Abstract
Postoperative cognitive dysfunction (POCD; cognitive change associated with anesthesia and surgery) is one of the most serious long-term postoperative complications that occur in elderly patients. Dexmedetomidine (DEX) has been shown to be beneficial for improving outcomes of postoperative cognitive function. However, the exact mechanism underlying this role requires is yet to be found. The present study aims to determine the pathways involved in the protective effects of DEX against POCD in C57BL/6 J aged mice. DEX was administered after POCD modeling in C57BL/6 J aged mice. The cognitive function was evaluated after DEX treatment using novel object recognition, open field, and Y-maze tests. We also assessed its effects on neuron apoptosis and production of TNF-α and IL-1β in mouse brain tissues as well as expression levels of DNA damage-related proteins p53, p21, and γH2AX. Interactions between early growth response 1 (EGR1) and p53, microRNA (miR)-381, and EGR1 were identified by ChIP and luciferase reporter assays, and gain- and loss-of-function experiments were performed to confirm the involvement of their interaction in POCD. DEX administration attenuated hippocampal neuron apoptosis, neuroinflammation, DNA damage, and cognitive impairment in aged mice. miR-381 targeted EGR1 and disrupted its interaction with p53, leading to a decline in hippocampal neuron apoptosis, DNA damage, neuroinflammation, and cognitive impairment. Furthermore, DEX administration resulted in the enhancement of miR-381 expression and the subsequent inhibition of EGR1/p53 to protect against cognitive impairment in aged mice. Overall, these results indicate that DEX may have a potential neuroprotective effect against POCD via the miR-381/EGR1/p53 signaling, shedding light on the mechanisms involved in neuroprotection in POCD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Postoperative cognitive dysfunction (POCD; cognitive change associated with anesthesia and surgery) is a common postoperative complication in patients that often exhibit acute cognitive dysfunction, impaired memory, and executive function deficits [1]. The elderly population is more prone to suffering from this disorder at some point during their lives [2]. Risk factors responsible for POCD include advanced age, systemic concomitant disease, and a lower level of education [3]. A multidisciplinary care bundle modality to risk factor stratification and reduction has been the most attractive current treatment approach; but up to now, no individual anesthetic, drug, or monitoring mode has been reported to help reduce POCD incidence [4]. Cognitive dysfunction following laparotomy is mainly attributed to neuroinflammation, tau phosphorylation, and DNA damage [5]. Hence, the identification of mechanisms involved in those processes may aid in developing alternative therapies for POCD.
Dexmedetomidine (DEX) is a highly selective α2 adrenergic receptor agonist and has been reported to play an efficient role on multi-organ protective mechanisms (such as the nervous system, heart, and lung) by reducing the inflammatory response in these organs, activating antiapoptotic signaling pathways [6]. A recent study reports that DEX can prevent cognitive decline by resolving high-mobility group box 1 protein-induced inflammation via a vagomimetic action in mice [7]. In addition, DEX protects PC12 cells from ropivacaine injury by upregulating the expression of microRNA-381 (miR-381) [8]. miRNAs are a class of noncoding RNAs that modulate post-transcriptional gene expression and have been implicated to participate in the pathogenesis of nervous system diseases [9]. miR-381 can alleviate neuropathic pain through targeting HMGB1 and CXCR4 [10]. Increased miR-381 favors the repair of nerve injury by negatively regulating LRRC4 through activating the SDF-1/CXCR4 signaling pathway in acute cerebral ischemia rats after cerebral lymphatic blockage [11]. In this study, StarBase database (http://starbase.sysu.edu.cn/) predicted the binding of miR-381 to the 3ʹ-untranslated region (UTR) of early growth response 1 (EGR1), a transcription factor acting as a leading mediator and regulator of synaptic plasticity and neuronal activity both physiologically and pathologically [12]. EGR1 is considered a potential therapeutic target to prevent postmenopausal cognitive decline by decreasing the expression of STAT3 [13]. Furthermore, upregulating EGR1 may promote p53 transcription [14] and targeting the transcription factor p53 is of great clinical significance for neuroprotection against severe cognitive decline induced by Alzheimer’s disease [15]. We hence speculated whether miR-381, EGR1, and p53 could participate in the regulation of DEX treatment in DNA damage and cognitive dysfunction. This study was designed in mice models of POCD to evaluate the potential effects of DEX treatment on POCD and to identify the mechanism involving miR-381, EGR1, and p53.
Materials and Methods
Ethics Statement
This study was carried out after being approved by the ethics committee of the First Hospital of China Medical University and was performed according to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. Extensive efforts were made to ensure minimal suffering of the animals used in the study.
Establishment of Mouse POCD Models
A total of 80 male C57BL/6 aged mice (18 weeks old) purchased from Shanghai Experimental Animal Center (Shanghai, China) were housed in a specific pathogen-free experimental animal center at 22 ± 1 °C under a 12-h light/dark cycle, with ad libitum food and water. Laparotomy was performed under sevoflurane anesthesia as previously reported [16]. Mice were placed on a heated pad and anesthetized using a rodent inhaler (MATRX, USA) with 4–5% sevoflurane (SEVO) (Sevorane™, Abbott, Switzerland), during which the oxygen flow rate was 0.8 L/min. A 2.5-cm longitudinal midline incision was made through the skin, abdominal muscle, and peritoneum. Approximately 10 cm of intestine was eviscerated and rubbed violently for 30 s. The intestinal loop was exposed outside the abdominal cavity for 1 min and then placed back into the abdominal cavity. A sterile suture (4–0, PS-2; Ethicon A) was used to suture the peritoneum, abdominal muscle, and skin. During the whole process, the rhythm and frequency of breathing and the color of claws were monitored. The whole procedure was completed under 15 min. The mice in the anesthetics group were anesthetized with SEVO at the same concentration and flow rate for 15 min.
Animal Treatment
Mice were treated with SEVO or intraperitoneally injected with DEX (0.5 mg/kg) with normal mice (without any treatment) serving as the control, 8 mice for each treatment, while 8 POCD-operated mice were non-treated, 8 POCD-operated mice treated with DEX, 8 POCD-operated mice intraventricularly injected with antagomir NC, or 8 POCD-operated mice intraventricularly injected with miR-381 antagomir after POCD, or 8 POCD-operated mice intraventricularly injected with miR-381 antagomir and intraperitoneally injected with DEX. The success rate of model construction was 82.4%. Mice were fixed in the stereotactic apparatus, and the stainless steel guide cannula (0.5 mm in diameter) with an insertion cannula (0.25 mm in diameter) was implanted into the right ventricle (approximately − 0.5 mm from bregma, ML = − 1.5 mm; DV = − 4.5 mm) for injection. miR-381 antagomir and antagomir NC were diluted to the appropriate concentration (2 μg dissolved in 50 μL PBS) and injected into mice 6 h before their scheduled operation [17]. DEX (SML0956, Sigma-Aldrich Chemical Company, St Louis, MO, USA) was dissolved in normal saline and diluted to 1 μM, 10 μM, and 100 μM.
Open Field Test
Spontaneous activity of mice was observed in an open field (40 × 40 × 40 cm) 11 days after the operation to assess exploratory locomotor activity. Each mouse was placed in the center of the field and left to roam around for 5 min. Their activity was automatically recorded using a video tracking system. Finally, the distance of mice moving in the open field and the time of exploring in the center were recorded.
