Abstract
Myelin, a lipid-enriched multi-layer membrane structure, allows for rapid long-distance saltatory conduction of neuronal impulses. Although glycolipids are the predominant types of lipids in the myelin bilayer, the role of glycolipid transfer protein (GLTP), which selectively mediates the transfer of various glycolipids between phospholipid bilayer, in myelin development and maintenance remains unknown at present. In this study, we identified Gltp as the key lipid metabolism gene in myelin-forming oligodendrocytes (OLs) through integrated omics analysis across independent transcriptomic and single-cell sequencing studies. Gene expression analysis revealed that Gltp is selectively expressed in the differentiated OLs. Functional study demonstrated that its expression is essential for the differentiation of OLs, and promotes the outgrowth of OL membrane. Moreover, we found that the expression of Gltp is regulated by OL-lineage transcriptional factors, such as NKX2.2, OLIG2, SOX10, and MYRF. These findings provide important insights into the unrecognized functions of Gltp in OL differentiation and maturation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
GLTP is a 24-kDa soluble protein which was first discovered in the bovine spleen and later identified in other species ranging from yeast to human [1,2,3,4,5,6]. The GLTP protein is highly conserved during evolution, and the amino acid sequences of porcine and bovine GLTP are 98% identical to that of human [4]. GLTP is localized in the cytoplasm and plasma membrane, but not in any cellular organelles [7]. Previous studies demonstrated that GLTP mainly functions in the transfer of glycolipids with a β-linked, but not α-linked sugar residue to its ceramide or glycerolipid backbone between two lipid membranes [8]. The substrates that can be transferred by GLTP include galactosylceramide (GalCer), lactosylceramide (LacCer), glucosylceramide (GlcCer), sulfatide, monosialoganglioside 1 (GM1), and monosialoganglioside 3 (GM3) [9]. The structure of GLTP is well studied, and four conserved hydrophobic residues in the middle part of GLTP protein are found to play a sensor role in gating function [10]. Alternation in the GLTP expression causes changes in lipid classes, including LacCer, GlcCer, and sphingomyelin, all of which are the components of myelin [7, 11,12,13,14], indicating the important function of GLTP in transferring glycolipids and myelin development.
Myelin structures are elaborated by OLs in the central nervous system (CNS) [15]. During development, OLs are derived from oligodendrocyte precursor cells (OPCs), which are highly proliferative and migratory bipolar cells [16]. Regulated by both extracellular cues and intrinsic molecular signals, OPCs withdraw from cell cycle and differentiate into mature OLs which form compacted myelin sheaths around neuronal axons [17]. Myelin expedites nerve conduction dramatically by reducing the transverse capacitance and increasing the transverse resistance of the axonal plasma membrane [18]. Compared to other plasma membranes, myelin contains higher lipid content and glycolipid including galactolipids, sulfatide, and 3-O-sulfogalactosylceramide [19,20,21]. These results suggest that GLTP may be involved in the transfer of glycolipid from endoplasmic reticulum to myelin sheath in myelinating OLs.
Here in, we identified that the Gltp gene is closely related to OL maturation through integrated multi-omics-based analysis. Our gene expression analyses revealed that Gltp is specifically expressed in differentiated mature OLs. Overexpression of Gltp in primary cultured OPCs accelerated the maturation process, while knocking down of Gltp hindered the maturation of OPCs. In addition, we found that the expression of Gltp is regulated by Nkx2.2 gene, as NKX2.2 protein dramatically increased the transcriptional activity of Gltp promoter.
Materials and Methods
Integrated Multi-Omics Analysis
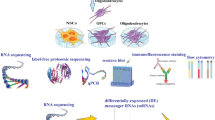
Three transcriptomic studies of cuprizone (CPZ)-induced demyelination were selected based on the following criteria: (1) The cuprizone-induced mouse models were used in the studies; (2) the samples were from primary tissues with normal controls; (3) experiments were run on the similar platform (microarray); (4) the studies were conducted by independent groups (Table S1). This accords with combining fold changes across studies to distinguish a meta-fold change for the meta-analysis, and the meta-p values were calculated by Fisher’s method. Top500 genes were selected when the meta-p value was less than 4.50E-5. Subsequently, membrane proteins specifically expressed by OLs were screened out through the Human Protein Atlas (HPA, https://www.proteinatlas.org/) and the LocDB (https://www.rostlab.org/) database as shown in the sketch of multi-omics analysis workflow (Fig. 1A).
