Abstract
Osterix (or Sp7) is an important transcription factor that promotes osteoblast differentiation by modulating the expression of a range of target genes. Although many studies have focused on Osterix/Sp7 regulatory mechanisms, the detailed functions have not been fully elucidated. Toward this end, in this study, we used CRISPR/Cas9 technology to knock out the zebrafish sp7 gene, and then analyzed its phenotype and biological function. Two knockout sp7 mutant lines were successfully obtained. The bone mineralization level was significantly reduced in the zebrafish sp7−/− homozygote, resulting in abnormal tooth development in the larvae. Quantitative real-time polymerase chain reaction showed that loss of sp7 led to down-regulated expression of the dlx2b and bglap genes related to tooth development and bone mineralization, respectively. Moreover, cell transfection experiments demonstrated that Sp7 directly regulates the expression of dlx2b and bglap through Sp7-binding sites on the promoter regions of these two genes. Overall, this study provides new insight into the role of Sp7 in bone mineralization and tooth development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Osterix (or Sp7) is an important zinc finger transcription factor that activates the transcription of downstream target genes through binding DNA, and is thus designated Sp7 protein binding sites by zinc finger domain (Koga et al. 2005; Zhang et al. 2008). To date, many genes have been identified as downstream targets of Sp7, including Col1a1(Ortuno et al. 2010), Col5a3 (Yun-Feng et al. 2010), Satb2 (Tang et al. 2011), Mmp13 (Nishimura et al. 2012) and sclerostin (Perez-Campo et al. 2016). Sp7 is mainly expressed in osteoblasts and plays an important role in the maturation of both osteoblasts and osteocytes (Nakashima et al. 2002). Recent studies have also pointed to a role of Sp7 in bone regeneration (Hosoya et al. 2013).
The majority of studies focused on the function of Sp7 have used different model organisms to knock out or knock-down the gene with various technologies. Knockout of Sp7 in mice resulted in severe skeletal loss, and the osteoblasts could not mature (Nakashima et al. 2002). Moreover, Sp7 mutant mice showed abnormal osteoblast formation and development, which was ultimately lethal at the prenatal stage (Nakashima et al. 2002). This study further supported an important role of Sp7 in determining the fate of skeletal development. In addition, the sp7 gene of Japanese medaka (Oryzias latipes) was knocked down via injection with antisense morpholino oligonucleotides (MO) to cause splicing and block translation, resulting in severe bone development abnormalities and a high larval mortality rate (Renn and Winkler 2014). In addition, ossification of more mature bones such as the operculum and cleithrum was delayed but recovered during further development. In the axial skeleton, formation of the neural arches and centra was strongly delayed (Yu et al. 2017). Similarly, knockout of sp7 in zebrafish caused reduced mineralization of the cranial bone and disturbance of the axial bone (Yu et al. 2017; Kawai et al. 2017). Recent studies also showed that the loss of sp7 in zebrafish can affect the formation and differentiation of teeth (Kague et al. 2018), suggesting a role of Sp7 in regulating tooth development; however, the precise role and mechanism remain unclear.
Thus, to further explore the role of the zebrafish sp7 gene in bone mineralization and tooth development, in the present study, we used CRISPR/Cas9 technology to knock out the zebrafish sp7 gene, which is recent genome editing technology that provides a faster and more convenient method for gene manipulation than conventional mutation generation (Ran et al. 2013). The confirmed mutants were analyzed with regard to the effect of sp7 loss on bone mineralization and tooth development during development. We further conducted cells transfection and rescue experiments to identify the underlying mechanism and identify target genes of Sp7. Together, these results should provide new insights into the role of sp7 in bone mineralization and tooth development.
2 Materials and methods
2.1 Fish maintenance
Adult zebrafish of the AB strain were raised in a recirculating water system under a 14-h/10-h light/dark (L/D) cycle at 28°C and fed three times per day. To produce embryos, male and female zebrafish were paired in the evening, and spawning occurred the next day within 1 h after the lights were turned on. The embryos were placed in 10-cm Petri dishes with egg water containing methylene blue (0.3 ppm) and raised in a light-controlled (14-h/10-h L/D) incubator at 28°C.