Novel Object Recognition (NOR) Test
The NOR test was carried out in an open field (30 × 30 × 30 cm) made of polyvinyl chloride, plywood and acrylic acid [18]. The task included adaptation, training, and maintenance for three consecutive days. On the first day, mice were allowed to adapt to the experimental area for 5 min without any behavioral stimulation. The next day, mice were free to explore two identical objects (black plastic cube, 5 × 5 × 5 cm) for 5 min. On the third day, one of the objects was replaced with a new one, and the shape and color of the object were changed compared with that of the previous day (gray plastic square pyramid, 5 × 5 × 7 cm). Mice were then left to adapt to the experimental area again for 5 min. Before the test, the experimental area was cleaned with 70% ethanol solution to remove any traces of odor. Mice explored objects by sniffing or touching objects at a distance of less than 2 cm with their noses or forepaws. Actions including sitting and turning were not considered an act of exploration. Finally, the exploratory behavior of each mouse was evaluated by observers who did not know the number and condition of the mice in a double-blinded fashion. The total exploration time indicated the time devoted to the location of two objects. Cognitive function was measured using the identification index, which was a comparison between the exploration time of two objects (training session) or new object (reserved conversation) and the total exploration time.
Y-Maze Test
The Y-maze test was used to evaluate aversion memory, that is, hippocampal-dependent learning and memory. The maze consisted of three plastic arms with 32 cm (length) × 10 cm (width) × 10 cm (height), of which two arms were black, with 3.175-mm stainless steel rods (8 mm away) at the bottom, and the other arm being the whiteboard. During the preoperative training, each mouse was allowed to explore any arm freely for 8 min. After that, the mice were placed in one of the black compartments and electric shocks (2 Hz, 10 s, 40 ± 5 V) were given until the mice entered the non-shock compartment within 10 s and stayed there for 30 s, which was the correct choice. The training was deemed successful when 9 consecutive correct choices were made after electric shock. In order to conduct postoperative tests 14 days after operation [5], each mouse was tested 10 times according to the same procedure as the training test. The number of wrong choices and the time required to entering the shockproof cabin were recorded.
Immunofluorescence
Brain tissues were fixed for 72 h, paraffin embedded, and cut into 6-μm-thick sections. The sections were then treated with 0.01 M citric acid buffer (pH 6.0) at 90 °C for 15 min and added with 10% goat serum to block any nonspecific binding. Afterwards, the sections were incubated overnight at 4 °C with primary antibodies against Iba1 (ab178846, 1:500, Abcam Inc., Cambridge, UK), CD68 (ab31630, 1:500, Abcam), p53 (ab90363, 1:200, Abcam), γH2AX (ab81299, 1:100, Abcam), and NeuN (ab177487, 1:200, Abcam). After sections were washed 3 times with PBS, they were further incubated with specific Alexa Fluor 568 or 488 secondary antibodies (Invitrogen) for 2 h. After washing, 3 μM 4ʹ,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) was added for nuclear counterstaining. The immunolabeled tissues were observed under a laser scanning confocal fluorescence microscope, equipped with ZEN software, and Z-stack images were obtained. All quantitative analyses were performed on at least four images obtained from at least four consecutive slices of each mouse. Each experiment was performed 3 times independently.
Terminal Deoxynucleotidyl Transferase-Mediated dUTP-Biotin Nick End Labeling (TUNEL) Assay
The TUNEL technique (In Situ Cell Death Detection Kit, TMR red; Roche Applied Sciences, Indianapolis, IN, USA) and NeuN (ab177487, 1:200, Abcam) co-staining were used to determine neuronal apoptosis in hippocampal tissues. The technique was used to label the cut ends of DNA fragments in apoptotic nuclei. The nuclei of all cells were stained with DAPI.
Enzyme-Linked Immunosorbent Assay (ELISA)
Tumor necrosis factor-α (TNF-α) and interleukin (IL)-1β were quantified using the customized Mouse ProcartaPlex™ Panel by following the manufacturers’ instructions (eBioscience, San Diego, CA, USA). MAGPIX (Luminex) was used for data acquisition. Each sample was measured twice.
Cell Culture and Treatment
HEK-293 T cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum and 1% penicillin–streptomycin solution. The primary hippocampal neurons were obtained from newborn mice (one day after birth). Mice were anesthetized with 5% isoflurane and the hippocampus were dissected and trypsinized. Polished Pasteur straws at three different sizes were then used to disperse cells 10 times for isolation. The cells were collected in the nerve basic medium (GIBCO, USA) and the impurities were filtered with a 40-mm filter. Hippocampal neurons were seeded on slides in an 8-well poly-l-lysine treatment chamber (30,000 cells per well) and then cultured in nerve basic medium (2% B27, 2 mM l-glutamine, 50 U/mL penicillin, and 50 U/mL streptomycin) at 37 °C and 5% CO2.
Neurons were transfected with sh-NC, sh-EGR1, miR-381 inhibitor, inhibitor NC, miR-381 mimic, and mimic NC plasmids, which were all synthesized and constructed by Shanghai GenePharma Co., Ltd. (Shanghai, China). The packaging plasmids pSPAX, pMD2.G, and the target gene vector were co-transfected into HEK-293 T cells. The target gene was constructed into pLKO.1 vector, and then cells were infected with lentivirus. After 48 h of cell culture, the supernatant was collected and the virus particles were filtered. After that, the virus titer was detected. The neurons were then treated with sh-NC, sh-EGR1#1, sh-EGR1#2, sh-EGR1#3, miR-381 inhibitor, inhibitor NC, miR-381 mimic, and mimic NC. Suspension with 4 × 105 cells/mL was seeded into 6-well plates, 2 mL per well, and cultured overnight at 37 °C for 48 h. Finally, the expressions of related genes in each group were detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
Etoposide (Sigma-Aldrich, a topoisomerase II inhibitor used to induce DNA damage) was dissolved in dimethyl sulfoxide (DMSO) and diluted to 15 μM and 30 μM to treat hippocampal neurons.
RNA Isolation and Quantitation
The total RNA content was extracted from mouse brain tissues using the TRIzol reagent (16,096,020, Thermo Fisher Scientific Inc., Waltham, MA, USA). The extracted RNA was reverse transcribed into complementary DNA (cDNA) using the reverse transcription kit (RR047A, Takara, Japan) for the detection of mRNA. The extracted RNA was reverse transcribed into cDNA with PolyA tail (containing universal PCR primer R and U6 universal PCR primer R) using the PolyA tailing kit (B532451, Shanghai Sangon Biotechnology Co. Ltd., Shanghai, China) for the detection of miRNA. Subsequently, the cDNA was subjected to real-time qPCR using the SYBR Premix Ex TaqTM II (Perfect Real Time) kit (DRR081, Takara, Japan) on an ABI 7500 instrument (Applied Biosystems, Foster City, CA, USA), with 3 duplicates set in each well. β-actin and U6 served as the internal reference and the fold changes were calculated using the method of 2−ΔΔCt. All primer sequences are listed in Table S1 .