Integrated multi-omics-based discovery and validation of Gltp as OL-specific gene. A Study outline of integrated multi-omics-based analysis. B The cell type-specific expression patterns of myelin markers and the differently expressed Gltp were analyzed based on the single-cell RNA-seq data from HPA. C Schematic diagram of the function of GLTP in the delivery of oligodendroglial glycolipid. GSL, glycosphingolipid, a main component of myelin. PLP, proteolipid protein, an important constitutive protein in myelin
Animals
All animal experiments were approved by the Committee of Laboratory Animals, Hangzhou Normal University, China. The wild-type (WT) mice were obtained from the Laboratory Animal Center of Hangzhou Normal University. Olig1-Cre mouse line was provided by Dr. Chuck Stiles, and the Nkx2.2fx mouse line was provided by Sussel [22].
In Situ RNA Hybridization (ISH)
Mouse spinal cord and brain tissues were isolated and fixed in 4% paraformaldehyde (PFA) overnight at 4 °C in phosphate-buffered saline (PBS). Tissues were then dehydrated by 30% sucrose in PBS overnight at 4 °C, embedded in OCT, and sectioned in a cryostat at 16-μm thickness. In situ hybridization with digoxigenin-labeled riboprobes was performed as described previously [23]. The digoxigenin-labeled RNA probe used in ISH corresponds to the 34–946 nt of mouse Gltp mRNA (NM_019821.2). Images were acquired using the Nikon Eclipse 90i microscope.
Immunofluorescent Staining
Tissue sections were subjected to ISH for Gltp, followed by antigen retrieval in citrate sodium buffer at 90 ℃ for 20 min. Sections were then examined by immunofluorescent staining. Procedures for immunofluorescent staining were described previously [24, 25]. The dilution ratio of primary antibodies is as follows: anti-mouse SOX10 (Oasisbiofarm, OB-PGP001, 1:500), anti-mouse CC1 (Abcam, ab16794, 1:500), anti-mouse GFAP (Millipore, MAB360, 1:2000), anti-mouse IBA1 (Wako, 019–19,741, 1:500), anti-GFP (Oasisbiofarm, OB-PGP003, 1:500), anti-Flag (Sigma, F1804, 1:1000), anti-CNP (Oasisbiofarm, OB-PRT006, 1:500), anti-GLTP (Proteintech, 10,850–1-AP, 1:200), anti-MBP (Abcam, ab7349, 1:500), anti-NeuN (Millipore, MAB377, 1:500), and anti-PLP (Oasisbiofarm, OB-PGP028, 1:200). Images of double immunofluorescent staining in tissues were achieved using the Zeiss LSM 710 confocal microscope, and others were obtained using the Nikon Eclipse 90i microscope.
Western Blotting
The proteins with equal amounts were loaded on 10% Bis–Tris/PAGE gel, then transferred onto PVDF membranes and blocked with 5% (wt/vol) milk dissolved in TBST (TBS, 0.05% Tween-20, pH 7.4) for 1 h at room temperature. The PVDF membranes were incubated with primary antibodies overnight at 4℃ and with secondary antibodies for 1 h at room temperature. Blots were developed using Pierce™ ECL western blotting substrate (Thermo Scientific, TD264271D) and imaged with chemiluminescent imaging system (ProteinSimple, FluorChem E). Primary antibodies are as follows: anti-GLTP (Proteintech, 10,850–1-AP, 1:1000), anti-β-actin (Huabio, EM21002, 1:10,000), anti-MBP (Abcam, ab7349, 1:3000), anti-NKX2.2 (DSHB, 74.5A5, 1:1000), anti-FASN (Proteintech, 66,591–1-lg, 1:5000), and anti-SREBF1 (Proteintech, 66,875–1-lg, 1:2000). Secondary antibodies were from goat anti-rabbit IgG (Invitrogen, 31,460, 1:5000), goat anti-mouse IgG (Invitrogen, 31,430, 1:5000), or goat anti-rat IgG (ABclonal, AS028, 1:5000).
