Abstract
Nkx2.2 is a homeobox transcription factor that is expressed in the pancreas, intestine and central nervous system (CNS) during embryogenesis and in the adult. In mice, global deletion of Nkx2.2 results in cell mis-specification in each of the tissues where it is expressed, and the null mice die as neonates with severe hyperglycemia. Although a whole body knockout demonstrates the importance of Nkx2.2 in cell specification and postnatal viability, it precludes assessment of the cell-autonomous and postnatal functions of Nkx2.2. In this study we report the generation and functional characterization of mice encoding a conditional allele of Nkx2.2. We demonstrate the functional integrity of the conditional Nkx2.2 allele and report successful in vivo deletion using a pancreas-specific Cre recombinase. The pancreas-specific deletion of Nkx2.2 results in similar defects found in the Nkx2.2 null pancreas and the mice die shortly after birth, demonstrating that the neonatal lethality associated with the null allele is caused by pancreatic islet dysfunction. The generation of a conditional Nkx2.2 allele provides an important tool for identifying the cell-autonomous and postnatal activities of Nkx2.2 in establishing and maintaining cell type identities and functions in the pancreas, intestine and CNS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nkx2.2 is a homeodomain-containing transcription factor expressed in the pancreas, intestine and central nervous system (CNS) (Arnes et al. 2012). In the pancreas, Nkx2.2 is expressed throughout the epithelium of the developing pancreatic buds, beginning at embryonic day (E) 8.75 (Jorgensen et al. 2007). As pancreatic development proceeds, Nkx2.2 remains expressed in a population of ductal cells, as well as specific endocrine cell subtypes. In the postnatal pancreas, Nkx2.2 becomes restricted to insulin-producing beta cells, glucagon-producing alpha cells, ghrelin-producing epsilon cells, and pancreatic polypeptide-producing PP cells (Arnes et al. 2012; Prado et al. 2004; Sussel et al. 1998).
Global deletion of Nkx2.2 in the mouse results in dramatic alterations to the complement of hormone-producing islet cells in the pancreas. Specifically, beta cells are absent, alpha and PP cells are reduced, and epsilon cells are concomitantly increased (Prado et al. 2004; Sussel et al. 1998). As a result of these defects, the mice are hyperglycemic at birth and die within the first week of postnatal life. In addition to the pancreatic defects, cell specification in the intestine and CNS are also affected by deletion of Nkx2.2. Similar to the pancreas, intestinal enteroendocrine cells are formed in altered ratios; ghrelin cells are dramatically increased with a corresponding decrease in many of the other enteroendocrine populations, including those expressing serotonin, somatostatin and GIP (Desai et al. 2008). Moreover, several neuronal cell populations are re-specified in the developing CNS (Briscoe et al. 1999; Qi et al. 2001). Due to the early postnatal lethality associated with the Nkx2.2 null allele, we were unable to determine the physiological consequences of the intestinal and CNS phenotypes. Furthermore, the early lethality associated with these mice precluded analysis of any postnatal functions of Nkx2.2 in each of the tissues where it is expressed.
To determine the tissue-specific and postnatal functions of Nkx2.2, we have used a gene targeting approach to generate Nkx2.2 conditional knockout mice. We have determined that the inducible allele does not disrupt normal Nkx2.2 function. Moreover, when the Nkx2.2 flox allele is homozygosed and combined with Pdx1-cre deleter mice (Hingorani et al. 2003), Nkx2.2 is efficiently deleted in the pancreas and the Nkx2.2 null islet phenotype is recapitulated. Furthermore, these mice die with hyperglycemia shortly after birth, confirming that the early postnatal lethality is primarily caused by the pancreatic defects.