Western Blot Analysis
The total protein content was extracted from tissues and cells using radio-immunoprecipitation assay (RIPA) lysis buffer (P0013B, Beyotime Biotechnology Co., Shanghai, China) containing phenylmethylsulfonyl fluoride (PMSF). The protein concentration was then determined with a bicinchoninic acid (BCA) kit (23,229, Thermo Fisher Scientific). The protein was transferred onto a polyvinylidene fluoride membrane after being separated by 8–12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Next, the membrane was treated with 5% skimmed milk powder or bovine serum albumin at room temperature for 1 h and then underwent overnight incubation at 4 °C with the following diluted primary rabbit antibodies purchased from Abcam Inc., Cambridge, UK: EGR1 (ab133695, 1:2000), p53 (ab131442, 1:1000), p21 (ab109520, 1:5000), γH2AX (ab81299, 1:1000), and β-actin (ab8227, 1:2000). The following day, the membrane was re-probed with horseradish peroxidase-labeled secondary goat anti-rabbit IgG (ab6721, 1:500, Abcam) at room temperature for 1 h. Afterwards, the membrane was visualized using enhanced chemiluminescence (ECL) solution (1,705,062, Bio-Rad Laboratories, Hercules, CA, USA). Protein bands were quantified by Image Quant LAS 4000C gel imager (GE Healthcare, Piscataway, NJ, USA), with β-actin serving as the loading control.
Flow Cytometry
After 48 h of transfection, the cells were digested with 0.25% trypsin without ethylenediaminetetraacetic acid (YB15050057, Shanghai Yubo Biotechnology Co., Ltd., Shanghai, China) and collected in a flow tube and centrifuged with the supernatant discarded. The cells were washed with cold PBS 3 times, and the supernatant was discarded after centrifugation. Annexin-V-FITC, propidium iodide (PI), and HEPES buffer solution were prepared into Annexin-V-FITC/PI staining solution at the ratio of 1:2:50 by following the instructions provided by the Annexin-V-fluorescein isothiocyanate (FITC) cell apoptosis detection kit (K201-100, BioVision, Milpitas, CA, USA). One hundred microliters of dye solution was later used to resuspend 1 × 106 cells, which was shaken, mixed, and incubated at room temperature for 15 min. Next, 1 mL of HEPES buffer solution (PB180325, Procell, Wuhan, China) was added to cells and mixed by shaking. The fluorescence of FITC and PI were detected at 525 nm and 620 nm by a 488-nm filter, which reflected cell apoptosis.
Chromatin Immunoprecipitation (ChIP)
Cells from two 100-mm culture dishes were fixed with 4% formaldehyde and terminated with 0.125 M glycine. Cells were resuspended in RIPA buffer (P0013B, Beyotime). The genomic DNA of adipose tissues was broken into 200–700-bp fragments by ultrasonic treatment. The DNA concentration of the sample was determined and divided into the input, EGR1 and IgG groups. The EGR1 and IgG groups were added with DynabeadsTM Protein G (Thermo Fisher Scientific) and corresponding antibodies against EGR1 (MA5-15,008, 1:200, Invitrogen) and IgG (ab190475, 1:200, Abcam) at 4 °C overnight. After incubation, the DNA samples were eluted and then extracted with a DNA extraction kit. The DNA was dissolved and stored at – 20 °C. RT-qPCR was used to detect the enrichment of DNA immunoprecipitated. The DNA primer sequences for this assay were as follows: F: 5ʹ-GGTTTCCACCCATTTTGCCC-3ʹ and R: 5ʹ -CAAGTCCTTCCGTGGGTTGA-3ʹ.
Dual-Luciferase Reporter Gene Assay
The complementary binding sites of miR-381 to the 3ʹ-UTR of target gene EGR1 were predicted by the StarBase software. psiCHECK2-EGR1-3ʹ-UTR-wild type (WT) and psiCHECK2-EGR1-3ʹ-UTR-mutant type (Mut) vectors were constructed by cloning EGR1-3ʹ-UTR-WT and EGR1-3ʹ-UTR-MUT into psiCHECK2 (Cat.# C8021; Promega Corp., Madison, Wisconsin, USA). HEK-293 T cells in the logarithmic growth phase were seeded into 6-well plates. After 24 h of conventional culture, EGR1-3ʹ-UTR-WT and EGR1-3ʹ-UTR-MUT were co-transfected with mimic NC and miR-381 mimic to the cells. For the luciferase activity measurement of p53 promoter, a 2000-bp fragment upstream of the transcription initiation side (TSS) of p53 was amplified to the psiCHECK vector by primer (F: 5ʹ-TCGTCCCACCGAAAGGGG-3ʹ and R: 5ʹ-GTCCCAATCCCAGCAACCC-3ʹ), followed by transfection into overexpressed (oe)-NC- and oe-EGR1-treated neurons respectively. After cells were transfected for 48 h, they were collected, lysed, and centrifuged for 3–5 min, after which the supernatant was obtained and detected with a luciferase detection kit (RG005, Beyotime) on a TD-20/20 Luminometer (E5311, Promega). The relative luciferase (RLU) activity was calculated as the RLU activity of firefly luciferase/RLU activity of renilla luciferase.
Statistical Analysis
All data were processed using the SPSS 21.0 statistical software (IBM Corp. Armonk, NY, USA). Measurement data were expressed as the mean ± standard deviation. Data between two groups were analyzed using an unpaired t-test while data among multiple groups were compared by one-way analysis of variance (ANOVA) with Tukey’s test. Data at different time points were analyzed using repeated measures of ANOVA followed by the Bonferroni post hoc test. In all statistical analyses, a value of p < 0.05 represents statistical significance.
Results
DEX Alleviates Cognitive Dysfunction and Inhibits Neuroinflammation and Neuron Apoptosis in POCD-Aged Mice
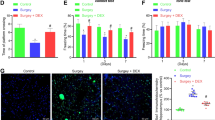
We selected aged mice to construct the POCD model (Fig. 1A) and then measured the bodyweight of mice at the 1, 3, 7, and 14 days after the POCD model was constructed. The results showed that mice treated with SEVO had a lower weight than that of the control mice, where POCD mice exhibited a more pronounced reduction in the weight. In addition, the weight of POCD mice treated with DEX was significantly decreased compared with that of the DEX-treated mice (p < 0.05, Fig. 1B). The results of the open field test revealed no significant differences in the dynamic distance (p < 0.05, Fig. 1C) and the time spent in the open field center (p < 0.05, Fig. 1D). Experimental data from the NOR test showed that the cognitive ability was reduced in POCD mice while it was increased following DEX treatment (p < 0.05, Fig. 1E). Moreover, the number of errors and latency period was increased in POCD mice while DEX nullified this, as revealed by the Y-maze test (p < 0.05, Fig. 1F, G).