OPC Isolation and Culture
Cerebral cortices from P0 SD rats were dissected out, minced, and digested in 0.25% trypsin (Genom, GNM25300) at 37 ℃ for 20 min. The digestion was terminated with Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Gibco, 21,331,020) containing 10% fetal bovine serum (FBS, Biological Industries, 04–001-1A). After centrifugation at 1400 rpm for 5 min, the digested cells were plated in a 75-cm2 tissue culture flask coated with 100 μg/ml poly-L-lysine (Sigma, P0899). The cell cultures were maintained in DMEM/F12 containing 20% FBS for 10 days, with a medium change every 3 days. The culture flasks were shaken on an orbital shaker model 420, Orbital Size 1.0 (Thermo Fisher, Waltham, MA, USA) for 1 h (200 rpm, 37℃) to remove microglial. The medium from the flasks was discarded and fresh DMEM/F12 with 10% FBS was added. The flasks were then shaken for 15–18 h (250 rpm, 37℃). On the next morning, the medium was collected and plated on tissue culture dishes for 30 min at 37 ℃ to deposit dead cells and microglial. The non-adherent cells (OPCs) were collected and centrifuged at 1400 rpm for 5 min, then replated in DMEM with 10% FBS. After attaching to the surface of culture dish, OPCs were cultured in DMEM/F12 supplemented with N2 (Invitrogen, 17,502,048), B27 (Invitrogen, 17,504,044), and 10 ng/ml of PDGF-AA (Peprotech, 100-13A-10).
Differentiation Assay
The shRNA targeting the sequence of Gltp was designed and synthesized as follows: sense, 5′-GATCCGCCCATCAAGGCAGACATAAGCTTCCTGTCAGACTTATGTCTGCCTTGATGGGCTTTTTG-3′ and antisense, 5′-AATTCAAAAAGCCCATCAAGGCAGACATAAGTCTGACAGGAAGCTTATGTCTGCCTTGATGGGCG-3′; BamHI and EcoRI digestion sites were added into the sense and antisense strands respectively. The annealed double-strand fragments were cloned into the pGreenPuro plasmid (System Biosciences, SI505A-1) to establish the pGreenPuro/Gltp shRNA or pGreenPuro/scramble lentiviral vectors. HEK293T cells for lentiviral production were purchased from ATCC and cultured in DMEM (Gibco, C11995500BT) supplemented with 10% FBS. The pCDH-CMV-MCS-EF1-copGFP (pCDH) empty vector (System Biosciences, CD511B-1) or pGreenPuro lentiviral vectors were co-transfected with pMD2.G (Addgene, 12,260) and psPAX2 (Addgene, 12,259) vectors into HEK293T cells by FuGENE. The culture medium was collected 48 h later, and centrifuged at 12,000 rpm for 1.5 h at 4℃. Then, the lentiviral sediment was resuspended with DMEM/F12 medium supplemented with N2, B27, and PDGF-AA. The titer of pCDH lentivirus was 2.50E + 07 TU/ml. The titer of pGreenpuro lentivirus was 5.50E + 07 TU/ml determined by infectious assay in HEK293T cells. The resuspended lentivirus was transferred to the dish which rat OPCs were cultured in. The protocol for rat OPC acquisition was described above. Six hours later, the OPC cultural medium was replaced by the fresh DMEM/F12 medium supplemented with N2, B27, and 40 ng/ml of T3 (Sigma, T2877) for 2 days. Finally, the differentiated OPCs were determined by immunofluorescent staining with anti-MBP antibody or anti-CNP antibody.
Luciferase Assay
A total of 2.5 kb, 1.5 kb, and 0.45 kb upstream sequences from mouse Gltp gene were cloned into pGL3-basic (pGL3b) vector (Promega). Flag-tagged Olig1, Olig2, Myrf, Sox10, and Nkx2.2 were cloned into pCDH empty vector. Each 2 μg of pGL3-basic mixed with 4 μg of either pCDH, pCDH-Olig1, pCDH-Olig2, pCDH-Myrf, pCDH-Sox10, or pCDH-Nkx2.2 and 1 μg of pRL-TK (Promega) was transfected into HEK293T cells by FuGENE. Luciferase activities were measured by Dual-Luciferase ® Reporter Assay System (Promega, E1910).
