Abstract
Purpose
The present research was undertaken to study probable genetic variations of MOV10L1 in 30 infertile men that had complete maturation arrest in their spermatocyte levels and 70 fertile men as the control group.
Methods
We performed polymerase chain reaction single strand conformation polymorphism (PCR-SSCP) on extracted DNAs and sequencing was used to confirm the results. Identified polymorphisms in the MOV10L1 were further subjected to a haplotype analysis.
Results
We identified eight single nucleotide polymorphisms (SNPs): one missense (rs2272837) and four nonsense polymorphisms (rs2272836, rs11704548, rs2272838, rs138271) in the exonic sequences and three polymorphisms (rs12170772, rs2272840, rs17248147) in the intronic regions. With the exception of rs2272838, there was a statistically significant association in all polymorphisms between study population (P < 0.05). The result of haplotyping analysis showed ten possible haplotypes, from which five were significantly increased in infertile patients compared with the control group.
Conclusions
Our results suggest that MOV10L1 gene polymorphisms in the studied infertile males with complete maturation arrest are linked to infertility.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Worldwide, approximately 10–15 % of reproductive age couples are affected by infertility, from which nearly 40–50 % of these cases are related to male infertility [1, 2]. Although there are many factors that contribute to male infertility, in 25 % of infertile men the cause is unknown. Genetic factors such as single-gene mutations are discussed in male infertility. Hundreds of genes involved in spermatogenesis have recently been identified [3, 4]. The Moloney leukemia virus 10-like 1 (MOV10L1) gene, as identified by Wang and colleagues in the mouse spermatogonia cells, is a germ cell-specific autosomal gene [5]. This gene has 27 exons and is located on chromosome 22 in human (http://www.ncbi.nlm.nih.gov/gene/54456). Based on mouse studies, four different transcripts have been determined: cardiac helicase activated by MEF2 protein (Champ), cardiac-specific isoform (Csm), Mov10l1, and E20-SV [6]. The Csm transcript starts from exon 16 of the Mov10l1 gene. This transcript is regulated by NKX2.5 in the embryonic heart [7]. Champ transcript starts from exon 14 of the Mov10l1 gene. It is suggested that CHAMP acts as a RNA helicase for RNA processing and/or transcriptional control [8, 9]. The E20-SV transcript is an additional transcript that starts at exon 20 as suggested by the EST database [6]. However, full expression of 27 exons in male germ cells specifically creates the MOV10L1 protein. This protein is a RNA helicase with specific expression in testicular germ cells that has been observed in males [5, 6].
Two research groups have produced mice that lack a functional Mov10l1 gene with the intent to assess the role of Mov10l1 in spermatogenesis. In one group, exon 20 of the Mov10l1 gene was removed from both male and female mice. This exon encodes a part of the helicase domain in the MOV10L1 protein. Although this genetic deletion caused infertility in male mice, female mice remained fertile [6]. The second group disrupted the putative RNA helicase domain of MOV10L1 when they removed a larger genetic region (deletion of exons 18 to 21) [10]. The results of these two studies were similar and showed that MOV10L1 protein is essential for reproduction of male mice. Spermatogenesis stops in the early stages of meiosis in Mov10l1-/- male mice. Defects in chromosomal synapse formation have also been observed in Mov10l1-/- mouse spermatocytes. The absence of a piRNA pathway in Mov10l1-/- mice testes causes spermatocyte apoptosis and complete maturation arrest in the pachytene stage [6]. It has been shown that the absence of a piRNA pathway in Mov10l1-/- mice reduces DNA methylation and a lack of inhibition genome transposon elements as a result of increasing transposons activity, spermatocytes experience apoptosis [10, 11].
Nearly 45 % of the human genome contains transposon elements [12]. Piwi-interacting RNAs (piRNAs) are noncoding RNAs that inhibit transposon elements in germ cells as an endogenous defense system [13–17]. The mammalian somatic cells lack piRNA pathway elements, this mechanism exists only in germ cells [6]. A few studies have researched piRNA biosynthesis [15, 17]. However recently the role of the MOV10L1 protein as a germ cell-specific RNA helicase has been proposed in piRNA biosynthesis and/or loading to Piwi proteins [6, 10]. MIWI, MIWI2 and MILI are members of the argonaute proteins and Piwi subfamily in mice. The Piwi proteins are present in the germline and play an essential role in piRNA production [18, 19]. According to research, the lack of MIWI, MIWI2 or MILI proteins in knockout mice causes meiotic arrest in spermatogenesis [18, 20]. Zheng et al. have determined that MOV10L1 interacts with MIWI, MILI and TDRD proteins. Interestingly, genetic disorders in the mouse MOV10L1 helicase domain lead to the elimination of MIWI2 and MILI in male germ cells [10].