Materials and methods
Generation of mice encoding a conditional allele of Nkx2.2
A loxP site (L83) and a FNFL cassette (FRT-Neo-FRT-LoxP) were engineered to flank exon 2 (E2) of the Nkx2.2 gene to generate the Nkx2.2 conditional targeting construct on a bacterial artificial chromosome (BAC). The gene targeting vector was generated by retrieving a 10 kb long homology arm (5′ end to L83), floxed sequence containing exon 2, FNFL cassette, and 2 kb short homology arm (end of FNFL to 3′ end) into a vector carrying the Diphtheria toxin alpha chain (DTA) negative selection marker. The FNFL cassette conferred G418 resistance during gene targeting in PTL1 ES cells (129/B6 hybrid; Precision Targeting Lab, LLC), and the DTA cassette provided autonomous negative selection to reduce random integration events during gene targeting. Five targeted ES cells were identified by PCR screening and clone 9G was injected into C57BL/6 blastocysts to generate chimeric mice. Two chimeric males conferred germline transmission. Male chimeras were bred to B6; SJL-Tg(ACTFLPe)9205Dym/J (ACTB FLPe/FLPe; Rodriguez et al. 2000) female mice on a C57BL/6 background to both transmit the floxed Nkx2.2 allele through the germline and to simultaneously remove the Neomycin cassette. Two founder lines of mice carrying the Nkx2.2 conditional allele (Nkx2.2 flox) were bred to determine the affect of homozygosity of the conditional allele.
Mice carrying the conditional allele were homozygosed by mating Nkx2.2 flox/+ animals, producing Nkx2.2 flox/flox mice. Both male and female heterozygous and homozygous animals were followed from birth to 12 weeks of age and subjected to metabolic study. Nkx2.2 flox/+ mice were also mated with Tg(Ipf1-cre)1Tuv (Pdx1-cre; Hingorani et al. 2003), and subsequently maintained as Nkx2.2 flox/+; Pdx1-cre on an outbred Black Swiss background (NTac:NIHBS, Taconic). To generate mutant animals, Nkx2.2 flox/+; Pdx1-cre animals were mated with either Nkx2.2 flox/+; Pdx1-cre or Nkx2.2 flox/flox animals. Nkx2.2 flox/flox; Pdx1-cre mutants and littermate controls were assessed at E15.5 and postnatal day (P) 1. All experiments involving mice were approved by the Columbia University Institutional Animal Care and Use Committee and performed in accordance with the National Institutes of Health guidelines for the care and use of animals.
For genotyping, a portion of embryonic or postnatal tail was removed, digested with proteinase K, and DNA extracted. Previously described conditions and primers were used to genotype for Pdx1-cre (Hingorani et al. 2003). Genotyping for the Nkx2.2 flox allele was carried out using two sets of primers: L83-forward: 5′-TCCTTTTAAAAATCTGCCCACGTCT-3′, L83-reverse: 5′-GGATTTGGAGCTCGAGTCTTGG-3′; FL146-forward 5′-GGGTTATCCAGACAGTGGAGGAGTG-3′, FL146-reverse 5′-GAGGTCAACTAGGCCTCAACTTGGT-3′. An initial denaturation at 94 °C for 5 min was used, followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 1 min, and a final extension of 72 °C for 5′. The L83 primer set produced two possible amplification products of 397 bp (wildtype) and 480 bp (Nkx2.2 flox); the FL146 primer set produced two possible amplification products of 420 bp (wildtype) and 566 bp (Nkx2.2 flox).
Metabolic analyses
Animals were maintained on a normal chow diet, and body weight was measured weekly using a standard scale. Blood glucose measurements were obtained with an Accu-Check Compact Plus glucose monitor (Model GT; Roche). Glucose tolerance tests (GTT) were performed after an overnight fast, as previously described (Banks et al. 2011).