Chromatin Immunoprecipitation (ChIP)-PCR Assay
Briefly, CG4 cells were infected by pCDH lentivirus (2.50E + 07 TU/ml) in 3.5-cm dish. One day later, the infected CG4 cells were collected and cultured in a 10-cm dish. After occupied all the space of 10-cm dish, the CG4 cells were treated by 1% formaldehyde in medium for 10 min at room temperature. Two washes with ice-cold PBS containing cocktail (Sangon Biotech, C600387) were performed, and these cells were dissociated by SDS lysis buffer (25 mM pH 7.4 Tris, 150 mM NaCl, 1% NP40, 1 mM EDTA). Then, the lysates were sonicated 15 times (10 s each) on ice. After centrifugation at 12,000 rpm for 20 min, the supernatant was diluted in ChIP dilution buffer (0.01% SDS, 1.1% TritonX-100, 1.2 mM EDTA, 16.7 mM pH8.0 Tris–Cl, 167 mM NaCl) and incubated overnight with anti-FLAG magnetic beads (Sigma, M8823) at 4 ℃. Samples were washed three times in TBS buffer (150 mM NaCl, 25 mM pH7.4 Tris), and the FLAG-tagged proteins were eluted with 120 μl TBS buffer containing 10 μg FLAG peptide (Sangon Biotech, T510060). After treated with magnetic separator, the supernatant was collected and disposed with proteinase K at 45℃ for 45 min. Coprecipitate DNAs were purified using DNA clean-up kit (CWBIO, CW2301). The immunoprecipitated DNA was eluted in 30 μl nuclease-free water and checked by PCR. Primer sequences were listed below: 5′-CCCGCAGACCGGCAGATCGA-3′; 5′-AAGAGGGATCTGGGCCGGTG-3′.
Determination of Cholesterol
For the first day, about 1.0E + 05 rat OPCs were cultured in 10-cm dish. At the second day, OPCs were infected by pCDH lentivirus (2.50E + 07 TU/ml). Six hours later, the OPC cultural medium was replaced by the fresh DMEM/F12 medium supplemented with N2, B27, and 40 ng/ml of T3 for 2 days. Eventually, all cells were digested in 0.05% trypsin, centrifuged at 1500 rpm for 10 min, and resuspended by PBS. The cell numbers were calculated by the automatic cell counter (BodBoge, JSY-FL-049). The content of cholesterol was determined by the total cholesterol assay kit (Nanjing Jiancheng Bioengineering Institute, A111-1–1). The unit for cholesterol in each group was μmol/million cells.
Statistical Analysis
All quantitative data were presented as the mean ± standard deviation (SD) and evaluated by a two-tailed Student’s t-test or two-way ANOVA. For each analysis, results from independent infections, transfections, cultures, or mice were treated as biological replicates (n ≥ 3).
Results
Identification of Gltp as OL-Specific Gene by Multi-Omics Analysis
To identify myelination-associated genes, we combined expression studies of CPZ-induced demyelination from the Gene Expression Omnibus (GEO) repository for a multiplex analysis. For each of the 35,000 transcripts tested, we calculated the meta-fold change by taking a linear combination of effect sizes (fold changes) weighted by the variance within each study, and the meta-p values across all studies by using Fisher’s method. Top500 genes were selected and then filtered through HPA and LocDB (Fig. 1A). We chose Gltp for further validation and characterization, as its expression appears to be OL-specific (Fig. 1B) and it is functionally related to lipid metabolism (Fig. 1C).
Gltp Is Expressed in Postnatal Spinal Cord and Brain Tissues
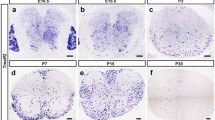
To investigate the possible function of Gltp in myelin development, we first examined the spatiotemporal pattern of Gltp expression in the developing mouse spinal cord and brain tissues. In situ RNA hybridization (ISH) revealed that expression of Gltp was not detected in embryonic mouse spinal tissues (Fig. 2A). Starting at postnatal day 0 (P0), a small number of Gltp-positive cells appeared in the ventral white matter of spinal cord (Fig. 2B). By P4, the expression of Gltp in the ventral and lateral white matter increased dramatically (Fig. 2C) and continued to accumulate at later stages (Fig. 2D-F). In the brain tissue, Gltp signal emerged slightly later as Gltp-positive cells first emerged in the corpus callosum at P4 (Fig. 3A) and gradually increased thereafter (Fig. 3B-D). In addition, Gltp expression was also detected in other brain regions including cortical plate and striatum (Fig. 3E-G).
The expression pattern of Gltp in the developing brain. A-D Transverse sections of the brain from P4 (A), P7 (B), P15 (C), and P30 (D) WT mice are subjected to ISH with Gltp riboprobes. Gltp is expressed in corpus callosum from P4. E–G Expression of Gltp in the right hemisphere of the brain. F and G are the higher magnifications of the cortex and striatum outlined in E. The Gltp-positive cells are represented by arrows
Gltp Is Exclusively Expressed in the Differentiated OLs
Since the white matter comprises OLs, astrocytes, and microglia [26], we next examined which cell type expressed Gltp. Double labeling experiments revealed that Gltp was only co-localized with SOX10 (Fig. 4A-D), a specific marker for OL lineage, but not with astrocyte marker GFAP (Fig. 4I-L) or microglial marker IBA1 (Fig. 4M-P). Further analysis demonstrated that Gltp staining was only detected in CC1 + , MBP + , and PLP + differentiated OLs (Fig. 4E-H, S1I-P), but not in Pdgfra + OPCs (Fig. S1E-H) or NeuN + neurons (Fig. S1A-D).
