Kumar M, Ansari AH, Rauthan R, Gulati S, Sharma N, Sinha D, Sharma S, Singh S, Acharya S, Sarkar S, Paul D, Kathpalia P, Aich M, Sehgal P, Ranjan G, Bhoyar RC, Singhal K, Lad H, Patra PK, Makharia G, Chandak GR, Pesala B, Chakraborty D, Maiti S (2021) Rapid and accurate nucleobase detection using FnCas9 and its application in COVID-19 diagnosis. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2021.113207
Phutela R, Gulati S, Kumar M, Maiti S, Chakraborty D (2022) FnCas9 editor linked uniform detection assay for COVID-19. Methods Mol Biol. https://doi.org/10.1007/978-1-0716-2395-4_11
Acknowledgements
We thank Dr. Apurva Agrawal for the constructive comments on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tewari, M., Rana, P. & Pande, V. Nanomaterial-Based Biosensors for the Detection of COVID-19. Indian J Microbiol (2024). https://doi.org/10.1007/s12088-024-01336-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12088-024-01336-0