Immunofluorescence
Immunofluorescence was performed according to previously published protocols, on pancreas sections from E15.5 embryos and postnatal pancreas tissue (P0 and P14) (Mastracci et al. 2011, 2013). In all cases, tissue was embedded in OCT, after fixation with 4 % PFA and cryopreservation in 30 % sucrose. Frozen sections (8 μm) were cut and mounted on glass slides. Sections were stained with goat α-ghrelin (1:800; Santa Cruz), guinea pig α-glucagon (1:1,000; Linco/Millipore, MA), guinea pig α-insulin (1:1,000; Millipore), rabbit α-insulin (1:1,000; Cell Signaling Technology), rabbit α-somatostatin (1:200; Phoenix Pharmaceuticals), rabbit α-pancreatic polypeptide (1:200; Zymed) and rabbit α-amylase (1:1,000; Sigma). Donkey α-guinea pig-Cy2, -Cy3 or -Cy5, α-rabbit-Cy2 or -Cy3, and α-goat Cy2 or -Cy5 secondary antibodies were used (1:400, Jackson ImmunoResearch). DAPI (1:1,000; Invitrogen) was applied for 30 min following secondary antibody incubation. Images were acquired on a Leica DM5500.
RNA in situ hybridization
RNA in situ hybridization was performed on 8 μm sections mounted on glass slides as previously described (Mastracci et al. 2013) using an antisense riboprobe transcribed from a linearized plasmid containing the full-length Nkx2.2 cDNA (Hartigan and Rubenstein 1996). RNA in situ hybridization was performed on pancreas tissue sections from Nkx2.2 flox/+, Nkx2.2 flox/flox and Nkx2.2 flox/flox; Pdx1-cre at E15.5.
Real time PCR
Pancreas was dissected from each embryo and stored in RNAlater (Ambion) until RNA was extracted using the NucleoSpin RNAII Kit (Clontech). Subsequently, cDNA was made with equal amounts of RNA for each sample (Superscript III Kit, Invitrogen, CA). Real time PCR was performed using TaqMan gene expression assays-on-demand (Applied Biosystems) for glucagon (Mm00801712_m1), ghrelin (Mm00445450_m1), insulin1 (Mm01950294_s1), insulin2 (Mm00731595_gH), pancreatic polypeptide (Mm00435889_m1) and somatostatin (Mm00436671_m1). CyclophilinB was used as a control housekeeping gene, and was assayed using a probe and primer set previously described (Anderson et al. 2009). Nkx2.2 was also assayed with a probe and primer set previously described (Anderson et al. 2009). Statistical analyses were conducted with Prism Software (GraphPad Software, La Jolla, CA) using a standard t test.
Results and discussion
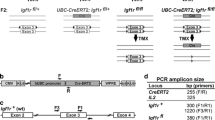
To generate a conditional allele of Nkx2.2, loxP sites were introduced into the Nkx2.2 genomic locus, flanking exon 2. The 5′ loxP site was positioned within intron 1 and the 3′ loxP site and Neomycin cassette were inserted into a non-conserved region of the 3′ UTR (Fig. 1a). The construct was electroporated into 129/B6 hybrid PTL1 ES cells. 192 colonies were screened by PCR, using primers from within the construct sequence to a genomic region located outside the sequences included in the construct (Fig. 1b). Five positive Nkx2.2 flox-targeted ES clones were identified and clone 9G was used to generate mice. Mating of chimeras for germline transmission and removal of the Neomycin cassette occurred simultaneously using mice expressing FLP recombinase on a B6 background (B6; SJL-Tg(ACTFLPe)9205Dym/J (ACTB FLPe/FLPe). Two founder lines were identified (CKO line A and CKO line B). Both lines of Nkx2.2 flox/+ animals were viable and fertile, and when the allele was homozygosed (Fig. 1c) no alteration to breeding/fertility was observed. CKO line A was used for all subsequent analysis.
