Abstract
The most severe type of male infertility is nonobstructive azoospermia (NOA), where there is no sperm in the ejaculate due to failure of spermatogenesis. The predictable frequency of NOA in the general population is one in 100 men. Genetic studies have recognized dozens of NOA genes. Most NOA aetiologies remain idiopathic. Monogenic mutations can be a reason for a part of idiopathic NOA cases. To address this, we studied the pedigree of a consanguineous family with three NOAs by a family-based exome sequencing. Our goal was to pinpoint the genetic variants responsible for idiopathic NOA to aid future clinical genetic diagnostics and treatment strategies. Bioinformatics analysis followed by Sanger sequencing revealed that NOA patients were homozygous for a rare novel missense variant in PNLDC1 (NM_173516:exon9:c.710G>A;p.Gly237Asp). In silico, single-cell RNA sequencing data analysis and protein modelling demonstrated that PNLDC1, Gly237Asp resided in the conserved region of the CAF1 domain which could lead to local instability in the structure and alteration of protein phosphorylation site. We conclude that the novel missense PNLDC1 variant may affect meiosis and spermatogenesis, leading to NOA and the genetic cause of this idiopathic NOA family. Our result helps genetic counselling for idiopathic NOA cases and provides the occasion for more efficient diagnosis in the clinical setting.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infertility is defined as a failure to achieve pregnancy at least one year after despite regular unprotected sexual intercourse (Pfeifer et al. 2015). One in six couples are infertile, and half of them have male factor infertility. The most common form of male factor infertility is azoospermia, where there is no sperm in the ejaculate. Azoospermia is classified as non-obstructive azoospermia (NOA) or obstructive azoospermia (OA). NOA is due to failure of spermatogenesis, while OA is caused by obstruction of the testicular and genital ductular system. The predictable frequency of NOA in the general population is one in 100 men (Kasak and Laan 2021). Also, the aetiology of azoospermia is categorized by the anatomical position of the cause: pretesticular azoospermia, post-testicular azoospermia, and testicular azoospermia. In contrast to post-testicular defects, NOA results from pretesticular and testicular azoospermia disturbing testis function (Jarvi et al. 2010). It occurs when there is a deficiency in spermatogenesis. Pretesticular azoospermia is characterized by insufficient stimulation of normal testicles and genital tract. Symptomatically, follicle-stimulating hormone (FSH) levels are proportionately (hypogonadotropic) low with insufficient stimulation of the testes to produce sperm. Pretesticular azoospermia is seen in about 2% of men with azoospermia. Testicular azoospermia indicates that the testes are abnormal, atrophic, or absent, leading to severely impaired or absent sperm production. FSH levels tend to be elevated (hypergonadotropic) due to the lack of feedback inhibition on FSH. Testicular failure comprises the absence of production, low production, and maturation arrest during spermatogenesis. Testicular azoospermia is seen in 49–93% of men with azoospermia (Jarvi et al. 2010; Tharakan et al. 2021). Genetic abnormalities related to male infertility affect about 15% of men with infertility. A recent systematic review and clinical validity assessment of male infertility genes revealed that 78 genes were linked to 92 male infertility phenotypes (Oud et al. 2019). Genetic causes of NOA consist of aberrations of sex chromosomes and rare damaging variants in genes considered for spermatogenesis. About 30–50% of male infertility cases are idiopathic, with no visible cause or related to female infertility. The genetic pathways resolve to discover spermatogenic failure's aetiologies and improve new treatments for male infertility (Kasak and Laan 2021). For example, mutations in STAG3, MSH4, Tex11, and CDK5RAP2 have been associated with NOA (Yatsenko et al. 2015; Akbari et al. 2021, 2022; Rahimian et al. 2023). Family-based whole-exome sequencing (WES) are accurate technique in genetic testing and has a qualified identification of novel candidate genes associated with idiopathic male infertility (Nagirnaja et al. 2021). Thus, penetrating a consanguineous family with multiple cases of NOA can be an appreciated effort.