The MOV10L1 gene is conserved in a few vertebrates such as H. sapiens, M. musculus, P. troglodytes, M. mulatta, C. lupus, and B. taurus. An 87 % homology exists between human and mice gene sequences. Removal of exons that encode functional domains of the MOV10L1 protein in male mice causes complete maturation arrest at the primary spermatocyte stage [10]. Because these exons encode functional domains of the MOV10L1 protein in humans, it clearly suggests that mutations and/or polymorphisms in the human MOV10L1 gene can be a reason for male infertility. This study assesses genetic changes in these exons with the intent to evaluate the association between MOV10L1 genetic changes and male infertility.
Materials and methods
Patients and control DNA samples
A total of 30 cases of non-obstructive azoospermia in Iranians with no consanguinity whose mean age was 34.46 years old, with complete maturation arrest in their spermatogenesis were chosen as the experimental group. The control group comprised 70 fertile males (mean age: 36.14 years) who had at least one child. For patient selection, we investigated the general clinical data of 800 azoospermia patients who referred to Royan Infertility Clinic, Tehran, Iran from 2005 to 2011. Of these, we identified 103 non-obstructive azoospermia patients with normal karyotype and normal AZF region. Testicular biopsies were performed to recover suitable spermatozoa for ICSI followed by histological examination of patients’ testicular tissues. Based upon pathology reports, we chose 30 cases that had complete maturation arrest. Pathology reports of the testis biopsies in these patients revealed that the testicular tissue was composed of different size seminiferous tubules with thick basement membrane that contained germ cells with maturation arrest at the spermatocyte level. We evaluated hormone profile in all selected patients, however, we didn’t find any significant changes in our study. In this study, we used only blood samples of study subjects and all donors signed an informed consent for participation in this research. This study was approved by the Ethics Committee of the Institute.
Polymerase chain reaction
Genomic DNA was extracted from peripheral blood lymphocytes using the PAXgene Blood DNA kit (Qiagene, Germany). Next, we measured the concentration of isolated DNA with a Nanodrop Spectrophotometer 2000. DNA was analyzed by electrophoresis on 2 % agarose gel for quality control. Primers were designed to amplify exons 17, 18, 19 and 21 of theM OV10L1 gene with Perlprimer v 1.1.14 software and a manually designed exon 20 (Table 1). Because removal of exons 18 to 21 of the Mov10l1 gene in the male mice causes complete maturation arrest at the primary spermatocyte stage [10], we have studied genetic changes in exons 18–21 from 30 infertile men with complete maturation arrest. In addition to computing the position of functional domains, it was determined that the p-loop NTPase (cl09099) domain of the MOV10L1 protein, which has an ATPase role, in the human genome starts at exon 17. Thus, genetic changes in this exon were also examined. Polymerase chain reaction (PCR) were performed as follows: initial denaturation for 5 min at 94 °C, 30 cycles for 45 s at 94 °C, 45 s at annealing temperature, 45 s at 72 °C, followed by a final extension for 5 min at 72 °C. PCR products were analyzed on 2 % agarose gel.
Single-strand conformation polymorphism
Single-strand conformation polymorphism (SSCP) was performed on the PCR products to detect probable mutations and/or polymorphisms. A total of 3 μl of PCR products were mixed with 8 μl of formamide deionized loading buffer, denatured at 95 °C and immediately cooled on ice for 10 min. Then, 6 μl of the samples were loaded on a 6 % polyacrylamide gel; electrophoresis was performed at 120 V for 16–18 h in 1xTBE electrophoresis buffer, after which the polyacrylamide gel was stained by silver nitrates. Normal sequences showed a specific conformational pattern, while altered sequences exhibited different patterns of electrophoretic mobility.
Sequencing analyses
All mobility shift bands in the SSCP results were classified and confirmed by sequencing. sequencing was performed using an ABI 3730 XL DNA Analyzer (Sequetech, Mountain view, CA, USA) with the Sanger sequencing method for 16.4 % of the samples as follows: exon 17 (n = 10), exon 18 (n = 18), exon 19 (n = 22), exon 20 (n = 30) and exon 21 (n = 2). The profile of SSCP and sequencing techniques performed for five selective exons in patients are shown in Table 2. FinchTV v.1.4.0 software and BLASTN at the NCBI Blast server were used to analyze and identify probable variations in comparison with the human MOV10L1 gene sequence at the NCBI-Gene database (NC_000022.10 was used as the reference sequence).