Nkx2.2 conditional allele construct. (a) A loxP site (L83) and a FNFL cassette (FRT-Neo-FRT-LoxP) were engineered to flank exon 2 (E2) of the Nkx2.2 gene to generate the Nkx2.2 conditional targeting construct on a bacterial artificial chromosome (BAC). The targeting construct was retrieved onto a vector carrying the diphtheria toxin alpha chain (DTA) negative selection marker. (b) Targeted ES cells were identified by PCR screening, producing a 2396 bp product if the construct had incorporated. Male chimeras were bred to B6; SJL-Tg(ACTFLPe)9205Dym/J (ACTB FLPe/FLPe; Rodriguez et al. 2000) female mice on a C57BL/6 background to both transmit the Nkx2.2 flox allele through the germline and to simultaneously remove the Neomycin cassette. (c) Mice carrying the Nkx2.2 conditional allele (Nkx2.2 flox) were genotyped for the presence of both loxP sites; the additional sequence added by the presence of the loxP site or the loxP and fragment of the FNFL cassette provided a sequence size difference in PCR product between the Nkx2.2 flox allele and the wildtype locus. Representative genotyping PCR amplification of wildtype, Nkx2.2 flox/+ and Nkx2.2 flox/flox mice, using both the L83 and FL146 primer sets
To ensure that the presence of flanking lox sites within the Nkx2.2 genomic locus did not adversely affect Nkx2.2 function, mice carrying the Nkx2.2 flox allele were mated for homozygosity and analyzed for pancreatic/metabolic function. Males and females were followed from weaning (3 weeks) to adult (12 weeks), and no change in body weight was observed in Nkx2.2 flox/flox compared with both Nkx2.2 flox/+ and wildtype mice (Fig. 2a, b). Moreover, males and females at weaning age (3 weeks) were fasted overnight and then subjected to an intraperitoneal GTT. Following glucose injection, blood glucose was measured at timed intervals over 120 min. Glucose clearance was unaltered in Nkx2.2 flox/flox mice compared with controls (Fig. 2c, d); area under the curve shows no significant difference (Fig. 2e). Mice subjected to GTT were of similar weight (Fig. 2f), and blood glucose before fasting was not different between genotypes (Fig. 2g).
Nkx2.2 flox heterozygous and homozygous mice have normal growth and glucose tolerance. Body weight was measured in female (a) and male (b) mice from 3 weeks (weaning) until 12 weeks of age, and the presence of the Nkx2.2 flox allele did not affect mass. Glucose tolerance tests were performed on female (c) and male (d) mice at 3 weeks of age, and the Nkx2.2 flox allele did not alter glucose tolerance. For these mice there was no significant difference in area under the curve (e), body weight (f) and ad lib blood glucose (g)
Pancreas tissue was analyzed during development and postnatally to determine that all hormone-expressing cells were present and islet morphology was unaltered. At E15.5, P0 and 2 weeks of age, the complement of hormone-expressing cells appeared unaltered between wildtype and Nkx2.2 flox/flox animals (Fig. 3). In addition, islet morphology displayed the expected structure of a core of insulin-producing beta cells surrounded by glucagon-, ghrelin-, somatostatin-, and PP-expressing cells (Fig. 3m–r).