DEX treatment mitigates cognitive dysfunction, suppresses neuroinflammation, and neuron apoptosis in POCD-aged mice. Control mice were used as controls, and the other mice were treated with POCD, SEVO, DEX, or POCD + DEX. A Experimental flowchart of the POCD model construction. B Mouse body weight at the 1st, 3rd, 7th, and 14th day following POCD model construction. C Dynamic distance of mice was analyzed by the open field test. D Time spent in the open field center was analyzed by the open field test. E The changes of cognitive index detected by the NOR test 14 days after the operation. F The number of errors of mice analyzed by the Y-maze test 14 days after the operation. G Latency period of mice analyzed by Y-maze test 14 days after the operation. H Quantitative analysis of Iba1-positive cells determined by immunofluorescence staining in the hippocampus tissues. I Co-expression of Iba1 and CD68 determined by immunofluorescence staining in mouse hippocampus CA1 14 days after the operation (200 ×). J Levels of TNF-α and IL-1β determined by ELISA in the hippocampus tissue. K Quantitative analysis of TUNEL + /NeuN + cells in mouse hippocampus CA1 14 days after the operation. n = 8 for mice upon each treatment. Asterisk “*” indicates p < 0.05. Data are shown as the mean ± standard deviation of three technical replicates and analyzed by unpaired t-test
The results of immunofluorescence staining showed that Iba1-positive cells in the hippocampus tissue were increased after POCD operation, which was annulled by DEX treatment (p < 0.05, Fig. 1H). Furthermore, co-expression of Iba1 and CD68 was activated in hippocampus CA1 of the POCD mice group, but it was inhibited in the presence of DEX (p < 0.05, Fig. 1I). The results of ELISA showed that the levels of TNF-α and IL-1β in the hippocampus tissue of the POCD mice were much higher than those of the control mice while DEX diminished TNF-α and IL-1β levels (p < 0.05, Fig. 1J). Analysis using TUNEL/NeuN double staining suggested an increase of the TUNEL + /NeuN + cells in hippocampus CA1 of POCD mice while the number was decreased in response to DEX treatment (p < 0.05, Fig. 1K). In summary, DEX could improve cognitive function and repress neuroinflammation and neuron apoptosis in POCD-aged mice.
DEX Suppresses POCD-Related DNA Damage in Hippocampus Tissues of Mice by Disrupting the p53 Signaling Pathway
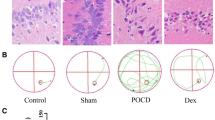
Next, we aimed to elucidate the mechanism of DEX in POCD. Drug PubChem database (https://pubchem.ncbi.nlm.nih.gov/) predicted the interaction network between DEX and chemical genes (Fig. 2A), and 8961 genes related to cognitive impairment were obtained from the GeneCards database (https://www.genecards.org/). A Venn diagram analysis of the aforementioned genes was conducted and 28 genes were included at the intersection (Fig. 2B). KEGG enrichment analysis using the clusterProfiler package in R (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) suggested that these 28 genes were associated with p53 and other signaling pathways (Fig. 2C). Additionally, Western blot analysis showed higher protein levels of p53, p21, and γH2AX in the hippocampus tissue of POCD mice, whereas DEX decreased these protein levels (p < 0.05, Fig. 2D). The fluorescence intensity of p53 and γH2AX was found to be enhanced in hippocampus CA1 of POCD mice while an opposing result was noted following DEX treatment (p < 0.05, Fig. 2E, F). These results indicated that POCD-related DNA damage in hippocampus tissues of mice could be inhibited by DEX.
DEX alleviates DNA damage in hippocampus tissues of POCD mice by inactivating the p53 signaling pathway. Control mice were used as controls, and the other mice were treated with POCD, SEVO, DEX, or POCD + DEX. A Interaction network of DEX and chemical genes predicted by the drug database PubChem. B Venn diagram of DEX target and cognitive dysfunction-related genes. C KEGG enrichment analysis of the 28 overlapped genes. Abscissa represents the number of genes involved, and ordinate represents the name of the enrichment pathway. D Western blot analysis of p53, p21, and γH2AX proteins in the mouse hippocampus tissue 14 days after the operation. E Quantitative analysis of fluorescence intensity of p53 and γH2AX in the mouse hippocampus tissue 14 days after the operation. F Representative images showing fluorescence intensity of p53 and γH2AX in the mouse hippocampus tissue 14 days after the operation (200 ×). n = 8 for mice upon each treatment. Asterisk “*” indicates p < 0.05. Data are shown as the mean ± standard deviation of three technical replicates and analyzed by an unpaired t-test
EGR1 Promotes Transcription of p53 and Elicits Apoptosis and DNA Damage of Neurons
We exploited the action of p53 in POCD and used GeneCards and MINT databases (https://mint.bio.uniroma2.it/) to predict upstream factors of the p53 signaling pathway, revealing 240 and 230 overlapped genes. In addition, the CTD database (http://ctdbase.org/; Inference Score > 10) predicted 933 genes related to cognitive impairment, and Venn analysis of these predicted genes presented with 22 genes at the intersection (p < 0.05, Fig. 3A). A co-expression network of 22 candidate genes was then obtained by the Coexpedia tool (http://www.coexpedia.org/), of which HSPA9, CDKN1A, GADD45A, and EGR1 scored higher in the website (score > 7) (p < 0.05, Fig. 3B). Moreover, the Jaspar website predicted the presence of a binding site of GGCGGGGGAGA in EGR1 and the 1253–1263 bp of p53 promoter (p < 0.05, Fig. 3C). Therefore, we speculated that EGR1 may regulate the cognitive impairment of POCD mice by the transcriptional regulation of p53 expression.
EGR1 is highly expressed in mice rendered with POCD and upregulates the expression of p53, inducing DNA damage of neurons. A Venn diagram of the intersected genes predicted by the GeneCards and MINT datasets and cognitive impairment of related genes predicted by the CTD database. B Co-expression network of 22 candidate genes was obtained by the Coexpedia database. C Binding site of EGR1 to the p53 promoter region predicted by the Jaspar website. D Expression of ERG1 determined by RT-qPCR and Western blot analysis in hippocampus CA1 of POCD mice treated with DEX. E Enrichment of EGR1 in the p53 promoter region determined by ChIP assay. F Luciferase activity of p53 promoter detected by dual-luciferase reporter gene assay in neurons treated with oe-EGR1. G Expressions of ERG1 and p53 determined by RT-qPCR and Western blot analysis in primary hippocampal neurons transfected with sh-EGR1. H Cell apoptosis measured by flow cytometry upon transfection with sh-EGR1. I Expression of ERG1 determined by RT-qPCR in neurons treated with 15 μM and 30 μM etoposide. J Western blot analysis of EGR1, p53, p21, and γH2AX in neurons treated with 15 μM and 30 μM etoposide. K Western blot analysis of EGR1, p53, p21, and γH2AX in neurons co-treated with sh-EGR1 and 30 μM etoposide. Asterisk “*” indicates p < 0.05. Data are shown as mean ± standard deviation of three technical replicates
Subsequent results of RT-qPCR and Western blot analysis showed that the expression of ERG1 was increased in hippocampus CA1 of POCD mice, while a decline was shown in response to DEX treatment (p < 0.05, Fig. 3D). EGR1 was found to be significantly expressed in the promoter region of p53 (p < 0.05, Fig. 3E), while the luciferase activity of the p53 promoter was increased in oe-EGR1-treated neurons (p < 0.05, Fig. 3F). To validate their interaction in vitro, hippocampal neurons were isolated from newborn mice. The expression of EGR1 was decreased in hippocampal neurons transfected with sh-EGR1#1, sh-EGR1#2, and sh-EGR1#3, among which sh-EGR1#1 had the lowest expression of EGR1 (p < 0.05, Fig. 3G). Furthermore, the expression of p53 was observed to be downregulated upon EGR1 silencing and similarly, sh-EGR1#1 showed the lowest expression of p53 (p < 0.05, Fig. 3G), which was thus used for subsequent analyses. The aforesaid findings indicated that the expression of p53 could be activated by EGR1.