Findings on the role of small RNAs and microRNAs in epigenetic regulations and spermatogenesis have prolonged understanding of these processes. P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs) are small noncoding RNAs that are found almost totally in germ cells and are essential for spermatogenesis (Özata et al. 2020). Pachytene piRNAs make up ~95% of the piRNAs in the adult testis. They regulate anomalous expression of transposable elements, and translation of repressed messenger RNAs (mRNAs) during differentiation of early post-meiotic spermatids and then remove mRNAs in the later spermatid stages (Gou et al. 2014; Dai et al. 2019). The biogenesis of piRNAs is not yet fully understood. A primary processing pathway is the only pathway used to produce pachytene piRNAs which occurs in the intermitochondrial cement and chromatoid bodies (Ding et al. 2018). Pachytene piRNAs result from 100 intergenic loci under the control of transcription factor MYBL1 (Li et al. 2013). A complex involving proteins PIWIL1, TDRKH, and PARN-like ribonuclease domain containing exonuclease 1 (PNLDC1) processes pre-piRNAs (with long lengths of 30 to 40 bases) (Saxe et al. 2013). Ubiquitination-deficient mutations in human PIWIL1 are associated with human male infertility (Wang and Liu 2022a). TDRKH is involved in NOA; gamete generation; and the piRNA metabolic process (Kherraf et al. 2022). Mutations in genes essential for meiosis such as MYBL1 lead to a meiotic arrest phenotype (Castañeda et al. 2018). Also, MOV10L1 is an RNA helicase essential for piRNA 5′ processing. MOV10L1 interacts with piRNA 5′ before PNLDC1. Conditions linked to MOV10L1 include azoospermia (Sarkardeh et al. 2014; Vourekas et al. 2015). The 3′ ends of pre-piRNAs are trimmed by the protein poly(A)-specific (PNLDC1) in an exonucleolytic style, resulting in mature piRNAs (with lengths of 21 to 35 bases) (Anastasakis et al. 2016). PNLDC1 is entirely expressed in spermatocytes. Mice lacking Pnldc1 have reduced testis size and are infertile (Ding et al. 2017; Zhang et al. 2017; Nishimura et al. 2018). Spermatogenesis arrests at meiotic and post-meiotic stages in Pnldc1 mt/mt mice, with reduced expression of PIWIL1, and longer and fewer piRNAs (Nishimura et al. 2018). A cohort study on infertile men by WES recently demonstrated that a single gene mutation in PNLDC1 causes NOA, OAT, oligozoospermia, and spermatogenic failure diseases (Nagirnaja et al. 2021; Wang and Liu 2022b).
Here, we report a novel missense variant in PNLDC1 within the CAF1 family RNase domain (PFAM: PF04857) in exon 9 (NM_173516:exon9:c.710G>A;p.Gly237Asp) in a consanguineous family with multiple cases of NOA. This variant is associated with NOA in the homozygous state.
Materials and methods
Patient selection
An infertile 33-year-old male, III-6, was enrolled at Royan Institute Infertility Clinic in Tehran, Iran. He has been married for 11 years and has suffered from infertility for 8 years. Genetic counselling revealed that he was born of a consanguineous marriage between first cousins with a history of infertility in two brothers, III-4, and III-7. III-4 with NOA had a child through embryo donation and in vitro fertilization (IVF). III-2 and III-3 were embryo donors. III-8 is single and did not participate in the project. Because of the consanguinity of his parents and the frequency of infertility in his brothers, we choose III-4 and III-6 as potential candidates for WES (figure 1). The routine diagnostic procedures failed to determine the cause of infertility, too. Each participant signed an informed written consent form. The Royan Institute Ethics Committee approved the study (IR.ACECR.ROYAN.REC.1400.019).
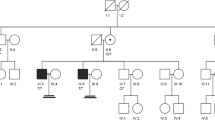
Pedigree of a consanguineous family with three NOA patients. A consanguineous marriage between first cousins has produced three infertile offspring. The NOA patients are represented by orange boxes. III-4 and III-6 were exome sequenced. All NOA patients were homozygous for a novel missense variant in PNLDC1 (NM_173516:exon9:c.G710A:p.G237D). ‘+’ denotes wild-type allele. The proband is marked with an arrow; III-4 has children by IVF; III-2 and III-3 were embryo donors; III-1 and III-8 did not participate in the project; III-8 is single; III-9 is fertile.
Clinical investigations
We analysed the semen of the infertile males (III-4, III-6, and III-7) by the guidelines of the World Health Organization (WHO) 2010 manual and recorded their reproductive history. The man with no clinical evidence of obstruction is the NOA patient. All infertile brothers did complete andrological examinations, investigating their medical histories, physical assessments, and gonadal ultrasonography. Serum concentrations of FSH, luteinizing hormone (LH), and testosterone were analysed by electrochemiluminescence immunoassay (ECLIA) using the COBAS E-601 and E-411 devices (Roche, Germany). For the standard genetic testing, we performed a karyotype of lymphocytes collected from the patient's peripheral blood samples and AZF microdeletions analysis tests by multiplex PCR assay (Totonchi et al. 2012).