2.2 Design of sp7 guide RNAs, and generation and genotyping of mutants
sp7 gRNAs were designed according to the methods described by Hsu et al. (2013). The purified gRNA (100 ng/μl each) was co-injected with Cas9 mRNA (300 ng/μl) into zebrafish embryos at the one-cell stage. Adult potential founders were genotyped by fin clipping and then crossed to obtain germ line mosaic F1 embryos. Adult genotyped F1 fish were then outcrossed with wild-type fish to obtain heterozygous F2 offspring. The phenotype of homozygous F3 carriers was then analyzed. For genotyping, larvae or adult fish were anaesthetized with 0.01% or 0.005% ethyl 3-aminobenzoate methanesulfonate (Tricaine; Sigma), respectively. Larvae or clipped caudal fin fragments were lysed individually in 50 μl of 50 mM NaOH and incubated at 95°C for 15 min. The samples were neutralized with 5 μl of 1 M Tris-HCl (pH 8.0). Stained embryos were fixed and washed with 1× phosphate buffered saline with 0.1% (v/v) Tween 20 (PBST) five times before genomic DNA isolation. Individual larvae were lysed with DNA lysis buffer (10 mM Tri-HCl pH 8.2, 50 mM KCl, 0.3% NP 40, 0.3% Tween) and Proteinase K (20 μg/ml; Sigma) at a 49:1 ratio and incubated at 55°C for 60 min and then at 90°C for 10 min. The supernatant contained genomic DNA for analysis. A 147-bp fragment was amplified (using primers GAGGAAACACGTTATGGATC and GTTCAGAAGTCATGCTGTAG) and digested with MspI (New England Biolabs). Mutants with undigested fragments were sequenced for confirmation.
2.3 Whole-mount skeletal staining
Alizarin red staining was performed on the larvae fish as previously described (Walker and Kimmel 2007). In brief, the zebrafish larvae were collected at 6, 10, and 12 days post-fertilization (dpf) and then fixed in 2% paraformaldehyde overnight. After washing with 10% glycerol/0.5% KOH, the larvae were stained overnight with 0.02% alizarin red stain/10% glycerol/0.5% KOH and then washed overnight twice with 50% glycerol/0.1% KOH. Images were acquired using a stereomicroscope (Leica, M165FC, Germany). Digital images were analyzed to quantify and compare the stained area and staining density. Three replicates were performed with 10 fish per replicate.
2.4 RNA isolation, cDNA synthesis, and reverse transcription-quantitative polymerase chain reaction (qRT-PCR)
Total RNA was extracted from more than 25 larvae by TRIzol (Invitrogen) reagent and reverse-transcribed into cDNAs using Superscript III Reverse Transcriptase (Invitrogen). qRT-PCR was then performed on an ABI Step-One Plus instrument using the SYBR (TaKaRa) system with a thermal profile of 40 cycles of 95°C for 10 s and 58°C for 30 s. Each qRT-PCR analysis was performed in triplicate on three independent biological samples, and β-actin served as the reference gene for relative quantification of expression levels, calculated using the 2−△△Ct method in Microsoft Office Excel. The qRT-PCR primers were designed based on the exon boundaries using Primer Express 3.0 software, and were checked for self-annealing, hetero-dimers, and hairpin structures with Oligo Analyzer 3.1. The primer sequences are listed in table 1.
2.5 Luciferase activity assay
A segment of the wild type (WT) about 3kb dlx2b and bglap promoter cloned into the pcDNA4.17 vector (Promega), and a mutant dlx2b and bglap-luc was generated by altering the predicted Sp7 binding site via a two-step PCR approach. HEK 293T cells were co-transfected with either a WT or mutant dlx2b and bglap-luc vector, and added Sp7 expression vector or not, cultured for 24 h, and then assessed to ascertain their exhibited luciferase activity using a dual-luciferase reporter assay system (Promega).
2.6 Statistical analysis
Three biological replicates and three technical replicates were performed for each experiment, and all experimental data are presented as mean ± standard deviation values. One-way analysis of variance was performed to determine the statistical significance, followed by post-hoc Dunnett’s or Turkey’s tests to independently compare the effects of sp7 knockout to wild-type fish. All statistical analyses were conducted using SPSS 16.0 software and values of p<0.05 were considered to be statistically significant.