Flow cytometry indicated that cell apoptosis was decreased upon silencing EGR1 (p < 0.05, Fig. 3H). Afterwards, etoposide was utilized to induce DNA damage in hippocampal neurons. The expression of EGR1 was much higher in cells treated with etoposide (p < 0.05, Fig. 3I) compared with that of DMSO-treated cells. Western blot analysis suggested that etoposide could diminish the protein expressions of EGR1, p53, p21, and γH2AX and 30 μM of etoposide had a more pronounced effect (p < 0.05, Fig. 3J). These results indicated that the expression of EGR1 in the cells with DNA damage induced by etoposide was increased at both mRNA and protein levels. Furthermore, silencing EGR1 greatly inhibited the protein expressions of EGR1, p53, p21, and γH2AX induced by 30 μM of etoposide (p < 0.05, Fig. 3K). These results suggested that EGR1 was highly expressed in mice rendered with POCD by promoting the expression of p53. Knockdown of EGR1 could inhibit neuron apoptosis; the increase of p53 mediated by EGR1 increased the DNA damage of neurons.
ERG1 Is a Target Gene of miR-381
After discovering the increased expression of ERG1 in POCD and its regulatory role in p53, we next aimed to investigate the potential upstream mechanism of EGR1. Bioinformatics databases StarBase (http://starbase.sysu.edu.cn/), microT (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/) and RNAInter (http://www.rna-society.org/rnainter/; Score > 0.58) predicted 8 candidates upstream miRNAs of EGR1, including miR-124-3p, miR-191-5p, miR-203-3p, miR-329-3p, miR-377-3p, miR-381, miR-362-3p, and miR-1198-5p (Fig. 4A). The results from RT-qPCR showed an enhancement of the following expressions of miR-124-3p, miR-191-5p, miR-203-3p, and miR-329-3p in the hippocampus tissue of POCD mice; on the contrary, the expressions of miR-381 and miR-377-3p were downregulated, with miR-381 exhibiting a more pronounced decline (p < 0.05, Fig. 4B) in expression. Besides, other miRNAs had no changes (p > 0.05, Fig. 4B). Binding sites of miR-381 to the 3ʹ-UTR of EGR1 were found in the StarBase database (Fig. 4C). Dual-luciferase reporter gene assay further confirmed that miR-381 mimic promoted the luciferase activity of EGR1-3ʹ-UTR-WT (p < 0.05) but had no effects on EGR1-3ʹ-UTR-MUT in HEK-293 T cells (p > 0.05, Fig. 4D). At the same time, the expression of EGR1 was decreased in the presence of miR-381 mimic (p < 0.05, Fig. 4E, F) in primary hippocampal neurons. The aforementioned data supported that miR-381 downregulated the expression of EGR1 by binding to its 3ʹ-UTR.
miR-381 targets EGR1 and downregulates its expression. A Venn diagram of the intersected upstream miRNAs of EGR1 predicted by the StarBase, microT, and RNAInter databases. B Expressions of miR-124-3p, miR-191-5p, miR-203-3p, miR-329-3p, miR-377-3p, miR-381, miR-362-3p, and miR-1198-5p determined by the expression of RT-qPCR in the hippocampus tissue of POCD mice. C Putative miR-381 binding sites to the 3ʹ-UTR of EGR1 in the StarBase database. D Binding of miR-381 to EGR1 confirmed by dual-luciferase reporter gene assay in HEK-293 T cells. E Expressions of miR-381 and EGR1 determined by RT-qPCR in primary hippocampal neurons transfected with miR-381 mimic. F Western blot analysis of EGR1 protein in primary hippocampal neurons transfected with miR-381 mimic. Asterisk “*” indicates p < 0.05. Data are shown as the mean ± standard deviation of three technical replicates
DEX Reduces Apoptosis and DNA Damage of Hippocampal Neurons by Upregulating miR-381 and Suppressing the EGR1/p53 Pathway
Next, we determined whether DEX could affect POCD through the miR-381/EGR1/p53 axis. The expression of miR-381 was upregulated in the hippocampus tissues of POCD mice treated with DEX, as revealed by RT-qPCR (p < 0.05, Fig. 5A). In hippocampal neurons, the expression of miR-381 was enhanced along with the increase of DEX concentration, the highest expression of which was found in the 100-μM DEX treatment group (p < 0.05, Fig. 5B). Furthermore, Western blot analysis showed that the expressions of EGR1 and p53 were decreased by increasing the concentration of DEX e, and the inhibition was the most significant under 100 μM of DEX treatment (p < 0.05, Fig. 5C).
DEX exerts an inhibitory role in the apoptosis and DNA damage of hippocampal neurons by regulating the miR-381/EGR1/p53 pathway. A The expression of miR-381 determined by RT-qPCR in the hippocampus tissue of POCD mice treated with DEX. B The expression of miR-381 determined by RT-qPCR in hippocampal neurons treated with 1 μM, 10 μM, and 100 μM of DEX. C Western blot analysis of EGR1 and p53 proteins in neurons treated with 1 μM, 10 μM, and 100 μM of DEX. D The expression of miR-381 determined by RT-qPCR in hippocampal neurons treated with DEX, miR-381 inhibitor, or both. E Apoptosis rate measured by flow cytometry in hippocampal neurons treated with DEX, miR-381 inhibitor, or both. F The expression of miR-381 determined by RT-qPCR in hippocampal neurons co-treated with 100 μM DEX and 30 μM etoposide. G Western blot analysis of EGR1, p53, p21, and γH2AX proteins in hippocampal neurons co-treated with 100 μM DEX and 30 μM etoposide. Asterisk “*” indicates p < 0.05. Data are shown as the mean ± standard deviation of three technical replicates
Moreover, the results of RT-qPCR demonstrated an uptrend of the expression of miR-381 in DEX-treated neurons, while it was significantly lower following the miR-381 inhibitor. The dual treatment with miR-381 inhibitor and DEX increased the expression of miR-381 (p < 0.05, Fig. 5D). Flow cytometric data showed a reduction in the apoptosis rate of DEX-treated neurons while an increase was observed in response to miR-381 inhibitor. Both miR-381 inhibitor and DEX decreased the apoptosis rate (p < 0.05, Fig. 5E). Furthermore, 30-μM etoposide treatment suppressed the expressions of miR-381, while the 100-μM DEX treatment group was found to abolish this effect (p < 0.05, Fig. 5F). Meanwhile, Western blot analysis showed elevated expressions of EGR1, p53, p21, and γH2AX upon 30-μM etoposide treatment, while DEX treatment annulled the results (p < 0.05, Fig. 5G). In summary, DEX has been shown to promote the expression of miR-381 and inhibit apoptosis and DNA damage of hippocampal neurons by activating miR-381 and inhibiting the EGR1/p53 pathway.