Statistical analysis
Data were processed and analyzed using SPSS software (version 15.0; SPSS, Chicago, IL). Qualitative data were expressed as frequency and percentage. We found either the χ2-test or Fisher’s exact test appropriate and applied either of them to evaluate each allele and genotype of the MOV10L1 polymorphisms between cases and controls. For all analyses, P < 0.05 was considered significant. Hardy-Weinberg equilibrium was assessed using the χ2-test for the observed genotype frequencies among cases and controls, separately. PLINK v1.07 software (http://pngu.mgh.harvard.edu/purcell/plink/) was used to reconstruct the haplotypes for each subject on the basis of the known genotypes.
Homologous sequences alignment
In this study we used Multiple Sequence Alignment as generated by MUSCLE version 3.6 (http://www.ncbi.nlm.nih.gov/pubmed/15034147) and compared human MOV10L1 amino acids in the location of the identified polymorphisms to homologous proteins in M. musculus, P. troglodytes, M. mulatta, C. lupus, and B. taurus.
Results
In order to determine the association between MOV10L1 and human male infertility we examined genetic changes in exons 17 to 21 and intronic regions that surrounded the exons in 30 non-obstructive azoospermia men with complete maturation arrest and 70 fertile men by PCR-SSCP and sequencing. PCR-SSCP results for exons 17 and 21 of the MOV10L1 gene showed similar bands in the experimental and control groups. There was no mutant band. To confirm the SSCP results, we randomly choose 12 samples for sequencing. No nucleotide changes, including mutations or polymorphisms in these exons and the surrounding intronic regions were observed. SSCP results showed the mutant bands in different groups for PCR products of exons 18, 19 and 20. A total of eight variations were found in screening these exons and the surrounding intronic regions (Table 3) which included a missense (50582626A>G) and four nonsense polymorphisms (50582550C>A, 50582630G>A, 50584201A>C, 50588131C>T) in the exonic sequences, and three polymorphisms (50584261C>A, 50588153G>A, 50588313G>A) in the intronic regions. These variations have all been previously reported as single nucleotide polymorphisms (SNPs) in the NCBI-SNP database. With the exception of 50584201A>C, we observed a statistically significant difference in all polymorphisms between both groups (P < 0.05).
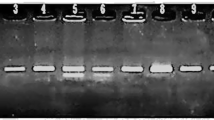
PCR-SSCP for exon 18 and the surrounding intronic regions showed two types of mutant bands in 6 out of the 30 patients compared to the 70 studied controls (Fig. 1). To confirm the results and determine the kinds of variations we sequenced 18 PCR products, 6 that included mutant bands and 12 with wild-type bands. Sequencing confirmed the results of SSCP. We observed two single nucleotide varations, 50582550C>A and 50582626A>G (variants 1 and 2), in the six patients. Moreover change 50582630G>A (variant 3) was detected in four out of these six patients. We detected no changes in the sequence of exon 18 in the control group.
A view of the wild-type and mutant bands that appeared on polyacrylamide gel in the PCR-SSCP technique for exon 18 of the MOV10L1 gene in non-obstructive azoospermia men with complete maturation arrest. Arrows indicate the positions of the mutant bands. Lane 10 is observed in two patients that the sequencing results showed three polymorphisms: 50582550C>A, 50582626A>G and 50582630G>A (variants 1, 2 and 3) in the homozygous forms, while the four patients, showed mutant band of lane 6. Sequencing results in two patients identified three polymorphisms 1, 2 and 3 in the different heterozygous forms and other two patients showed two polymorphisms 1 and 2 in the heterozygous forms. Sequences of lanes 1–5, 7–9 and 11 were normal
Table 3 shows the profile of the genetic alterations in exon 18. Variant 2 is a missense polymorphism that causes the glutamine amino acid at position 820 to convert to arginine at its protein level. Two other genetic changes, variants 1 and 3, are nonsense polymorphisms (arginine at position 795 and proline at position 821, respectfully). There were statistically significant differences between the frequency of these three variants in the patient and control groups (P < 0.05). Assuming MOV10L1 plays an essential role in spermatogenesis, variant 2 may be a candidate for involvement in male infertility.