3 Results
3.1 Generation of sp7 zebrafish mutants using the CRISPR/CAS9 system
To obtain sp7 mutants, CRISPR/CAS9 technology was used to knock-out the sp7 gene in zebrafish. The process of mutation generation and selection is outlined in figure 1. The gRNA target site was exon 3, and the restriction enzyme MspI was chosen to identify the mutation close to the PAM sites. To confirm the activity of the candidate gRNAs generated, different concentrations of gRNA were mixed Cas9, and capped mRNA was injected single-fertilized eggs. The efficiency of the gRNA was 62%, 50%, 48%, 45%, and 40% when mixed at concentrations of 100, 50, 25, 15, and 10 ng, respectively. Uncleaved bands were extracted from the gel and cloned into the pMD-19T vector. The sequencing results demonstrated a difference of mutant type generated by non-homologous recombination repair. Ultimately, two homologous sp7 mutants were identified in the F2 generation. Bioinformatics analysis further revealed that these two mutants encode 124- and 45-amino acid truncated proteins, which both result in nonsense mutations. Most of the subsequent experiments were performed using the 11-bp-deletion mutant.
Generation of the sp7 mutant line using CRISPR/Cas9. (A) The target site of gRNA selection in sp7 exons 3; the MspI restriction enzyme cutting site was located in the PAM site to facilitate selection of mutant zebrafish. (B) Different concentrations of gRNA were mixed with 300 μg/L Cas9-capped mRNA, and then injected into single-cell zebrafish fertilized eggs. At 24 h post-fertilization, genomic DNA was extracted to check the gRNA efficiency. (C) Sequencing results showing different mutants generated by non-homologous recombination repair. (D) Two mutant types were ultimately chosen in the F1 generation of zebrafish. Neither mutant encodes Sp7 proteins. The mutant with an 11-bp deletion was chosen for subsequent experiments.
The homozygous sp7 mutants developed into adults fish, but showed an abnormal body morphology such as a defected gill and crooked spine. Compared with wild-type (WT) zebrafish, the homozygous sp7 mutant also had a lower body length and body weight (data no shown). Importantly, the homozygous sp7 mutant was not able to spawn. However, when eggs out from the sp7 mutant were removed and artificially fertilized with WT sperm, the eggs developed normally. These results suggested that sp7 likely also plays a role in fertilization.
3.2 Bone mineralization and tooth development were defective in the homozygous sp7 mutant
Alizarin Red staining performed at 6, 10, and 12 dpf showed that the area of stained bone tissue significantly decreased in the sp7 mutant compared with WT zebrafish (figure 2). In particular, there was barely any mineralization detected in the craniofacial bones in the mutant, with only a 20% stained area compared with that of WT fish.
Whole-mount skeletal staining showing decreased bone mineralization lack of tooth development in the sp7 mutant homozygous zebrafish. (A–C) Alizarin Red staining of WT fish at 6, 10, and 12 dpf. (D–F) Alizarin Red staining in sp7 mutant fish at 6, 10, and 12 dpf. (G–I) Digital image analysis of the stained area and staining density of mineralized tissue via fluorescence intensity. Mean values are plotted (n = 10) and Student’s t-test was performed to determine statistical significance.
3.3 Genes related to tooth development and bone mineralization were down-regulated in the sp7 mutant
As shown in figure 3, compared to the WT fish, the expression levels of the tooth development genes dlx2b, map, pthlha, and smoc2 significantly decreased in the sp7 mutant zebrafish; however, the levels of acvr1l and ssvb2rs1 significantly increased, and there was no difference in the expression level of ercc2. The expression levels of the bone mineralization genes bglap, sparc, spp1, col1a1a, and col10a1a significantly decreased in the sp7 mutant compared with those of WT zebrafish.
Tooth development- and bone mineralization-related genes were down-regulated in the sp7 mutant zebrafish at 6 dpf. (A–G) Tooth development gene expression patterns in the sp7 mutant zebrafish. (H–L) Bone mineralization gene expression patterns in the sp7 mutant zebrafish. Student’s t-test was performed to determine statistical significance.
3.4 Sp7 positively regulates dlx2b and bglap
Bioinformatics promoter analysis identified an Sp7-binding site in the 2-kb promoter region of dlx2b (figure 4A), which is known to play a role in tooth morphogenesis in mice and zebrafish. Moreover, cell transfection experiments showed that Sp7 significantly up-regulated dlx2b expression; however, when the Sp7-binding site was mutated, this up-regulation was no longer observed (figure 4B).
Sp7 positively regulates tooth and bone mineralization-genes (dlx2b and bglap, respectively). (A) Promoter analysis of dlx2b identifying one Sp7-binding site. (B) Cell transfection experiment showing that Sp7 regulates dlx2b expression via direct interaction with the promoter. (C) Promoter analysis of bglap identifying two sp7-binding sites. (D) Cell transfection experiment showing that Sp7 regulates bglap by diret interaction with the promoter. SBS: sp7 binding site. Three independent experiments were performed. One-way ANOVA was performed to calculate statistical significance followed by post-hoc Dunnett or Turkey tests.