Islet cell hormone expression and morphology in the Nkx2.2 flox pancreas. The presence of hormone-expressing endocrine cells was assessed by immunofluorescence staining on pancreas tissue sections from Nkx2.2 flox/flox mice and wildtype control littermates. At E15.5 and P0, the presence of cells expressing glucagon (a, d, g, j), insulin (a–l), ghrelin (b, e, h, k), somatostatin or pancreatic polypeptide (c, f, i, l) appeared unchanged between genotypes. By 2 weeks of age (P14) there continued to be no difference in islet hormone expression and islet morphology appeared identical between wildtype (m–o) and Nkx2.2 flox/flox (p–r) mice
To test that Nkx2.2 could be deleted efficiently, Nkx2.2 flox mice were bred to Pdx1-cre transgenic mice to produce mice with a deletion of Nkx2.2 in all Pdx1-expressing cells, which includes the entire pancreatic domain. Embryos were harvested at E15.5 and the pancreas was analyzed using RNA in situ hybridization for the presence of Nkx2.2 transcript. Whereas the expected expression pattern of Nkx2.2 was observed in wildtype and Nkx2.2 flox/flox pancreas, pancreatic tissue from Nkx2.2 flox/flox; Pdx1-cre embryos was devoid of Nkx2.2 (Fig. 4a–e). Moreover, Nkx2.2 flox/flox; Pdx1-cre animals were born but died within the first week of life; pups analyzed at P1 displayed elevated blood glucose levels (Fig. 4g), as reported for the Nkx2.2 null animals, which are devoid of insulin-producing beta cells (Sussel et al. 1998). Given this similarity, Nkx2.2 flox/flox; Pdx1-cre mutant embryos were further analyzed to determine if the islet cell phenotype of the Nkx2.2 null was recapitulated in the Nkx2.2 flox/flox; Pdx1-cre mutants. As expected, ghrelin (Ghr) was dramatically increased in the Nkx2.2 flox/flox; Pdx1-cre compared with wildtype (Fig. 5a). Nkx2.2 was expectedly decreased, along with insulin1 (Ins1), insulin2 (Ins2), glucagon (Gcg), and pancreatic polypeptide (Ppy) transcript; somatostatin (Sst) expression was unchanged (Fig. 5a). Moreover, pancreas tissue from E15.5 embryos was analyzed by immunofluorescence and the observed cellular phenotype recapitulated the expression data and phenocopied the Nkx2.2 null embryos (Fig. 5b–e).
Nkx2.2 flox/flox; Pdx1-cre mutants recapitulate the Nkx2.2 null pancreatic phenotype. Using immunofluorescent staining for the expression of glucagon, the presence of alpha cells was observed in the pancreas from Nkx2.2 flox/+ (a) and Nkx2.2 flox/flox (b) embryos at E15.5, whereas a significant decrease of alpha cells was observed in the Nkx2.2 flox/flox; Pdx1-cre pancreas (c). The known pattern of expression for Nkx2.2 was observed in the control pancreas sections (d, e) compared with the efficient loss of Nkx2.2 in the mutant (f) due to Pdx1-cre mediated recombination of the Nkx2.2 flox allele. Similar to the phenotype observed in the Nkx2.2 null mice, pups were born and had severely elevated blood glucose (g). DAPI marks nuclei (a–c). Images are ×20
Pancreatic hormone expression in the Nkx2.2 flox/flox; Pdx1-cre mutants is similar to Nkx2.2 null mice. (a) The relative expression of islet hormones was assessed in the Nkx2.2 flox/flox; Pdx1-cre mice and wildtype control littermates at P0. The expression of ghrelin (Ghr), insulin1 (Ins1), insulin2 (Ins2), glucagon (Gcg), pancreatic polypeptide (Ppy) and somatostatin (Sst) appeared similar to the expected pancreas phenotype of the Nkx2.2 null mice. Pdx1-cre mediated recombination of the Nkx2.2 flox allele was efficient in reducing Nkx2.2 transcript. E15.5 pancreas tissue from wildtype (b, c) and Nkx2.2 flox/flox; Pdx1-cre (d, e) embryos was subject to immunofluorescence for glucagon, ghrelin, insulin and amylase. DAPI marks nuclei. Images are ×40
Altogether these data demonstrate that the Nkx2.2 flox allele does not disrupt normal Nkx2.2 function and successfully allows for the efficient deletion of Nkx2.2 in the presence of a Cre recombinase. In addition, Nkx2.2 flox/flox; Pdx1-cre mutant animals recapitulate the pancreatic phenotype associated with the whole body deletion of Nkx2.2 (Prado et al. 2004; Sussel et al. 1998). These studies also confirm that the neonatal lethality associated with the Nkx2.2 null mice is due to loss of Nkx2.2 activity in the pancreas. The generation of this conditional allele of Nkx2.2 facilitates further analysis of the cell-autonomous and stage-specific functions of Nkx2.2 in the pancreas, intestine and CNS.