SSCP for 30 patients in exon 19 showed four types of different bands, of which three types were observed in the control group. These results were confirmed by sequencing. The results showed a nonsense SNP 50584201A>C in exon 19 (threonine at position 863) and an intronic SNP 50584261C>A in intron 19 (variants 4 and 5, respectively). In three patients, variant 4 was in the homozygous form; the heterozygous form was observed in the other six patients. Additionally, this variant was found to be heterozygous in 21 individuals from the control group whereas only one was homozygous. There was no significant difference between patient and control groups in this polymorphism. The observed genotype frequencies agreed with the Hardy-Weinberg equilibrium for variant 4 (χ2 test; case group: P = 0.121, control group: P = 0.738, P > 0.05). Variant 5 was observed to be heterozygous in four patients, however this variant was not observed in the control group. Variant 5 was significantly different between both groups (P < 0.05).
SSCP results for exon 20 in 30 patients revealed six types of different bands; there were four types of different bands visualized on the gel for the 70 controls. Three SNPs were identified using sequencing in this genomic region: 50588131C>T (variant 6) in exon 20, 50588153G>A and 50588313G>A (variants 7 and 8) in the sequence of intron 20. Variant 6 was noted in three patients to be homozygous, whereas it was not detected in the controls. This nucleotide change is a nonsense polymorphism (aspartic acid in position 905) that does not impact protein structure. Variant 7 was observed in 17 patients, from which there were 9 homozygous and 8 heterozygous patients. Of controls, this variation was present in homozygous form in 15 males and the heterozygous form in 6 males. Variant 8 was observed in ten patients heterozygously and in one patient it was homozygous. In the control group, two males had the homozygous form. The observed genotype frequencies among the case subjects agreed with the Hardy-Weinberg equilibrium (χ2 test; P = 0.974). The statistical data showed a statistically significant difference between the two groups in terms of these three variations (variant 6: P = 0.025, variant 7: P = 0.01, variant 8: P = 0.000, P < 0.05).
Haplotype analysis
In this study, we analyzed haplotyping by PLINK v1.07 software. When we combined the eight loci together and performed the haplotype inference, there were ten possible haplotypes derived from the observed genotypes. The frequency of each haplotype in the patient and control groups was determined separately. Haplotype distribution between cases and controls are shown in Table 4. Haplotype 1 was the most common haplotype in the affected and control populations that accounted for more than 50 % of alleles (38.6 % in patients and 64.3 % in controls). We identified seven haplotypes (1, 2, 3, 6, 7, 9 and 10) which were statistically significant between cases and controls. Our results showed that the frequencies of two haplotypes, 1 and 10, significantly decreased (χ2 test; haplotype 1: P = 0.001; haplotype 10: P = 0.023), whereas five haplotypes (2, 3, 6, 7 and 9) had a significant increase in infertile patients compared with controls according to the χ2 test (haplotype 2: P = 0.001, haplotype 3: P = 0.001, haplotype 6: P = 0.003, haplotype 7: P = 0.024 and haplotype 9: P = 0.001, P < 0.05).
Discussion
We studied genetic changes of the MOV10L1 gene in a population of infertile Iranian males with complete maturation arrest. Screening of genomic regions of exons 17 and 21 revealed that these regions have a high degree of stability. Assessment of exons 18, 19, 20 and adjacent intronic regions identified eight polymorphisms—a missense and four nonsense polymorphisms in the exonic sequences and three polymorphisms in the intronic regions (Table 3). There were six patients who had variant 2 in exon 18. This nucleotide change causes the polar uncharged glutamine residue to convert to a positively charged arginine residue at the protein level (Glu820Arg). This variation in the position of the helicase domain of the MOV10L1 protein could potentially alter the structure and function of the MOV10L1 enzyme. It is not known if this amino acid change could the protein’s structure and ultimately its function. Additional studies should be conducted to determine how this missense polymorphism affects MOV10L1 protein and spermatogenesis. Given that the change was not observed in the control group and the significant relationship between the two patient and control groups (P = 0.000), this was presumed to be strongly related to infertility in the studied patient population. Additional studies with larger population would be needed to confirm the effect of Gln820Arg in azoospermia. Results of a comparison of the human MOV10L1 sequence to homologous proteins in M. musculus, P. troglodytes, M. mulatta, C. lupus, and B. taurus showed that the glutamine residue in this position was not conserved; instead, in the mouse lysine and in other mammals, arginine, while the two amino acids in positions 795 and 821 were conserved in all aforementioned organisms (Fig. 2).
