Abstract
Association between serum creatinine (sCr) and amyotrophic lateral sclerosis (ALS) has been reported in previous observational studies, but results are at risk of confounding bias and reverse causation. Therefore, whether such association is casual remains unclear. Herein, we performed a two-sample Mendelian randomization study to evaluate the causal relationship between sCr and ALS in both European and East Asian populations. Our analysis was conducted using summary statistics from genome-wide association studies with 358,072 individuals for sCr and 80,610 individuals for ALS in European population, and 142,097 individuals for sCr and 4,084 individuals for ALS in East Asian population. The inverse-variance weighted method was used to estimate the casual-effect of sCr on ALS in both populations, and other MR methods were also performed as sensitivity analyses. We found evidence that genetically predicted sCr was inversely associated with risk of ALS (OR, 0.92; 95% CI, 0.85–0.99; P = 0.028) in European population. However, there was no strong evidence for a causal relationship between sCr and ALS in East Asian population (OR, 0.92; 95% CI, 0.84–1.01; P = 0.084). This study provides evidence that sCr protects against ALS in European population but not in East Asian population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amyotrophic lateral sclerosis (ALS) is a neuro-degenerative disease that is characterized by progressive motor neuron degeneration, with death from respiratory failure within 2–3 years after disease onset [1]. The standardized incidence of ALS is 1–2 per 100,000 person-years [2]. The number of ALS patients will increase by ~ 70% in the following decades, resulting in enormous social and economic burden across the globe [3]. Therefore, it is important to identify casual risk factors of ALS.
To date, no agreed environmental risk factors for ALS have been identified. Recently, several observational studies have found that serum creatinine (sCr) is reduced in ALS patients compared to controls [4,5,6,7,8], suggesting that sCr may protects against ALS. However, the baseline sCr in these studies were measured after disease onset, and it is unclear whether the sCr in ALS patients has been reduced before disease onset. Therefore, it still remains unclear whether sCr can reduce the risk of ALS. Given that ALS is rare in the population, it is very difficult to conduct longitudinal studies for investigating the effect of sCr on ALS. In addition, most previous studies are observational, and they are easily biased by confounding factors. Therefore, whether the causal relationship between sCr and ALS is needed to further investigate.
Mendelian randomization (MR) is a new approach using genotypes as instrumental variables to estimate the causal effect of an exposure on an outcome [9, 10]. MR can reduce the bias from confounding factors and reverse causation that are often exist in observational studies, because the genotypes from parents to offspring are random allocated, and are unchanged through a lifetime [10]. To our knowledge, MR approach has not been used to investigate the causal relationship between sCr and ALS. Therefore, we performed this two-sample MR study to assess the causal relationship between sCr and ALS in both European and East Asian populations.
Materials and Methods
Data Sources and Instrument Selection
All relevant ethics approval and informed consent are from original genome-wide association studies (GWASs) [11,12,13,14].
For the European population, a total of 485 independent SNPs (r2 < 0.1) associated with (P < 5 × 10−8) sCr were selected from a GWAS of 358,072 European individuals in UK Biobank [11]. The mean (SD) sCr concentration of whole UK biobank participants is 72.4 (18.5) umol/L. In the East Asian population, 64 independent autosomal SNPs (r2 < 0.1) associated with sCr were selected from a GWAS in 142,097 Japanese individuals [12]. The mean (SD) sCr concentration of Japanese participants is 0.77 (0.22) mg/dL. The proportion of variance explained by each SNP was evaluated using R2 [15], and the F statistic was calculated to assess the instrument strength. The F statistic above 10 is considered as an indicator of strong instruments [16].
In the European population, the summary statistics for ALS were obtained from a GWAS with 80,610 European individuals (20,806 cases and 59,804 controls) [13]. For the East Asian population, we obtained the summary statistics of ALS from an East Asian GWAS in 4084 Chinese individuals (1234 cases and 2850 controls) [14].
Statistical Analysis
All MR analyses were conducted using R version 4.0.2, “TwoSampleMR” [17], and “MR-PRESSO” packages [18].