DNA isolation and exome sequencing (WES)
We extracted genomic DNA from peripheral blood samples of the available family members (II-2, III-4, III-5, III-6, III-7, and III-9), by using the salting-out procedure (figure 1). High-quality DNA samples from individuals III-4 and III-6 were selected for WES. DNA integrity, purity, and quality were verified through gel electrophoresis and A260/280 and A260/230 ratios by NanoDrop (Thermo Fisher Scientific, USA). For whole-exome analysis, we used SureSelect Human All Exon V5 kit for library construction and capturing target regions (Agilent, USA), followed by paired-end 150-bp sequencing on the Hiseq2500 sequencing system (Illumina, USA). The exome sequence data were 150-base paired-end reads generated from the Illumina HiSeq 2500 platform. DNA fragments were barcoded and sequenced on lanes of a fl2500 p. Reads were mapped to the UCSC reference human genome assembly GRCh37 (hg19) using the Burrows–Wheeler Aligner (BWA) package v. 0.7.17-r1188 (Li and Durbin 2009). Mapped files in SAM format were converted to the BAM format and sorted by SAM tools v. 0.1.18. Marking PCR duplicates was performed by Picard tools (https://broadinstitute.github.io/picard/) v. 1.107. GATK 4.1.1.1 was used for the processing steps, including base quality score recalibration (McKenna et al. 2010). These steps built appropriate BAM files that underwent variant calling by HaplotypeCaller (DePristo et al. 2011). VCF files were annotated and filtered by ANNOVAR, the efficient software tool to utilize update-to-date information to functionally annotate genetic variants detected from diverse genomes (Wang et al. 2010). Shortlisted candidates were prepared according to the American College of Medical Genetics and Genomics (ACMG) guidelines (Richards et al. 2015). As a result, uncommon or new variations, except synonymous variations, were selected for analysis. In silico assessment of missense variants was conducted with BayesDel_addAF, BayesDel_noAF, MutPred, MetaRNN, FATHMM-MKL, M-CAP, MutationAssessor, MutationTaster, SIFT, SIFT4G, PolyPhen-2, LRT, PROVEAN (Chun and Fay 2009; Sundaram et al. 2018).
Segregation analysis (Sanger sequencing)
The gold standard for genotyping is Sanger sequencing. Oligonucleotide primers flanking the candidate variant were designed by PerlPrimer-1.1.21 and used to amplify the region of interest in the pedigree members by polymerase chain reaction (PCR): 5′-TCAGTGGCTCCATTGTTTCTC-3′ (forward primer) and 5′-CTAAAGACTGCCTTTAGGCCA-3′ (reverse primer). The amplicons were sequenced through Sanger Sequencing with Macrogen (Seoul, Korea), and the generated sequence was aligned with the human reference genome (hg19/b37) and analysed by Chromas.
In silico single-cell RNA sequencing data analysis
We investigated the expression level of PNLDC1 in the integrated scRNA-Seq data of spermatogenesis cells from our previous research (Hermann et al. 2018). We generated the data based on the GEO datasets of GSE109037 (Wang et al. 2010) and GSE106487 (Butler et al. 2018). We used the Seurat3.2 R package for data analysis (Berman et al. 2000).
Protein modelling
The 3D structure of PNLDC1 was not presented in the protein data bank (PDB), which led us to predict the 3D structure of this protein (The UniProt 2021). The FASTA sequence of PNLDC1 was retrieved from the UniProtKB database (UniProtKB ID: Q8NA58) (Yang and Zhang 2015). The I-TASSER server was used for protein modelling (Humphrey et al. 1996). The FASTA sequence of this protein was submitted as input to the I-TASSER server, and its models were built from the most significant templates in the threading alignments. Then structural models were clustered based on the pair-wise structural similarity. Finally, the best structural model with the highest TM-score and C-score based on structural similarity and structural quality was selected for further analysis. The G237D mutation was implemented in the best model of PNLDC1 with the Psfgen plugin in VMD1.9.3. Also, the structural model was shown by VMD 1.9. (Ng and Henikoff 2003).
This study was approved by the Royan Institute Ethics Committee (IR.ACECR.ROYAN.REC.1400.019). Each participant signed an informed written agreement form.
Result
Clinical findings
Pedigree analysis revealed that the three brothers were infertile, and genetic counselling demonstrated that they were born of a consanguineous marriage between first cousins (figure 1). Physical examination of the patients showed normal external male genitalia, palpable vas deferens, reduction in testicular volume, and no varicocele. Further clinical investigations revealed no history of infection, chronic disease, vasectomy, or surgical scars. No obstructions were reported by transrectal and scrotal ultrasonography. The pathology report on the obtained diagnostic testicular biopsy mentioned bilateral maturation arrest (MA) up to primary spermatocytes (SPC). Karyotype analysis indicated no clinically detectable abnormalities. Y chromosome microdeletion genetic tests that included azoospermia factors a, b, and c (AZFa, AZFb, and AZFc) were negative (table 1). Semen analysis of all affected cases indicated NOA in cases III-4, III-6, and III-7. The serum levels of FSH, LH, and testosterone were within the normal range in these individuals (table 2). The proband (III-6) and his brother (III-7) attempted testicular sperm extraction (micro-TESE) without success. III-8 is single and did not participate in the project. III-9 is fertile and has a child (table 1).