Gltp is specifically expressed in the differentiated OLs. The transverse sections of spinal cord from P4 mice are first subjected to ISH with Gltp riboprobe (A, E, I, M), followed by immunofluorescent labeling with SOX10 (B), CC1 (F), GFAP (J), or IBA1 (N) antibodies. The ISH signal of Gltp is converted to green by photoshop, and merged with SOX10 (D), CC1 (H), GFAP (L), or IBA1 (P). The cellular nuclei were counter-stained with DAPI (C, G, K, O)
To further confirm that Gltp is selectively upregulated in the differentiated OLs, we carried out western immunoblotting with the spinal cord and cerebral tissues isolated from wild-type mice aged from P0 to P60. In the spinal tissue, the 25 kDa GLTP protein first appeared at P7, increased sharply at P15, and peaked at P30 (Fig. 5A). In the cerebrum, the signal was clearly visible at P15, and also plateaued at P30 (Fig. 5B). In parallel, MBP was used as a marker protein for differentiated OLs. Consistently, we found that the temporal sequence of GLTP expression in the spinal and cerebral tissues was nearly identical to that of MBP (Fig. 5A-D).
Developmental expression profile of Gltp correlates with the maturation of OLs. A-D Western blot analyses comparing the expression profiles of GLTP and MBP in the spinal cord and cerebrum of P0, P4, P7, P15, P30, or P60 WT mouse. Quantitative analyses of expression levels of GLTP and MBP relative to β-actin in the spinal cord and cerebrum are shown on the right (B, D). PD, postnatal day
Gltp Promotes the Differentiation and Maturation of OLs
We next wonder whether Gltp is functionally involved in the regulation of OL differentiation and maturation. To determine the direct effect of Gltp on OL differentiation, we knocked down the expression of Gltp in primary rat OPCs with shRNA, and found the number of MBP + (Fig. 6A-H, S) and CNP + (S2A-H, Q) OLs markedly decreased. On the contrary, when we overexpressed Gltp in the OPCs with lentivirus, it was found that Gltp overexpression significantly increased the number of MBP + (Fig. 6I-P, T) and CNP + (Fig. S2I-P, R) differentiated OLs. Furthermore, the overexpression of Gltp increased the length of OL membrane branch strongly (Fig. S3A, C), and improved the content of cholesterol (a main ingredient of membrane) per million differentiated OLs (Fig. S3D). Although enhancing the transferring of glycolipid to cytoplasmic membrane, the overexpressing Gltp had mild effect on the upstream lipid synthesis. Because the expression of lipid enzyme like FASN slightly increased, the lipid regulatory transcription factor, such as SREBP1, received no remarkable change by the overexpressing Gltp (Fig. S4). These results suggest that Gltp is essential for the differentiation and maturation of OPCs.
Gltp is essential for the differentiation of OPCs in vitro. A-H The shRNA-mediated knockdown of Gltp attenuated the differentiation of primary cultural OPCs significantly. I-P The overexpression of Gltp promoted the differentiation of OPCs in vitro. Q-R Western blot analysis of GLTP protein after the infection of lentivirus in CG4 cells. These cells were treated with T3 for 2 days to induce the expression of Gltp (Q). Short arrow indicated the endogenous GLTP, while the long arrow showed the exogenous 3xFlag-tagged one. S-T The percentage of MBP-positive cells ever infected by lentivirus in subpopulation which also underwent infection. The data of cell counting originate from independent cultures (n ≥ 3). The field under 20 × lens was randomly selected for each culture, and the cells within this field ever infected by lentivirus were counted
NKX2.2 Enhances Gltp Promoter Activity
As Gltp is specifically expressed by the differentiated OLs, we next investigated the molecular mechanism underlying the transcriptional regulation of Gltp expression in these cells. In this study, genomic DNA fragments containing the Gltp transcription start site together with its 2.2 kb, 1.3 kb, or 0.1 kb upstream sequence were subcloned into the luciferase reporter vector (pGL3-basic) (Fig. 7A). The resultant Gltp promoter-luciferase plasmids were co-transfected with the expression vector (pCDH) carrying a specific transcription factor into HEK293T cells. Several candidate OL-specific transcription factors such as OLIG1, OLIG2, SOX10, MYRF, and NKX2.2 were tested in this experiment. Measurements of the luciferase activity revealed that NKX2.2 markedly increased Gltp promoter activity by up to tenfold or 30-fold as compared with the control and other TF-encoding constructs (Fig. 7B-D), indicating that NKX2.2 is specific activator for Gltp transcription. To determine the specific binding of NKX2.2 to the Gltp promoter, we analyzed the sequence of Gltp promoter and found a potential NKX2.2 binding site in both mouse and rat (Fig. 7E). Subsequently, 3xFlag-tagged NKX2.2 was expressed in CG4 (a rat OPC cell line) cells and ChIP-PCR was performed with anti-FLAG beads. The predicted lane appeared specifically in 3xFLAG-tagged NKX2.2 group (Fig. 7F), which confirmed the binding specificity of NKX2.2 to the Gltp promoter.