Similarly, two Sp7-binding sites was identified were identified in the promoter region of bglap (figure 4C), which encodes osteocalcin secreted by osteoblasts, and is thus implicated in bone mineralization and calcium ion homeostasis. In addition, cell transfection experiments showed that Sp7 up-regulated bglap expression, which was not observed with mutation of the two Sp7-binding sites (figure 4D).
3.5 Whole-body sp7 expression rescued tooth development and bone mineralization in sp7 mutant zebrafish
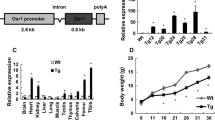
Transgenic zebrafish were generated that express sp7 throughout the entire body. The plasmid was generated via a Tol2 backbone, and crystailin beta promoter-driven EGFP was clonde into this vector as a screening tag. EF1α then triggered sp7 to express Sp7 in the whole body (figure 5B). qRT-PCR experiments analysis showed that whole-body sp7 expression rescued the decrease of dlx2b and bglap expression from the homozygous mutation. Whole-body Alizarin staining at 12 dpf further demonstrated that whole-body sp7 expression rescued both bone mineralization and tooth development in the mutant.
Whole-body expression Sp7 in the sp7 mutant zebrafish rescued tooth development and bone mineralization. (A) Construction of a whole-body expression sp7 plasmid. (B) Whole-body expression of Sp7 in transgenic zebrafish was identified via the naked eye by EGFP fluorescence. (C) Whole-body expression of Sp7 in the sp7 mutant zebrafish rescued dlx2b expression. (D) Whole-body expression of Sp7 in the sp7 mutant zebrafish rescued bglap expression. (E) Whole-body expression of Sp7 in the sp7 mutant zebrafish rescued the tooth development and bone mineralization phenotype. Three independent experiments were performed. One-way ANOVA was performed to calculate statistical significance followed by post hoc Dunnett’s or Turkey’s tests.
4 Discussion
Sp7 is a zinc finger transcription factor that regulates the expression of many genes by binding to corresponding sites on the DNA sequence, and plays an important role in bone development (Koga et al. 2005; Nishimura et al. 2012). In this study, we used CRISPR/Cas9 technology to successfully construct sp7 homozygous zebrafish mutants, and confirmed its role in regulating bone development.
sp7 mutants have been generated in other model organisms. In mice, the loss of sp7 has a lethal effect before birth due to abnormal development of the bone and osteoblasts (Nakashima et al. 2002; Baek et al. 2009). Similarly, less than 1% of medaka sp7 mutants survived beyond 1 month of age, and none survived up to 2 months (Yu et al. 2017). By contrast, in zebrafish, all of the homozygous sp7 mutants could survive to adulthood (Kague et al. 2018), confirming previous studies indicating that the function of zebrafish sp7 slightly differs from that of mammals and medaka. Establishment of an adult sp7 mutant also provides the unique opportunity to study the function of Sp7 in adult fish. However, our homozygous sp7 mutant could not successfully spawn, although in vitro-fertilized eggs developed successfully. Thus, the bone damage caused by the loss of sp7 also appears to affect the ovulation process.
Overall, loss of sp7 resulted in significant damage to bone mineralization and tooth development. The zebrafish dentition is fully established at around 26 days, comprising 11 teeth positioned on every fifth ceratobranchial (Jackman et al. 2004; Wise and Stock 2010; Yuan et al. 2017). In the early stage, zebrafish tooth development is similar to those of mammals, and the process of zebrafish bone formation is very similar to that of humans, with the same osteocyte type, bone structure, and extracellular matrix, making it a suitable animal model for human translation (Arnold et al. 2008; Slavkin et al. 1992). So the zebrafish is a good model to research the tooth pathobiology.