Novel PNLDC1 variant detection and impact
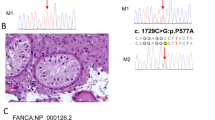
We performed WES on proband (III-6) and his brother (III-4). A minimum of 94% of the total reads passed the initial quality filter (Q ≥30), generating 11.4 Gb of sequence data. The patients were born after two generations of consanguineous marriage between first cousins. The family inheritance pattern was autosomal recessive. We achieved a shortlist after the deletion of intronic, intergenic, and synonymous single-nucleotide variants (SNVs). Also, we prioritized homozygous deleterious variants in genes involved in NOA, meiosis, and spermatogenesis. These genes exhibited elevated or distinct levels of expression in the testicular tissue and were common among the individuals affected, warranting further examination. Then, WES analysis showed a rare homozygous nonsynonymous SNV in the PNLDC1 gene located on chromosome 6 (NM_173516:exon9:c.710G>A;p.Gly237Asp) in the proband (III-6) and his brother (III-4) with NOA. This variant in PNLDC1 is classified as ‘likely pathogenic’ based on the PP2, PM2, and PP3 criteria of the ACMG/AMP guidelines (Richards et al. 2015). The candidate mutation was termed deleterious by pathogenic computational verdict based on 13 pathogenic predictions from BayesDel_addAF, BayesDel_noAF, MutPred, MetaRNN, FATHMM-MKL, M-CAP, MutationAssessor, MutationTaster, SIFT, SIFT4G, PolyPhen-2, LRT, PROVEAN (Chun and Fay 2009; Sundaram et al. 2018) (table 3). For conservation assessment, GERP++ was used which ranked this variant as highly conserved with a high score of 5.56 out of a maximum score of 6.17 (Dong et al. 2015). PhyloP100way was 5.91, which is less than 6.8. According to gnomAD, the candidate mutation had never been reported in homozygous and heterozygous states. Also, the variant is absent in the Iranome database consisting of 800 healthy Iranian individuals. The identified mutation was located in exon 9 of the PNLDC1 gene, which participates in encoding the CAF1 family RNase domain (PFAM: PF04857) of the PNLDC1 protein. This mutation causes the substitution of a glycine residue with aspartate near tyrosine 121, one of two phosphorylation sites of the CAF1 domain (figure 2a). Multiple alignments of residues encoded by exon 9 of the human PNLDC1 with several species from different kingdoms demonstrated substantial conservation as the glycine residue (figure 2c). Subsequent Sanger sequencing confirmed the homozygous status (−/−) of the c.G710A mutation in the azoospermic proband (III-6) and his brother (III-4). The PNLDC1 mutation co-segregated within this family such that the other affected brother with the NOA phenotype (III-7) was homozygous (−/−) for the c.G710A. In contrast, the unaffected brother (III-9), and one of the sisters (III-5) were homozygous (+/+) wild-type. The proband’s mother (II-2) was heterozygous (+/−) for the c.G710A (figure 2d). These findings were consistent with the autosomal recessive pattern of inheritance. Genotyping results of the full pedigree are available in figure 1.
Identification of novel PNLDC1 variant in the homozygous state leading to NOA. (a) PNLDC1 is composed of 19 exons encoding a protein with 520 amino acid residues. (b) Here is a novel missense variant in PNLDC1 within the CAF1 family RNase domain in exon 9 (NM_ 173516:exon9:c.G710A:p.G237D). (c) Orthologous alignment showing that the amino acid residue is highly conserved across mammals, https://www.uniprot.org. (d) Sanger sequencing validated WES results. Hetero, heterozygous; Homo, homozygous for the variant; WT, homozygous for the wild-type allele.
Population study
PNLDC1 mutation (c.G710A) was studied in 30 Iranian men with idiopathic azoospermia who had normal physical examinations, normal karyotype, and no Y chromosome microdeletions to evaluate whether this mutation can explain the failure in sperm counts. No homozygous patients with PNLDC1 (c.G710A) were found.
In silico single-cell RNA sequencing data analysis
The expression levels of PNLDC1 in spermatogenesis cells were assessed based on our previous research on generating an integrated dataset of spermatogenesis cells. The clustering and marker gene expression showed different cell types in the spermatogenesis dataset (Hermann et al. 2018). The PNLDC1 analysis depicted this gene's high expression and presentence in leptotene spermatocytes, early-round spermatid cells, and round spermatid cells 1 (figure 3a). Also, some expressions of this gene were detected in the undifferentiated spermatogonial cells and differentiating spermatogonial cells.
In silico single-cell RNA sequencing data analysis and protein modelling. (a) The expression pattern of PNLDC1 in spermatogenesis scRNAseq data. (b) The 3D structural model of PNLDC1. The piRNA binding site, phosphorylation sites, reported mutagenesis sites and G237D mutation are highlighted. The adjacency of G237 and G237D with T121 is shown in inset figures.