NKX2.2 enhances Gltp promoter activity. A The upstream regulatory region of Gltp. Genomic DNA fragment of the mouse Gltp transcriptional start site was cloned into a luciferase vector (pGL3-basic). B-D The luciferase reporter assay showed that NKX2.2 protein dramatically increased the promoter activity of Gltp gene compared to other oligodendrocyte lineage transcription factors. Two-way ANOVA was performed to determine the statistical significance among different groups. E The mouse Gltp promoter contained a potential binding site (tagged by the red) of NKX2.2. The homologous comparison of sequence around this binding site was analyzed by the DNAMAN software between mouse and rat. The fully conserved nucleotide was highlighted in black boxes, while the non-conserved one was marked in blue. F The upper figure showed the expression effect of exogenous Nkx2.2 and the specific adhesion ability of anti-FLAG beads (checked by anti-NKX2.2 antibody). The lower one indicated that ChIP-PCR assay confirmed the binding specificity of NKX2.2 to the Gltp promoter in CG4 cells. The bands pointed by long black arrows suggested the rat sequence (from + 101 to + 213 bp)
Disruption of Nkx2.2 Has No Effects on the Expression of Gltp in Adult Mouse
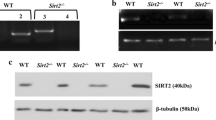
In order to determine whether Nkx2.2 also regulates Gltp expression during in vivo development, we generated the Olig1Cre/+;Nkx2.2fx/fx conditional mutant mice in which Nkx2.2 is specifically deleted in OL-lineage cells. It was found that conditional ablation of Nkx2.2 dramatically reduced the number of Gltp + cells in the white matter of the spinal cord in mice at P7 (Fig. 8A-C). Our previous studies showed that Nkx2.2 controls the timing of OL differentiation, but myelin gene expression returns to a normal level at about P21 in Nkx2.2 cKO mice [27]. Consistently, the number of Gltp + cells showed no difference between the control and Nkx2.2 cKO groups (Fig. 8D-I). Similarly, there is no difference in the level of GLTP protein between these two groups (Fig. S5). These results from conditional mutants are somewhat contradictory to our in vitro observation that NKX2.2 protein promotes reporter gene expression from the Gltp promoter sequence, suggesting that additional regulatory factors, such as MYRF, SOX10, or OLIG2, may also be involved in the regulation of Gltp expression in vivo.
Genetic deletion of Nkx2.2 in vivo does not affect the expression of Gltp eventually. A-I The expression of Gltp in the spinal cord of control (Olig1Cre/+; Nkx2.2fx/+) and Nkx2.2 cKO (Olig1Cre/+; Nkx2.2fx/fx) mice at P7, P35, and P60 as shown by ISH. Statistical analyses of Gltp-positive cells per section in the spinal cord of control and Nkx2.2 cKO at P7, P35, and P60 are shown on C, F, and I
Discussion
Multi-Omics Approach Increases Confidence of Gltp as Myelin-Associated Genes
We have applied a multi-omics approach to developing a workflow from discovery of OL-specific candidates to validation of functional molecules of myelination (Fig. 1A and B). Through this approach of integrating quantitative analysis, we have identified and validated Gltp as a functional molecule involved in OL maturation. GLTPs are ubiquitous proteins characterized by their ability to accelerate the intermembrane transfer of glycolipids (Fig. 1C), implying that GLTP possibly acts as a regulator to assist the production of lipids required for the formation of compact myelin structures in myelinating OLs.