DEX Inhibits DNA Damage and Apoptosis of Hippocampal Neurons and Alleviates POCD in Mice by miR-381/EGR1/p53
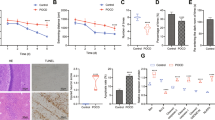
In order to further explore whether DEX can alleviate cognitive dysfunction in POCD mice through miR-381/EGR1/p53 pathway, we injected 100 ng miR-381 antagomir/antagomir NC into mouse brains 6 h before the operation, with the construction model shown in Fig. 6A. On the 1st, 3rd, 7th, and 14th day after the operation, the bodyweight of POCD mice was subsequently reduced while DEX treatment increased mice’s weight (p < 0.05, Fig. 6B). In addition, the cognitive index of POCD mice was lowered, whereas it was enhanced following DEX treatment. miR-381 antagomir impaired the cognitive index while further DEX treatment potentiated the index (p < 0.05, Fig. 6C). The results of the Y-maze test showed that the number of errors and latency period of POCD mice was increased, which was negated by DEX. miR-381 antagomir led to an increase in the number of errors and latency period while DEX reduced the aforementioned indicators (p < 0.05, Fig. 6D, E).
DNA damage, neuroinflammation, and neuron apoptosis were attenuated by DEX through the miR-381/EGR1/p53 pathway in POCD mice. Control mice were used as controls, and POCD mice were treated with DEX, miR-381 antagomir, or both. A Experimental flow chart of the POCD mice treated with miR-381 antagomir. B Mouse body weight on the 1st, 3rd, 7th, and 14th day. C The changes of cognitive index detected by the NOR test 14 days after the operation. D The number of errors of mice analyzed by the Y-maze test 14 days after the operation. E Latency period of mice analyzed by the Y-maze test 14 days after the operation. F Quantitative analysis of Iba1-positive cells determined by immunofluorescence staining in the mouse hippocampus tissue 14 days after the operation. G Expressions of TNF-α and IL-1β determined by ELISA in mouse hippocampus CA1 14 days after the operation. H Quantitative analysis of TUNEL + /NeuN + cells in mouse hippocampus tissues. I The expression of miR-381 determined by RT-qPCR in mouse hippocampus tissues. J Western blot analysis of EGR1, p53, p21, and γH2AX proteins in mouse hippocampus tissues. K Fluorescence intensity of γH2AX in mouse hippocampus tissues determined by immunofluorescence staining. n = 8 for mice upon each treatment. Asterisk “*” indicates p < 0.05. Data are shown as the mean ± standard deviation
Iba1-positive cells were increased, microglia in hippocampus CA1 was activated, and fluorescence intensity of CD68 and Iba1+ co-expression were increased in POCD mice, but such effects were undermined by DEX. The results demonstrated increased Iba1-positive cells and activated microglia in response to treatment with miR-381 antagomir, while DEX counteracted the changes caused by miR-381 antagomir (p < 0.05, Fig. 6F). Meanwhile, levels of TNF-α and IL-1β were also increased by miR-381 antagomir; the effects of which were blocked by DEX (p < 0.05, Fig. 6G). TUNEL staining showed that DEX could impede the increased apoptosis of neurons induced by POCD. POCD mice treated with miR-381 antagomir also showed increased cell apoptosis, but additional treatment with DEX did not increase cell apoptosis significantly more (p < 0.05, Fig. 6H). Meantime, the expression of miR-381 was decreased in hippocampus tissues of POCD mice but was elevated following DEX treatment. miR-381 antagomir reduced the expression of miR-381, which was restored by additional DEX treatment (p < 0.05, Fig. 6I). Data from Western blot analysis suggested an ascending trend in the protein expressions of EGR1, p53, p21, and γH2AX in hippocampus tissues of POCD mice, while mice treated with DEX exhibited reduced expression levels of the mentioned proteins. As expected, miR-381 antagomir could upregulate the protein expressions of EGR1, p53, p21, and γH2AX, which, however, was reversed by DEX (p < 0.05, Fig. 6J). Moreover, the fluorescence intensity of γH2AX was increased in neurons of POCD mice, which was diminished by DEX treatment. miR-381 antagomir increased the γH2AX intensity but treatment with DEX inhibited this increase (p < 0.05, Fig. 6K). Overall, these findings suggested that DEX could inhibit DNA damage, neuroinflammation, and neuron apoptosis through the regulation of the miR-381/EGR1/p53 pathway, hence alleviating POCD in vivo.
Discussion
Emerging literature suggests DEX may provide a protective effect on brain function injury [19, 20]. Our data demonstrated that DEX could potentially alleviate POCD in mice, in which DEX elevated miR-381 and attenuated EGR1-mediated p53 activation, thus relieving DNA damage, neuroinflammation, and neuron apoptosis and ultimately attenuating cognitive dysfunction.
Microglia normally reside in a quiescent state in the central nervous system. When activated, these cells exhibit diverse functions either beneficial or harmful depending on the situation [21]. DEX has been revealed to exert anti-inflammatory effects by upregulating the expression of miR-340 in lipopolysaccharide (LPS)-stimulated microglia BV-2 cells [22]. Meanwhile, in a rat spinal cord injury model, DEX can reduce the microglia-mediated neuroinflammation via upregulation of programmed cell death protein 1 [23]. Previous results have also documented the ability of DEX to improve outcomes of early-stage POCD in aged mice [24]. In addition, DEX administration results in a decline in hippocampal neuron apoptosis and attenuation in cognitive impairment in mice with Alzheimer’s disease by elevating the expression of miR-129 [25]. Moreover, POCD is well known as a common central nervous system complication after surgical anesthesia. The central neurotransmitter system includes a variety of excitatory neurotransmitters and inhibitory neurotransmitters. Published literature has shown that DEX can inhibit the potential of a single synapse and slow down spinal nerve root reflex in a concentration-dependent manner [26]. In addition, another study revealed that DEX could enhance the inhibitory postsynaptic transmission within the superficial dorsal horn without altering excitatory synaptic transmission or evoking direct postsynaptic membrane currents [27]. DEX has also been confirmed to selectively inhibit high-intensity excitatory postsynaptic potential [28]. The aforementioned findings from previous studies concur with the present results that DEX could enhance cognitive function and repress neuroinflammation and neuron apoptosis in aged mice.
Our study also provided evidence suggesting that DEX suppressed the POCD-associated DNA damage by downregulating p53 expression in mice. Treatment with DEX contributes to the disruption of p53 activation in mice suffering from acute kidney injury [29]. In addition, DEX has been demonstrated to alleviate POCD through regulating the expression of p53 in aged rats [30]. DNA damage is a consequence of radiation and chemotherapy and contributes to cellular apoptosis or senescence as outcomes of the DNA damage response (DDR), thus being a determinant for the aging process; in the presence of DNA damage, p53 was found to be stabilized by DDR signaling [31]. Inhibition of p53 has a potentially preventive effect against DNA damage-induced apoptosis in mouse primary cortical neurons [32]. However, how DEX regulates p53 still requires further explanation.
Another critical observation obtained from the present study was that EGR1 promoted the transcription of p53 and potentiated the apoptosis and DNA damage of neurons. Accumulating studies highlighted the positive correlation between EGR1 and p53 gene transcription [33, 34]. EGR1 is highly expressed e in the hippocampus tissues and neurons of rats following isoflurane anesthesia and conversely inhibited the expression of EGR1 and repressed hippocampal neuron apoptosis, thus improving the spatial learning and memory ability of rats [35]. p21 gene, a DNA damage-related protein [36], was found to be transactivated by EGR1 [37]. In this study, the luciferase reporter assay identified that miR-381 was bound to the 3ʹ-UTR of EGR1 mRNA and thus negatively regulated its expression. The expression of miR-381-3p was detected to be significantly lower in plasma samples of patients with Alzheimer’s disease than that in the control individuals [38]. Moreover, miR-381 is decreased in chronic constriction injury rats while its upregulation reduces neuroinflammation by downregulating the expressions of IL-6, IL-1β, and TNF-α [39]. By targeting Hes1, miR-381 promotes neural stem cell proliferation and drives their differentiation to neurons [40]. Meanwhile, miR-381 was discovered to be upregulated after DEX treatment [41], which is consistent with our findings. Furthermore, our data demonstrated that DEX could inhibit DNA damage, neuroinflammation, and neuron apoptosis through upregulation of miR-381 and inhibiting the EGR1/p53 pathway, therefore alleviating POCD. The regulatory roles of miR-381 on DNA damage still remain unknown, although miRNAs have been suggested to play a crucial role in the regulation of DNA damage response [42].