Protein modelling
The 3D structural model of PNLDC1 in the wild-type form was designed, which showed the closest structural similarity (RMSD: 0.51) with the structure of mouse PARN (PDB ID: 3D45:A) and a 27.2% and 70.6%, identity and coverage, respectively. The piRNA binding with PNLDC1 is essential for trimming and transposon silencing during spermatogenesis in mice (Wu et al. 2005). The K326 in human PARN showed a crucial role in binding to the poly(A) substrate, equivalent to K294 in PNLDC1 (Yang and Zhang 2015). So, the piRNA binding site of PNLDC1 (LYS294) was assessed in the 3D structure far from the G237D variation point (figure 3b). The structural survey of recently reported variants in PNLDC1 R47G, P84S, P95S, M259T, M270T, E381K, P452T, R463T, and R476W depicted P84S in a 7.9 Å of G237D variation point (figure 3b) (Nagirnaja et al. 2021; Sha et al. 2022; Wang and Liu 2022b). On the other hand, the phosphorylation sites of PNLDC1, S209, and T121 were extracted from the PhosphoSitePlus web server (https://www.phosphosite.org/homeAction), that are detected with Phospho-Tyrosine Mouse mAb (P-Tyr-100) antibodies. Our structural analysis showed that phosphorylation sites of PNLDC1, and T121 are at a 3.2 Å and 2.3 Å distance from the G237D and P84S variation points, respectively (figure 3b).
Discussion
One of the major trials for genetic research on male infertility is to understand and demonstrate a fundamental role for rare missense variants found in patients with unexplained male infertility. We studied a special pedigree of a consanguineous family with three idiopathic NOA by family-based exome sequencing.
The inability to suppress retrotransposons in the germline is usually related to infertility in most species and LINE1 is the most copious retrotransposon type in the mammalian genome. LINE1 motivation in spermatocytes causes massive DNA damage, which affects the inactivation of the meiotic checkpoint and the following apoptosis of spermatocytes (Yang and Wang 2016). Mammalian germ cells employ multiple mechanisms to silence retrotransposon activity, including small noncoding piRNAs (Ding et al. 2017).
The rare novel nonsynonymous SNV discovered here for NOA is described as PNLDC1: (NM_173516:exon9:c.710G>A;p.Gly237Asp). We hypothesize that this novel variation severely disrupts the post-transcriptional modification of PNLDC1 and affects piRNA maturation.
Structurally, PNLDC1 gene contains 19 exons (figure 2a). PNLDC1 had two isoforms that differ in their N-terminal ends because of alternative splicing of humans: isoform-1, with 520 amino acids, 61.3 kD, and isoform-2, 60.1 kD. Both human isoforms have an N-terminal CAF1 nuclease domain with the DEDD motif for the active site and a C-terminal transmembrane domain (figure 2b) (Anastasakis et al. 2016). PNLDC1 is conserved in mice and humans (figure 2c) (Ding et al. 2017). In previous studies, human single-cell RNA sequencing data showed that PNLDC1 is absent in somatic testicular cells. However, in spermatogonia stem cells, the leptotene and zygotene stages in spermatocytes, PNLDC1 was in the highest quantity (Nagirnaja et al. 2021). Given that the testicular histologic findings in our patient with NOA consistently revealed error-prone meiosis and spermatogenic arrest, characterized by round spermatids and the most advanced population of germ cells (table. 1). Also, our PNLDC1 single-cell RNA sequencing data revealed high expression of PNLDC1 in leptotene spermatocytes, early-round spermatid cells, and round spermatid cells. Additionally, some expressions of this gene were detected in the undifferentiated spermatogonial cells and differentiating spermatogonial cells (figure 3a). According to the PNLDC1 single-cell RNA sequencing data and the testicular histologic findings in our patient, the lack of PNLDC1 expression could lead to spermatogenic arrest, especially in round spermatid cells.