Gltp Is Essential for the Differentiation and Maturation of OLs
In this study, we systematically investigated the developmental expression of Gltp in the developing CNS tissue. For the first time, we demonstrated that Gltp is specifically expressed in CC1 + , MBP + , and PLP + OL population (Fig. 4E-H and S1I-P). The protein level of GLTP co-fluctuated with that of MBP at postnatal stages (Fig. 5A-D) in both spinal cord and cerebrum. It is well documented that CC1, MBP, or PLP label both newly differentiated OLs in early postnatal CNS tissues and differentiated myelinating OLs in adult tissues. The specific expression pattern of Gltp suggests that this gene may play an important role in the OL differentiation, myelin formation, and maintenance, as myelin sheaths contain a fairly large amount of glycolipids. We found that the knocking down of Gltp inhibited the differentiation of primary cultured OPCs into MBP + (Fig. 6A-H, S) and CNP + (Fig. S2A-H, Q) OLs; conversely, the enforced expression of Gltp promoted the OPC differentiation (Fig. 6I-P, T and Fig. S2I-P, R). The more interesting thing is that the overexpression of Gltp significantly enhanced the outgrowth of OL membrane (Fig. S3), indicating that Gltp is very likely to participate in the formation of myelin. Besides the function in transferring of glycolipid, we also determined the effect of Gltp on the synthesis of lipid in vitro. The proteins involved in the synthesis of lipid showed no great change by the overexpression of Gltp (Fig. S4), suggesting that Gltp has mild effect on this process. However, elucidation of its in vivo role in myelin development and maintenance requires the generation of conditional Gltp genetic mutants.
Nkx2.2 Regulates the Expression of Gltp
As Gltp is found to be expressed in differentiated OLs, we searched for OL-lineage transcription factors that are possibly involved in regulating the expression of this gene. In this study, we have screened several candidates including OLIG1, OLIG2, SOX10, MYRF, and NKX2.2 that are highly expressed in differentiating or mature OLs. Among these factors, OLIG2, SOX10, and MYRF slightly enhanced reporter gene expression from the Gltp promoter sequence, while NKX2.2 drastically increased the reporter gene expression from the Gltp promoter sequence (Fig. 7B-D). We further confirmed the specific binding of NKX2.2 to the Gltp promoter by ChIP-PCR assay (Fig. 7F). However, contrary to this result, the expression of Gltp is not dramatically altered in adult Nkx2.2 cKO mice (Fig. 8G-I and S5). These in vivo results suggest that Nkx2.2 is not absolutely necessary for the expression of this gene, inferring that additional transcriptional factors may participate in regulating Gltp expression. A previous study showed that functional SP1 transcription factor can bind to four GC boxes in the Gltp promoter sequence and enhance its promoter activity [28]. Moreover, Sp1 is selectively expressed in mature OLs [29]. Therefore, it is conceivable that NKX2.2, SP1, OLIG2, SOX10, and MYRF may co-operate to regulate the expression of Gltp during OL differentiation and myelin formation.
Data Availability
The datasets supporting the conclusions of this article are included within the article and its supplementary files.
References
Metz RJ, Radin NS (1980) Glucosylceramide uptake protein from spleen cytosol. J Biol Chem 255(10):4463–4467
Abe A, Yamada K, Sasaki T (1982) A protein purified from pig brain accelerates the inter-membranous translocation of mono- and dihexosylceramides, but not the translocation of phospholipids. Biochem Biophys Res Commun 104(4):1386–1393
Yamada K, Sasaki T (1982) Rat liver glycolipid transfer protein. A protein which facilitates the translocation of mono- and dihexosylceramides from donor to acceptor liposomes. J Biochem 92(2):457–64
Li XM et al (2004) Human glycolipid transfer protein: probing conformation using fluorescence spectroscopy. Biochemistry 43(31):10285–10294
West G et al (2008) Identification of a glycosphingolipid transfer protein GLTP1 in Arabidopsis thaliana. FEBS J 275(13):3421–3437
Saupe S et al (1994) Inactivation of the Podospora anserina vegetative incompatibility locus het-c, whose product resembles a glycolipid transfer protein, drastically impairs ascospore production. Proc Natl Acad Sci U S A 91(13):5927–5931
Tuuf J, Mattjus P (2007) Human glycolipid transfer protein–intracellular localization and effects on the sphingolipid synthesis. Biochim Biophys Acta 1771(11):1353–1363