The extremely low degree of bone mineralization in zebrafish sp7 mutants is consistent with previous results in both zebrafish and medaka (Ortuno et al. 2010; Renn and Winkler 2014). Among bone mineralization-related genes, loss of sp7 decreased the expression levels of bglap, sparac, spp1, col1a1a, and col10a1a in our results. The latter two genes were previously identified as direct targets of sp7 (Ortuno et al. 2010). spp1 encodes osteopontin (Reinholt et al. 1990), sparc encodes osteonectin (Termine et al. 1981), and bglap encodes osteocalcin (Weinreb et al. 1990); thus, these three genes play an important regulatory role in bone mineralization, especially bglap. The finding that sp7 can directly regulate bglap expression by binding to the bglap gene promoter further supports that mineralization of the zebrafish bone is achieved through the interaction of Sp7 with bglap (figure 6). However, some of the bones could still be mineralized in the sp7 mutants, suggesting that there may be additional pathways that also play a role in bone mineralization (figure 6).
The Dlx gene family is associated with many developmental processes, such as mandibular and limb development (Verreijdt et al. 2006; Merlo et al. 2000), in which dlx2 is an important transcription factor in tooth mineralization (Thomas et al. 1995). Previous studies have found that retinoic acid signaling pathway regulates the expression of dlx2b in pharyngeal ridge and affects the development of zebrafish teeth (Gibert et al. 2015). And dlx5 is directed regulated by sp7 (Lee et al. 2003). In our experiments, we found that Sp7 can positively regulate the development of zebrafish teeth though regulate the expression of dlx2b, and Sp7 overexpression can also rescue dlx2b expression and tooth loss phenotype, which indicates that Sp7 can regulate tooth development through dlx2b (figure 6).
In conclusion, zebrafish Sp7 can directly regulate the expression of the dlx2b and bglap genes through Sp7-binding sites of the promoter regions. This study thus provides new insights into the role of Sp7 in bone mineralization and tooth development.
References
Arnold WH, Naumova KI, Naumova EA and Gaengler P 2008 Comparative qualitative and quantitative assessment of biomineralization of tooth development in man and zebrafish (Danio rerio). Anat. Rec. 291 571–576
Baek WY, Lee MA, Jung JW, Kim SY, Akiyama H, de Crombrugghe B and Kim JE 2009 Positive regulation of adult bone formation by osteoblast-specific transcription factor osterix. J. Bone Miner. Res. 24 1055–1065
Gibert Y, Samarut E, Pasco-Viel E, Bernard L, Borday-Birraux V, Sadier A, Labbe C, Viriot L and Laudet V 2015 Altered retinoic acid signalling underpins dentition evolution. Proc. Biol. Sci. 282 https://doi.org/10.1098/rspb.2014.2764.
Hosoya A, Yukita A, Ninomiya T, Hiraga T, Yoshiba K, Yoshiba N, Kasahara E and Nakamura H 2013 Localization of SUMOylation factors and Osterix in odontoblast lineage cells during dentin formation and regeneration. Histochem. Cell Biol. 140 201–211
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G and Zhang F 2013 DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31 827–832
Jackman WR, Draper BW and Stock DW 2004 Fgf signaling is required for zebrafish tooth development. Dev. Biol. 274 139–157
Kague E, Witten PE, Soenens M, Campos CL, Lubiana T, Fisher S, Hammond C, Brown KR, Passos-Bueno MR and Huysseune A 2018 Zebrafish sp7 mutants show tooth cycling independent of attachment, eruption and poor differentiation of teeth. Dev. Biol. 435 176–184
Kawai S, Yamauchi M and Amano A 2017 Zinc-finger transcription factor Odd-skipped related 1 regulates cranial bone formation. J. Bone Miner. Metab. 36 640–647
Koga T, Matsui Y, Asagiri M, Kodama T, de Crombrugghe B, Nakashima K and Takayanagi H 2005 NFAT and Osterix cooperatively regulate bone formation. Nat. Med. 11 880–885
Lee MH, Kwon TG, Park HS, Wozney JM and Ryoo HM 2003 BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem. Biophys. Res. Commun. 309 689–694
Merlo GR, Zerega B, Paleari L, Trombino S, Mantero S and Levi G 2000 Multiple functions of Dlx genes. Int. J. Dev. Biol. 44 619–626
Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR and de Crombrugghe B 2002 The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108 17–29