In summary, our study demonstrates the protective roles of DEX in the treatment against POCD. After the treatment, DEX attenuated DNA damage, neuroinflammation, neuron apoptosis, and cognitive dysfunction, all of which is associated with an increase of miR-381 expression and blockade of EGR1-mediated p53 activation (Fig. 7). These findings may provide evidence for the possible epigenetic effects of DEX on neuroprotection in POCD, as well as novel insights into the development of new therapies for the prevention of POCD. Further research is necessary to fully elucidate the pathogenesis of POCD and to apply these findings to targeted therapies in clinics.
A schematic representation of the potential molecular mechanisms of DEX in POCD. miR-381 was downregulated in mice rendered with POCD and targeted EGR1, which in turn promoted p53 transcription and induced neuron apoptosis and DNA damage. DEX alleviated the DNA damage caused by EGR1-mediated p53 activation by upregulating miR-381 and reduced neuroinflammation and neuron apoptosis, ultimately attenuating cognitive dysfunction in POCD
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Devinney MJ, Mathew JP, Berger M (2018) Postoperative delirium and postoperative cognitive dysfunction: two sides of the same coin? Anesthesiology 129(3):389–391. https://doi.org/10.1097/ALN.0000000000002338
Austin CA, O’Gorman T, Stern E, Emmett D, Sturmer T, Carson S, Busby-Whitehead J (2019) Association between postoperative delirium and long-term cognitive function after major nonemergent surgery. JAMA Surg 154(4):328–334. https://doi.org/10.1001/jamasurg.2018.5093
Hood R, Budd A, Sorond FA, Hogue CW (2018) Peri-operative neurological complications. Anaesthesia 73(Suppl 1):67–75. https://doi.org/10.1111/anae.14142
Needham MJ, Webb CE, Bryden DC (2017) Postoperative cognitive dysfunction and dementia: what we need to know and do. Br J Anaesth 119(suppl_1):i115–i125. https://doi.org/10.1093/bja/aex354
Huang C, Chu JM, Liu Y, Chang RC, Wong GT (2018) Varenicline reduces DNA damage, tau mislocalization and post surgical cognitive impairment in aged mice. Neuropharmacology 143:217–227. https://doi.org/10.1016/j.neuropharm.2018.09.044
Bao N, Tang B (2020) Organ-protective effects and the underlying mechanism of dexmedetomidine. Mediators Inflamm 2020:6136105. https://doi.org/10.1155/2020/6136105
Hu J, Vacas S, Feng X, Lutrin D, Uchida Y, Lai IK, Maze M (2018) Dexmedetomidine prevents cognitive decline by enhancing resolution of high mobility group box 1 protein-induced inflammation through a vagomimetic action in mice. Anesthesiology 128(5):921–931. https://doi.org/10.1097/ALN.0000000000002038
Xue Y, Xu T, Jiang W (2020) Dexmedetomidine protects PC12 cells from ropivacaine injury through miR-381/LRRC4 /SDF-1/CXCR4 signaling pathway. Regen Ther 14:322–329. https://doi.org/10.1016/j.reth.2020.03.001
Omahen DA (2011) MicroRNA and diseases of the nervous system. Neurosurgery 69(2):440–454. https://doi.org/10.1227/NEU.0b013e318215a3b3
Zhan LY, Lei SQ, Zhang BH, Li WL, Wang HX, Zhao B, Cui SS, Ding H et al (2018) Overexpression of miR-381 relieves neuropathic pain development via targeting HMGB1 and CXCR4. Biomed Pharmacother 107:818–823. https://doi.org/10.1016/j.biopha.2018.08.053
Piao JM, Wu W, Yang ZX, Li YZ, Luo Q, Yu JL (2018) MicroRNA-381 favors repair of nerve injury through regulation of the SDF-1/CXCR4 signaling pathway via LRRC4 in acute cerebral ischemia after cerebral lymphatic blockage. Cell Physiol Biochem 46(3):890–906. https://doi.org/10.1159/000488821
Duclot F, Kabbaj M (2017) The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front Behav Neurosci 11:35. https://doi.org/10.3389/fnbeh.2017.00035
Cong J, Wang C, Pu D, Liu J, Hu G, Gao C, Wu J (2014) Expression of early growth response 1 affects miR-106a/signal transducer and activator of transcription 3 regulating cognitive impairment in ovariectomized mice. Menopause 21(10):1143–1150. https://doi.org/10.1097/GME.0000000000000234
Ao H, Liu B, Li H, Lu L (2019) Egr1 mediates retinal vascular dysfunction in diabetes mellitus via promoting p53 transcription. J Cell Mol Med 23(5):3345–3356. https://doi.org/10.1111/jcmm.14225
Jazvinscak Jembrek M, Slade N, Hof PR, Simic G (2018) The interactions of p53 with tau and Ass as potential therapeutic targets for Alzheimer’s disease. Prog Neurobiol 168:104–127. https://doi.org/10.1016/j.pneurobio.2018.05.001
Qiu LL, Ji MH, Zhang H, Yang JJ, Sun XR, Tang H, Wang J, Liu WX et al (2016) NADPH oxidase 2-derived reactive oxygen species in the hippocampus might contribute to microglial activation in postoperative cognitive dysfunction in aged mice. Brain Behav Immun 51:109–118. https://doi.org/10.1016/j.bbi.2015.08.002
Chen Y, Sun JX, Chen WK, Wu GC, Wang YQ, Zhu KY, Wang J (2019) miR-124/VAMP3 is a novel therapeutic target for mitigation of surgical trauma-induced microglial activation. Signal Transduct Target Ther 4:27. https://doi.org/10.1038/s41392-019-0061-x
Bevins RA, Besheer J (2006) Object recognition in rats and mice: a one-trial non-matching-to-sample learning task to study ‘recognition memory.’ Nat Protoc 1(3):1306–1311. https://doi.org/10.1038/nprot.2006.205
Deiner S, Luo X, Lin HM, Sessler DI, Saager L, Sieber FE, Lee HB, Sano M et al (2017) Intraoperative infusion of dexmedetomidine for prevention of postoperative delirium and cognitive dysfunction in elderly patients undergoing major elective noncardiac surgery: a randomized clinical trial. JAMA Surg 152(8):e171505. https://doi.org/10.1001/jamasurg.2017.1505
Wang SL, Duan L, Xia B, Liu Z, Wang Y, Wang GM (2017) Dexmedetomidine preconditioning plays a neuroprotective role and suppresses TLR4/NF-kappaB pathways model of cerebral ischemia reperfusion. Biomed Pharmacother 93:1337–1342. https://doi.org/10.1016/j.biopha.2017.06.051
Boche D, Perry VH, Nicoll JA (2013) Review: activation patterns of microglia and their identification in the human brain. Neuropathol Appl Neurobiol 39(1):3–18. https://doi.org/10.1111/nan.12011