Functionally, PNLDC1 is exonucleolytic cleavage reaction of poly(A) to 5′-AMP. It means a 3-prime-to-5-prime exonuclease that functions as a trimming enzyme for pre-piRNAs to generate mature piRNAs (Anastasakis et al. 2016; Ding et al. 2017, 2018). Pnldc1 is a pre-piRNA 3-prime trimming enzyme restricted in the mitochondrial segment and interacts with the Tudor protein Papi (TDRKH) on the mitochondrial surface. Papi bound Piwi proteins to call up Pnldc1 for trimming. Knockdown of Papi and Pnldc1 created a 3-prime addition and reduced piRNAs in cells. 3-prime trimming of pre-piRNAs steady with 2-prime-O-methylation and mature piRNAs targets LINE1 elements (Izumi et al. 2016). Also, mouse knockdown Pnldc1 embryonic stem cells show Pnldc1 pathways in the cell cycle, reprogramming, and translational regulation (Anastasakis et al. 2016). Male Pnldc1−/− mice had normal growth, smaller testis than wild-type, and normal germ cells without normal spermatozoa in seminiferous tubules (Ding et al. 2017). Another model of the male Pnldc1−/− mice had oligoasthenoteratozoospermia. In azoospermia patients, expression of PNLDC1 cooperative proteins like PIWIL1, PIWIL4, and MYBL in pachytene spermatocytes and spermatogonia diminished and TDRKH exhibited reduced expression levels from the spermatogonia stage to the late-stage pachytene spermatocytes. Transcription levels confirmed these changes too (Nagirnaja et al. 2021; Wang and Liu 2022a).
As the offspring were the products of a consanguineous marriage, an autosomal recessive inheritance pattern was expected to prioritize homozygous or compound heterozygous variants shared among the affected subjects for further investigation. In our study, WES analysis in a consanguineous family showed a rare homozygous nonsynonymous SNV in the PNLDC1 gene in the proband (III-6) with a homozygous state in NOA. Subsequent Sanger sequencing confirmed the homozygous status (−/−) of the c.G710A mutation in the azoospermic proband (III-6) and his brother (III-4). The PNLDC1 mutation co-segregated within this family such that the other affected brother with the NOA phenotype (III-7) was homozygous (−/−) for the c.G710A. In contrast, the unaffected brother (III-9) was homozygous wild-type (+/+). Their mother had a heterozygous PNLDC1 variant (+/−), so these findings were consistent with the autosomal recessive pattern of inheritance (figures 1 and 2d).
The 3D structural protein model of PNLDC1 in the wild-type form was designed, which showed the closest structural similarity (RMSD: 0.51) with the structure of mouse PARN (PDB ID: 3D45:A), as well as a 27.2% and 70.6%, identity and coverage, respectively. Our structural analysis showed that the phosphorylation site of T121 is at a 3.2 Å distance from the G237D variation point, and the recently reported pathogenic variant (Nagirnaja et al. 2021), PNLDC1 P84S is at a 7.9 Å of the G237D variation point and 2.3 Å of the phosphorylation site of T121 (figure 3b). Therefore, our novel variation PNLDC1 G237D is close to the reported variant, PNLDC1 P84S and both of them are near the PNLDC1 phosphorylation site of T121. Thus, we could imagine the same pathogenic pathway for both PNLDC1 P84S and PNLDC1 G237D. About the PNLDC1 P84S (SCV001760919), this variant maps to a CAF1 domain common among mRNA deadenylases and disrupts a site that is conserved in mammals. Testicular histologic findings in a sample from NOA patient carrying PNLDC1 P84S mutation consistently showed error-prone meiosis and spermatogenic arrest with round spermatids and the most advanced population of germ cells, where they were as same as our histopathology for patients (III-4 and III-6) (table 1). Furthermore, gene and protein expression of PNLDC1 P84S, the piRNA-processing proteins PIWIL1, PIWIL4, MYBL1, and TDRKH, were lost or significantly reduced in cells of testes. Additionally, the length distribution of piRNAs and the number of pachytene piRNAs were significantly altered in patients carrying PNLDC1 P84S mutation (Nagirnaja et al. 2021). In the p.G237D variation point, the glycine as the minor amino acid changed to the aspartic acid as a negatively charged amino acid. The amino acid type and size change can lead to local instability in the protein structure. Posttranslational modification is a general regulatory mechanism of protein functions. So, this instability near the phosphorylation site of T121 can cause interference in the PNLDC1 phosphorylation process in posttranslation modification and lead to protein inactivation. As for deadenylases, only two types of modifications have been identified by studies: proteolysis and phosphorylation. For example, changes in the phosphorylation status of PARN have been observed in several stress disease-related conditions, such as acute leukaemia (Maragozidis et al. 2012).
For this reason, PNLDC1 could not interact with the piRNA-processing proteins and thus did not leave the piRNA poly (A) tail to produce mature piRNA. The enzyme known as HEN methyltransferase 1 (HENMT1) was found to be incapable of attaching a 2′-O-methyl group to the 3’-end of processed piRNA molecules. Therefore, immature piRNA did not methylate and could not pass into the cell nucleus. Finally, expression of LINE1 elements was not suppressed and led to genome instability in germ cells and spermatogenesis arresting in the pachytene stage.