Mattjus P (2009) Glycolipid transfer proteins and membrane interaction. Biochim Biophys Acta 1788(1):267–272
Yamada K, Abe A, Sasaki T (1985) Specificity of the glycolipid transfer protein from pig brain. J Biol Chem 260(8):4615–4621
Ochoa-Lizarralde B et al (2018) Structural analyses of 4-phosphate adaptor protein 2 yield mechanistic insights into sphingolipid recognition by the glycolipid transfer protein family. J Biol Chem 293(43):16709–16723
Yoshihara T et al (2018) Lactosylceramide synthases encoded by B4galt5 and 6 genes are pivotal for neuronal generation and myelin formation in mice. PLoS Genet 14(8):e1007545
Meixner M et al (2011) Myelination in the absence of UDP-galactose:ceramide galactosyl-transferase and fatty acid 2 -hydroxylase. BMC Neurosci 12:22
Kjellberg MA et al (2014) Alternation in the glycolipid transfer protein expression causes changes in the cellular lipidome. PLoS ONE 9(5):e97263
Nakai Y et al (2000) Lysenin-sphingomyelin binding at the surface of oligodendrocyte lineage cells increases during differentiation in vitro. J Neurosci Res 62(4):521–529
Bradl M, Lassmann H (2010) Oligodendrocytes: biology and pathology. Acta Neuropathol 119(1):37–53
Hardy R, Reynolds R (1991) Proliferation and differentiation potential of rat forebrain oligodendroglial progenitors both in vitro and in vivo. Development 111(4):1061–1080
Bergles DE, Richardson WD (2015) Oligodendrocyte development and plasticity. Cold Spring Harb Perspect Biol 8(2):a020453
Nave KA, Werner HB (2014) Myelination of the nervous system: mechanisms and functions. Annu Rev Cell Dev Biol 30:503–533
Norton WT, Poduslo SE (1973) Myelination in rat brain: changes in myelin composition during brain maturation. J Neurochem 21(4):759–773
Coetzee T et al (1996) Myelination in the absence of galactocerebroside and sulfatide: normal structure with abnormal function and regional instability. Cell 86(2):209–219
Saravanan K et al (2004) Specific downregulation and mistargeting of the lipid raft-associated protein MAL in a glycolipid storage disorder. Neurobiol Dis 16(2):396–406
Mastracci TL, Lin CS, Sussel L (2013) Generation of mice encoding a conditional allele of Nkx2.2. Transgenic Res 22(5):965–72
Cai J et al (1999) Expression and regulation of the chicken Nkx-6.2 homeobox gene suggest its possible involvement in the ventral neural patterning and cell fate specification. Dev Dyn 216(4–5):459–68
Zhu Q et al (2011) Dorsally-derived oligodendrocytes in the spinal cord contribute to axonal myelination during development and remyelination following focal demyelination. Glia 59(11):1612–1621
Huang H et al (2018) Interactive repression of MYRF self-cleavage and activity in oligodendrocyte differentiation by TMEM98 protein. J Neurosci 38(46):9829–9839
Sharma R, Sekhon S, Cascella M (2021) White matter lesions. In: StatPearls. StatPearls Publishing, Treasure Island. https://www.ncbi.nlm.nih.gov/books/NBK562167. Accessed 5 Sep 2022
Zhu Q et al (2014) Genetic evidence that Nkx2.2 and Pdgfra are major determinants of the timing of oligodendrocyte differentiation in the developing CNS. Development 141(3):548–55
Zou X et al (2011) Human glycolipid transfer protein gene (GLTP) expression is regulated by Sp1 and Sp3: involvement of the bioactive sphingolipid ceramide. J Biol Chem 286(2):1301–1311
Henson J, Saffer J, Furneaux H (1992) The transcription factor Sp1 binds to the JC virus promoter and is selectively expressed in glial cells in human brain. Ann Neurol 32(1):72–77
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province (grants LQ23C090003) and the National Natural Science Foundation of China (grants 32170969).
Author information
Authors and Affiliations
Contributions
Z.CF, CZ, and TZ performed the experiments; Z. CF and TZ designed the study; Z. CF and TZ wrote the manuscript; Q. MS, W. XP, and ZD revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
All institutional and national guidelines for the care and use of laboratory animals were followed during the experiments. All procedures performed in this study observed the ethical standards of the Second Xiangya Hospital of Central South University and Hangzhou Normal University.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, C., Chen, Z., Zhang, D. et al. Role of Gltp in Maturation of Oligodendrocytes Under the Regulation of Nkx2.2. Mol Neurobiol 60, 4897–4908 (2023). https://doi.org/10.1007/s12035-023-03383-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-023-03383-y