References
Anderson KR, White P, Kaestner KH, Sussel L (2009) Identification of known and novel pancreas genes expressed downstream of Nkx2.2 during development. BMC Dev Biol 9:65
Arnes L, Leclerc K, Friel JM, Hipkens SB, Magnuson MA, Sussel L (2012) Generation of Nkx2.2:lacZ mice using recombination-mediated cassette exchange technology. Genesis 50(8):612–624
Banks AS, Kim-Muller JY, Mastracci TL, Kofler NM, Qiang L, Haeusler RA, Jurczak MJ, Laznik D, Heinrich G, Samuel VT, Shulman GI, Papaioannou VE, Accili D (2011) Dissociation of the glucose and lipid regulatory functions of FoxO1 by targeted knockin of acetylation-defective alleles in mice. Cell Metab 14(5):587–597
Briscoe J, Sussel L, Serup P, Hartigan-O’Connor D, Jessell TM, Rubenstein JL, Ericson J (1999) Homeobox gene Nkx2.2 and specification of neuronal identity by graded Sonic hedgehog signalling. Nature 398(6728):622–627
Desai S, Loomis Z, Pugh-Bernard A, Schrunk J, Doyle MJ, Minic A, McCoy E, Sussel L (2008) Nkx2.2 regulates cell fate choice in the enteroendocrine cell lineages of the intestine. Dev Biol 313(1):58–66
Hartigan DJ, Rubenstein JL (1996) The cDNA sequence of murine Nkx-2.2. Gene 168(2):271–272
Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA, Kawaguchi Y, Johann D, Liotta LA, Crawford HC, Putt ME, Jacks T, Wright CV, Hruban RH, Lowy AM, Tuveson DA (2003) Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4(6):437–450
Jorgensen MC, Ahnfelt-Ronne J, Hald J, Madsen OD, Serup P, Hecksher-Sorensen J (2007) An illustrated review of early pancreas development in the mouse. Endocr Rev 28(6):685–705
Mastracci TL, Wilcox CL, Panea C, Golden JA, May CL, Sussel L (2011) Nkx2.2 and Arx genetically interact to regulate pancreatic endocrine cell development and endocrine hormone expression. Dev Biol 359(1):1–11
Mastracci TL, Anderson KR, Papizan JB, Sussel L (2013) Regulation of Neurod1 contributes to the lineage potential of Neurogenin3+ endocrine precursor cells. PLoS Genet 9(2):e1003278. doi:10.1371/journal.pgen.1003278
Prado CL, Pugh-Bernard AE, Elghazi L, Sosa-Pineda B, Sussel L (2004) Ghrelin cells replace insulin-producing beta cells in two mouse models of pancreas development. Proc Natl Acad Sci USA 101(9):2924–2929
Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M (2001) Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development 128(14):2723–2733
Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM (2000) High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet 25(2):139–140
Sussel L, Kalamaras J, Hartigan-O’Connor DJ, Meneses JJ, Pedersen RA, Rubenstein JL, German MS (1998) Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development 125(12):2213–2221
Acknowledgments
The authors acknowledge the previous efforts of Dr. Lei Zhang and Aimee Bernard in their attempts to generate a conditional allele of Nkx2.2. Support for this study was provided by National Institutes of Health (NIH) Beta Cell Biology Consortium (BCBC) grants U01 DK072504 and U01 DK089523 (LS); NIH grant R01 DK082590 (LS); Naomi Berrie Fellowship in Diabetes Research and Juvenile Diabetes Research Foundation (JDRF) Postdoctoral Fellowship (TLM). Additional support was provided by the Columbia University DERC (P30 DK63608), the Columbia Cancer Center Transgenic Mouse/ES Cell Core Facility, and Precision Targeting Lab, LLC.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mastracci, T.L., Lin, CS. & Sussel, L. Generation of mice encoding a conditional allele of Nkx2.2 . Transgenic Res 22, 965–972 (2013). https://doi.org/10.1007/s11248-013-9700-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-013-9700-0