Conclusion
One of the primary challenges in genetic research on male infertility is elucidating the significance of rare missense variants in individuals with unexplained male infertility. In this study, we investigated a specific consanguineous family with three brothers affected by idiopathic NOA through family-based exome sequencing. Our results reveal a novel mutation in the PNLDC1 gene that may lead to spermatogenic failure in homozygous NOA individuals by affecting the phosphorylation site of PNLDC1. These findings not only contribute to a deeper comprehension of the genetic basis of NOA but also have implications for enhancing diagnostic and therapeutic strategies in this context.
References
Akbari A., Padidar K., Salehi N., Mashayekhi M., Almadani N., Sadighi Gilani M. A. et al. 2021 Rare missense variant in MSH4 associated with primary gonadal failure in both 46, XX and 46, XY individuals. Hum. Reprod. 36, 1134–1145
Akbari A., Zoha Tabatabaei S., Salehi N., Padidar K., Almadani N., Ali Sadighi Gilani M. et al. 2022 Novel STAG3 variant associated with primary ovarian insufficiency and non-obstructive azoospermia in an Iranian consanguineous family. Gene 821, 146281
Anastasakis D., Skeparnias I., Shaukat A.-N., Grafanaki K., Kanellou A., Taraviras S. et al. 2016 Mammalian PNLDC1 is a novel poly (A) specific exonuclease with discrete expression during early development. Nucleic Acids Res. 44, 8908–8920
Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H. et al. 2000 The Protein Data Bank. Nucleic Acids Res. 28, 235–242
Butler A., Hoffman P., Smibert P., Papalexi E. and Satija R. 2018 Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420
Castañeda J. M., Miyata H., Ikawa M. and Matzuk M. M. 2018 Sperm defects. In Encyclopedia of reproduction (ed. M. K. Skinner), 2nd edition. pp. 276–281, Academic Press, Oxford.
Chun S. and Fay J. C. 2009 Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561
Dai P., Wang X., Gou L. T., Li Z. T., Wen Z., Chen Z. G. et al. 2019 A Translation-activating function of MIWI/piRNA during mouse spermiogenesis. Cell 179, 1566–1581
DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C. et al. 2011 A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498
Ding D., Liu J., Dong K., Midic U., Hess R. A., Xie H. et al. 2017 PNLDC1 is essential for piRNA 3′ end trimming and transposon silencing during spermatogenesis in mice. Nat. Commun. 8, 1–10
Ding D., Liu J., Dong K., Melnick A. F., Latham K. E. and Chen C. 2018 Mitochondrial membrane-based initial separation of MIWI and MILI functions during pachytene piRNA biogenesis. Nucleic Acids Res. 47, 2594–2608
Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K. and Liu X. 2015 Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 24, 2125–2137
Gou L. T., Dai P., Yang J. H., Xue Y., Hu Y. P., Zhou Y., Kang J. Y. et al. 2014 Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 24, 680–700
Hermann B. P., Cheng K., Singh A., Roa-De La Cruz L., Mutoji K. N., Chen I. C. et al. 2018 The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 25, 1650–1667
Humphrey W., Dalke A. and Schulten K. 1996 VMD: visual molecular dynamics. J. Mol. Graph. 14(33–38), 27–38
Izumi N., Shoji K., Sakaguchi Y., Honda S., Kirino Y., Suzuki T. et al. 2016 Identification and Functional Analysis of the Pre-piRNA 3’ Trimmer in Silkworms. Cell 164, 962–973
Jarvi K., Lo K., Fischer A., Grantmyre J., Zini A., Chow V. et al. 2010 CUA Guideline: The workup of azoospermic males. Can. Urol. Assoc J. 4, 163–167
Kasak L. and Laan M. 2021 Monogenic causes of non-obstructive azoospermia: challenges, established knowledge, limitations and perspectives. Hum. Genet. 140, 135–154
Kherraf Z.-E., Cazin C., Bouker A., Fourati Ben Mustapha S., Hennebicq S., Septier A. et al. 2022 Whole-exome sequencing improves the diagnosis and care of men with non-obstructive azoospermia. Am. J. Hum. Genet. 109, 508–517
Li H. and Durbin R. 2009 Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760
Li X. Z., Roy C. K., Dong X., Bolcun-Filas E., Wang J., Han B. W. et al. 2013 An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell. 50, 67–81
Maragozidis P., Karangeli M., Labrou M., Dimoulou G., Papaspyrou K., Salataj E. et al. 2012 Alterations of deadenylase expression in acute leukemias: evidence for poly(a)-specific ribonuclease as a potential biomarker. Acta Haematol. 128, 39–46
McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. et al. 2010 The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303
Nagirnaja L., Mørup N., Nielsen J. E., Stakaitis R., Golubickaite I., Oud M. S. et al. 2021 Variant PNLDC1, defective piRNA processing, and azoospermia. N. Engl. J. Med. 385, 707–719