Nishimura R, Wakabayashi M, Hata K, Matsubara T, Honma S, Wakisaka S, Kiyonari H, Shioi G, Yamaguchi A, Tsumaki N, Akiyama H and Yoneda T 2012 Osterix regulates calcification and degradation of chondrogenic matrices through matrix metalloproteinase 13 (MMP13) expression in association with transcription factor Runx2 during endochondral ossification. J. Biol. Chem. 287 33179–33190
Ortuno MJ, Susperregui AR, Artigas N, Rosa JL and Ventura F 2010 Osterix induces Col1a1 gene expression through binding to Sp1 sites in the bone enhancer and proximal promoter regions. Bone 52 2013 548–556
Perez-Campo FM, Santurtun A, Garcia-Ibarbia C, Pascual MA, Valero C, Garces C, Sanudo C, Zarrabeitia MT and Riancho JA 2016 Osterix and RUNX2 are transcriptional regulators of sclerostin in human bone. Calcif .Tissue Int. 99 302–309
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y and Zhang F 2013 Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154 1380–1389
Reinholt FP, Hultenby K, Oldberg A and Heinegard D 1990 Osteopontin–a possible anchor of osteoclasts to bone. Proc. Natl. Acad. Sci. USA 87 4473–4475
Renn J and Winkler C 2014 Osterix/Sp7 regulates biomineralization of otoliths and bone in medaka (Oryzias latipes). Matrix Biol. 34 193–204
Slavkin HC, Hu CC, Sakakura Y, Diekwisch T, Chai Y, Mayo M, Bringas P Jr, Simmer J, Mak G, Sasano Y, et al. 1992 Gene expression, signal transduction and tissue-specific biomineralization during mammalian tooth development. Crit. Rev. Eukaryot. Gene Expr. 2 315–329
Tang W, Li Y, Osimiri L and Zhang C 2011 Osteoblast-specific transcription factor Osterix (Osx) is an upstream regulator of Satb2 during bone formation. J. Biol. Chem. 286 32995–33002
Termine JD, Kleinman HK, Whitson SW, Conn KM, McGarvey ML and Martin GR 1981 Osteonectin, a bone-specific protein linking mineral to collagen. Cell 26 99–105
Thomas BL, Porteus MH, Rubenstein JL and Sharpe PT 1995 The spatial localization of Dlx-2 during tooth development. Connect. Tissue Res. 32 27–34
Verreijdt L, Debiais-Thibaud M, Borday-Birraux V, Van der Heyden C, Sire JY and Huysseune A 2006 Expression of the dlx gene family during formation of the cranial bones in the zebrafish (Danio rerio): differential involvement in the visceral skeleton and braincase. Dev. Dyn. 235 1371–1389
Walker MB and Kimmel CB 2007 A two-color acid-free cartilage and bone stain for zebrafish larvae. Biotech. Histochem. 82 23–28
Weinreb M, Shinar D and Rodan GA 1990 Different pattern of alkaline phosphatase, osteopontin, and osteocalcin expression in developing rat bone visualized by in situ hybridization. J. Bone Miner. Res. 5 831–842
Wise SB and Stock DW 2010 bmp2b and bmp4 are dispensable for zebrafish tooth development. Dev. Dyn. 239 2534–2546
Yu T, Graf M, Renn J, Schartl M, Larionova D, Huysseune A, Witten PE and Winkler C 2017 A vertebrate-specific and essential role for osterix in osteogenesis revealed by gene knockout in the teleost medaka. Development 144 265–271
Yuan Q, Zhao M, Tandon B, Maili L, Liu X, Zhang A, Baugh EH, Tran T, Silva RM, Hecht JT, Swindell EC, Wagner DS and Letra A 2017 Role of WNT10A in failure of tooth development in humans and zebrafish. Mol. Genet. Genomic Med. 5 730–741
Yun-Feng W, Matsuo N, Sumiyoshi H and Yoshioka H 2010 Sp7/Osterix up-regulates the mouse pro-alpha3(V) collagen gene (Col5a3) during the osteoblast differentiation. Biochem. Biophys. Res. Commun. 394 503–508
Zhang C, Cho K, Huang Y, Lyons JP, Zhou X, Sinha K, McCrea PD and de Crombrugghe B 2008 Inhibition of Wnt signaling by the osteoblast-specific transcription factor Osterix. Proc. Natl. Acad. Sci. USA 105 6936–6941
Acknowledgements
This work was supported by Grants from Suzhou people’s Welfare and Science Items (SYS201659).
Author information
Authors and Affiliations
Corresponding author
Additional information
Editorial Responsibiility: BJ Rao.
Corresponding editor: BJ Rao
Rights and permissions
About this article
Cite this article
Chen, Z., Song, Z., Yang, J. et al. Sp7/osterix positively regulates dlx2b and bglap to affect tooth development and bone mineralization in zebrafish larvae. J Biosci 44, 127 (2019). https://doi.org/10.1007/s12038-019-9948-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12038-019-9948-5