Bao Y, Zhu Y, He G, Ni H, Liu C, Ma L, Zhang L, Shi D (2019) Dexmedetomidine attenuates neuroinflammation in LPS-stimulated BV2 microglia cells through upregulation of miR-340. Drug Des Devel Ther 13:3465–3475. https://doi.org/10.2147/DDDT.S210511
He H, Zhou Y, Zhou Y, Zhuang J, He X, Wang S, Lin W (2018) Dexmedetomidine mitigates microglia-mediated neuroinflammation through upregulation of programmed cell death protein 1 in a rat spinal cord injury model. J Neurotrauma 35(21):2591–2603. https://doi.org/10.1089/neu.2017.5625
Qian XL, Zhang W, Liu MZ, Zhou YB, Zhang JM, Han L, Peng YM, Jiang JH et al (2015) Dexmedetomidine improves early postoperative cognitive dysfunction in aged mice. Eur J Pharmacol 746:206–212. https://doi.org/10.1016/j.ejphar.2014.11.017
Sun W, Zhao J, Li C (2020) Dexmedetomidine provides protection against hippocampal neuron apoptosis and cognitive impairment in mice with Alzheimer’s disease by mediating the miR-129/YAP1/JAG1 axis. Mol Neurobiol 57(12):5044–5055. https://doi.org/10.1007/s12035-020-02069-z
Kobayashi T, Otsuguro K, Yamaguchi S, Ito S (2015) Contribution of alpha2A-adrenoceptor subtype to effect of dexmedetomidine and xylazine on spinal synaptic transmission of mice. Eur J Pharmacol 761:321–329. https://doi.org/10.1016/j.ejphar.2015.06.020
Funai Y, Pickering AE, Uta D, Nishikawa K, Mori T, Asada A, Imoto K, Furue H (2014) Systemic dexmedetomidine augments inhibitory synaptic transmission in the superficial dorsal horn through activation of descending noradrenergic control: an in vivo patch-clamp analysis of analgesic mechanisms. Pain 155(3):617–628. https://doi.org/10.1016/j.pain.2013.12.018
Faber ES, Chambers JP, Evans RH (1998) Depression of NMDA receptor-mediated synaptic transmission by four alpha2 adrenoceptor agonists on the in vitro rat spinal cord preparation. Br J Pharmacol 124(3):507–512. https://doi.org/10.1038/sj.bjp.0701873
Liang H, Liu HZ, Wang HB, Zhong JY, Yang CX, Zhang B (2017) Dexmedetomidine protects against cisplatin-induced acute kidney injury in mice through regulating apoptosis and inflammation. Inflamm Res 66(5):399–411. https://doi.org/10.1007/s00011-017-1023-9
Cao C, Deng F, Hu Y (2020) Dexmedetomidine alleviates postoperative cognitive dysfunction through circular RNA in aged rats. 3 Biotech 10(4):176. https://doi.org/10.1007/s13205-020-2163-0
Ou HL, Schumacher B (2018) DNA damage responses and p53 in the aging process. Blood 131(5):488–495. https://doi.org/10.1182/blood-2017-07-746396
Brochier C, Dennis G, Rivieccio MA, McLaughlin K, Coppola G, Ratan RR, Langley B (2013) Specific acetylation of p53 by HDAC inhibition prevents DNA damage-induced apoptosis in neurons. J Neurosci 33(20):8621–8632. https://doi.org/10.1523/JNEUROSCI.5214-12.2013
Gong D, Zeng Z, Yi F, Wu J (2019) Inhibition of histone deacetylase 11 promotes human liver cancer cell apoptosis. Am J Transl Res 11(2):983–990
Liu S, Yao X, Zhang D, Sheng J, Wen X, Wang Q, Chen G, Li Z et al (2018) Analysis of transcription factor-related regulatory networks based on bioinformatics analysis and validation in hepatocellular carcinoma. Biomed Res Int 2018:1431396. https://doi.org/10.1155/2018/1431396
Yang W, Guo Q, Li J, Wang X, Pan B, Wang Y, Wu L, Yan J et al (2019) microRNA-124 attenuates isoflurane-induced neurological deficits in neonatal rats via binding to EGR1. J Cell Physiol 234(12):23017–23032. https://doi.org/10.1002/jcp.28862
Abella N, Brun S, Calvo M, Tapia O, Weber JD, Berciano MT, Lafarga M, Bachs O et al (2010) Nucleolar disruption ensures nuclear accumulation of p21 upon DNA damage. Traffic 11(6):743–755. https://doi.org/10.1111/j.1600-0854.2010.01063.x
Kim SJ, Kim JM, Shim SH, Chang HI (2014) Shikonin induces cell cycle arrest in human gastric cancer (AGS) by early growth response 1 (Egr1)-mediated p21 gene expression. J Ethnopharmacol 151(3):1064–1071. https://doi.org/10.1016/j.jep.2013.11.055
Zhou Q, Luo L, Wang X, Li X (2019) Relationship between single nucleotide polymorphisms in the 3’UTR of amyloid precursor protein and risk of Alzheimer’s disease and its mechanism. Biosci Rep 39(5):BSR20182485. https://doi.org/10.1042/BSR20182485
Xia LX, Ke C, Lu JM (2018) NEAT1 contributes to neuropathic pain development through targeting miR-381/HMGB1 axis in CCI rat models. J Cell Physiol 233(9):7103–7111. https://doi.org/10.1002/jcp.26526
Shi X, Yan C, Liu B, Yang C, Nie X, Wang X, Zheng J, Wang Y et al (2015) miR-381 regulates neural stem cell proliferation and differentiation via regulating Hes1 expression. PLoS ONE 10(10):e0138973. https://doi.org/10.1371/journal.pone.0138973
Zhang Y, Wang X, Liu Z, Yu L (2018) Dexmedetomidine attenuates lipopolysaccharide induced acute lung injury by targeting NLRP3 via miR-381. J Biochem Mol Toxicol 32(11):e22211. https://doi.org/10.1002/jbt.22211
Wan G, Mathur R, Hu X, Zhang X, Lu X (2011) miRNA response to DNA damage. Trends Biochem Sci 36(9):478–484. https://doi.org/10.1016/j.tibs.2011.06.002
Author information
Authors and Affiliations
Contributions
Yi-Liang Wang designed the study. Da-Sheng Cai was involved in data collection. Ying Zhang and Da-Sheng Cai performed the statistical analysis and preparation of figures. Yi-Liang Wang and Ying Zhang drafted the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Statement
This study was carried out after being approved by the ethics committee of the First Hospital of China Medical University and was performed according to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. Extensive efforts were made to ensure minimal suffering of the animals used in the study.
Conflicts of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, YL., Zhang, Y. & Cai, DS. Dexmedetomidine Ameliorates Postoperative Cognitive Dysfunction via the MicroRNA-381-Mediated EGR1/p53 Axis. Mol Neurobiol 58, 5052–5066 (2021). https://doi.org/10.1007/s12035-021-02417-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-021-02417-7