Ng P. C. and Henikoff S. 2003 SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814
Nishimura T., Nagamori I., Nakatani T., Izumi N., Tomari Y., Kuramochi-Miyagawa S. and Nakano T. 2018 PNLDC 1, mouse pre-pi RNA Trimmer, is required for meiotic and post-meiotic male germ cell development. EMBO Rep. 19, e44957
Oud M. S., Volozonoka L., Smits R. M., Vissers L., Ramos L. and A. Veltman J. 2019 A systematic review and standardized clinical validity assessment of male infertility genes. Hum. Rep. 34, 932–941
Özata D. M., Yu T., Mou H., Gainetdinov I., Colpan C., Cecchini K. et al. 2020 Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat. Ecol. Evol. 4, 156–168
Pfeifer S., Butts S., Dumesic D., Fossum G., Gracia C., La Barbera A. et al. 2015 Diagnostic evaluation of the infertile female: a committee opinion. Fertil. Steril. 103, e44–e50
Rahimian M., Askari M., Salehi N., Riccio A., Jaafarinia M., Almadani N. et al. 2023 A novel missense variant in CDK5RAP2 associated with non-obstructive azoospermia. Taiwan. J. Obstet. Gynecol. 62, 830–837
Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J. et al. 2015 Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–423
Sarkardeh H., Totonchi M., Asadpour O., Sadighi Gilani M. A., Zamani Esteki M., Almadani N. et al. 2014 Association of MOV10L1 gene polymorphisms and male infertility in azoospermic men with complete maturation arrest. J. Assist. Reprod. Genet. 31, 865–871
Saxe J. P., Chen M., Zhao H. and Lin H. 2013 Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. EMBO J. 32, 1869–1885
Sha Y., Li L. and Yin C. 2022 Defective piRNA processing and azoospermia. N. Engl. J. Med. 386, 1675
Sundaram L., Gao H., Padigepati S. R., McRae J. F., Li Y., Kosmicki J. et al. 2018 Predicting the clinical impact of human mutation with deep neural networks. Nat. Genet. 50, 1161–1170
Tharakan T., Luo R., Jayasena C. N. and Minhas S. 2021 Non-obstructive azoospermia: current and future perspectives. Fac. Rev. 10, 7
The UniProt C. 2021 UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489
Totonchi M., Mohseni Meybodi A., Borjian Boroujeni P., Sedighi Gilani M., Almadani N. and Gourabi H. 2012 Clinical data for 185 infertile Iranian men with Y-chromosome microdeletion. Assist. Reprod. Genet. 29, 847–853
Vourekas A., Zheng K., Fu Q., Maragkakis M., Alexiou P., Ma J. et al. 2015 The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 29, 617–629
Wang X., Gou L. T. and Liu M. F. 2022a Noncanonical functions of PIWIL1/piRNAs in animal male germ cells and human diseases. Biol. Reprod. 107, 101–108
Wang X., Tan Y. Q. and Liu M. F. 2022b Defective piRNA processing and azoospermia. N. Engl. J. Med. 386, 1674–1675
Wang Y., Stojiljković N. and Jehle J. 2010 Cloning of complete genomes of large dsDNA viruses by in vitro transposition of an F factor containing transposon. J. Virol. Methods 165, 95–99
Wu M., Reuter M., Lilie H., Liu Y., Wahle E. and Song H. 2005 Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 24, 4082–4093
Yang F. and Wang P. J. 2016 Multiple LINEs of retrotransposon silencing mechanisms in the mammalian germline. Semin. Dev. Biol. 59, 118–125
Yang J. and Zhang Y. 2015 I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 43, W174–W181
Yatsenko A. N., Georgiadis A. P., Röpke A., Berman A. J., Jaffe T., Olszewska M. et al. 2015 X-Linked TEX11 Mutations, meiotic arrest, and azoospermia in infertile men. N. Engl. J. Med. 372, 2097–2107
Zhang Y., Guo R., Cui Y., Zhu Z., Zhang Y., Wu H. et al. 2017 An essential role for PNLDC1 in piRNA 3’ end trimming and male fertility in mice. Cell Res. 27, 1392–1396
Acknowledgements
We are greatly thankful to the personnel of the Department of Genetics at Royan Institute for Reproductive Biomedicine, the Department of Genetics at Islamic Azad University, Marvdasht branch, the Department of Environmental, Biological and Pharmaceutical Sciences and Technologies (DiSTABiF), Università degli Studi della Campania “Luigi Vanvitelli”, Caserta, Italy. This work was financially supported in part by a grant from Islamic Azad University, Marvdasht branch and a grant from Royan Institute for reproductive biomedicine, Tehran, Iran. We thank the patients and their family members for participating in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Ashwin Dalal
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rahimian, M., Askari, M., Salehi, N. et al. A novel missense variant in PNLDC1 associated with nonobstructive azoospermia. J Genet 103, 28 (2024). https://doi.org/10.1007/s12041-024-01478-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12041-024-01478-6