We identified three polymorphisms in intronic regions 19 and 20 that showed statistically significant relationships between the two groups. Several studies have proven the role of intronic polymorphisms in mRNA splicing. By checking the positions of the reported intronic polymorphisms, we noted that none of these polymorphisms were located in splice site areas. Therefore we rejected the hypothesis that these polymorphisms were implicated in mRNA splicing of MOV10L1.
In this study, three nonsense polymorphisms and three intronic polymorphisms associated with infertility were identified. Obviously, they could not directly affect the structure and function of the protein. SNPs do not commonly function individually, rather, the combination of SNP markers work together to cause a disease condition [21]. In order to determine the role of these polymorphisms we performed haplotyping analysis for eight locis. This analysis identified ten possible haplotypes, of which two (1 and 10) were associated with reduced risk azoospermia and five (2, 3, 6, 7 and 9) accounted for the increased risk of non-obstructive azoospermia with complete maturation arrest (Table 4). Haplotype 2 showed the highest frequency of variations. This haplotype (AGACCCAG) included five variants: variants 1, 2 (the missense SNP), 3, 4, and 7. This haplotype was observed in 7.1 % of patients’ alleles, but it was not observed in the control alleles. Therefore, this haplotype could have a strong association with increased risk of infertility.
Taken together, several variations such a missense SNP (Glu820Arg) in coding region of MOV10L1 gene showed statistically significant differences between the two groups. Furthermore, association of five studied haplotypes with complete maturation arrest offers that MOV10L1 gene polymorphisms in the infertile males with complete maturation arrest are linked to infertility.
We have shown that MOV10L1 SNPs may predispose men to spermatogenesis defects, although the mechanism of involvement of the SNPs in azoospermia remains unclear. Our results may be useful for a better understanding of meiotic arrest as a cause for azoospermia. Studies on larger populations and in other ethnic groups can confirm the results as a reason for male infertility.
References
de Kretser DM. Male infertility. Lancet. 1997;349:787–90.
Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4(Suppl):s41–9.
Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–213.
Poongothai J, Gopenath TS, Manonayaki S. Genetics of human male infertility. Singap Med J. 2009;50:336–47.
Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–6.
Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby, et al. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci U S A. 2010;107:11847–52.
Ueyama T, Kasahara H, Ishiwata T, Yamasaki N, Izumo S. Csm, a cardiac-specific isoform of the RNA helicase Mov10l1, is regulated by Nkx2.5 in embryonic heart. J Biol Chem. 2003;278:28750–7.
Liu ZP, Nakagawa O, Nakagawa M, Yanagisawa H, Passier R, et al. CHAMP, a novel cardiac-specific helicase regulated by MEF2C. Dev Biol. 2001;234:497–509.
Liu ZP, Olson EN. Suppression of proliferation and cardiomyocyte hypertrophy by CHAMP, a cardiac-specific RNA helicase. Proc Natl Acad Sci U S A. 2002;99:2043–8.
Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci U S A. 2010;107:11841–6.
Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–17.
Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703.
Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:283–90.
Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108.
Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–39.
Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–68.
Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–76.
Kuramochi-Miyagawa S, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–49.
Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–7.
Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, et al. Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108:121–33.
Singh M, Singh P, Juneja PK, Singh S, Kaur T. SNP-SNP interactions within APOE gene influence plasma lipids in postmenopausal osteoporosis. Rheumatol Int. 2011;31:421–3.
Acknowledgments
We thank the study participants for their participation in this study. We are grateful to Dr. M. Akhond and Dr. M. shamsipour for statistical analyses. This thesis was supported by a grant provided by Royan Institute.
Author information
Authors and Affiliations
Corresponding author
Additional information
Capsule Genetic disruption of MOV10L1 gene in a mouse model halts spermatogenesis during meiosis I causing azoospermia due to complete meiotic arrest. Therefore, it is expected that this gene may also play a role in human male infertility. We identified eight SNPs in the infertile men that had complete maturation arrest in their spermatocytes. The result of haplotyping analysis showed ten possible haplotypes, from which five were significantly increased in infertile patients compared with the control population.
Rights and permissions
About this article
Cite this article
Sarkardeh, H., Totonchi, M., Asadpour, O. et al. Association of MOV10L1 gene polymorphisms and male infertility in azoospermic men with complete maturation arrest. J Assist Reprod Genet 31, 865–871 (2014). https://doi.org/10.1007/s10815-014-0240-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-014-0240-1