The main analyses were performed with the inverse-variance weighted method, which can provide an unbiased estimate if the MR assumptions are met and the horizontal pleiotropy is balanced. Other MR methods were also performed as sensitivity analyses, including the following: (1) Mendelian randomization robust adjusted profile score (MR-RAPS) method, which accounts for weak instruments [19], (2) weighted median method, which can provide a consistent estimate when at least 50% of instrumental variables are valid [20], and (3) MR-Egger method, which can adjust for bias from directional pleiotropy, at the cost of reduced statistical power [21], The MR-Egger is based on the “NO measurement error assumption (NOME),” and this assumption was evaluated by the regression dilution I2GX statistic [22]. If I2GX was below 90% (indicating the violation of NOME assumption), MR-Egger with simulation extrapolation correction (SIMEX) was used [22]. The Mendelian Randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO) was also applied to identify potential pleiotropic outlier (P < 0.1) [18].
The pleiotropy in MR estimates were assessed using three approaches: (1) The Corchan’s Q, which can assess the heterogeneity across all individual SNPs in IVW estimates. A significant Corchan’s Q test (P < 0.05) suggests the presence of horizontal pleiotropy; (2) MR-Egger intercept, which provides an indicator of directional pleiotropy. A significant MR-Egger intercept test (P < 0.05) suggests the presence of directional pleiotropy, and MR-Egger estimates were used to validate the results from IVW estimates; and (3) Leave-one-out (LOO) analysis, in which IVW estimates were re-calculated removing each SNP in turn, to evaluate whether a single SNP drove the association. We evaluated whether each SNP is associated with potential risk factors for ALS (P < 5 × 10−8): smoking (The GWAS & Sequencing Consortium of Alcohol and Nicotine use) [23, 24]; drinking (The GWAS & Sequencing Consortium of Alcohol and Nicotine use) [23, 25]; and low-density lipoprotein (Global lipids Genetics Consortium) [26, 27]. These potential pleiotropic SNPs were removed, and the IVW estimates were re-calculated to rule out potential pleiotropic effects.
Finally, MR Steiger test was conducted to test whether the SNPs for sCr explained more variances in sCr than ALS (the opposite indicate reverse causation) [28]. Power calculations were performed with the mRnd online tool at https://shiny.cnsgenomics.com/mRnd/ [29]. Odds ratios (ORs) with 95% confidence intervals (95% CIs) were scaled per SD unite increase in sCr concentrations. A P value < 0.05 was considered as statistically significant.
Results
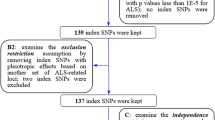
In the European population, a potential SNP outlier rs34674752 identified by MR-PRESSO was excluded in the analysis. Therefore, a total of 484 independent SNPs associated with sCr, which all were available in the ALS dataset, were included in the analysis. These 484 SNPs explained 8.2% phenotypic variation of sCr concentration (Supplementary Table 1). For these SNPs, the median F statistic is 43 (ranging from 30 to 749) with an overall F statistic of 66, suggesting that weak instruments was unlikely to bias the estimates of casual effects. In the East Asian population, MR-PRESSO did not identify any outliers. A SNP rs533988 for sCr was not available in the ALS dataset, and no proxy SNP (r2 > 0.8) can be used to replace it. Hence, a total of 63 SNPs were included in the primary analysis. These 63 SNPs explained 2.4% phenotypic variation of sCr concentration (Supplementary Table 2). For these SNPs, the median F statistic is 42 (ranging from 31 to 239) with an overall F statistic of 55, indicating that weak instruments was also unlikely to bias the estimates of casual effects in the East Asian population.
sCr and ALS in the European Population
In the European population, we found evidence that genetically predicted 1-SD unit increase in sCr concentration was inversely associated with risk of ALS (IVW OR, 0.92; 95% CI, 0.85–0.99; P = 0.028; Figs. 1 and 2). Other MR estimates yield similar effect estimates (Figs. 1 and 2), although with wider CI due to lower statistical power. The I2GX for MR-Egger was 98.4%, indicating that SIMEX correction was not required to apply. The Corchan’s Q statistic suggested substantial heterogeneity (Corchan’s Q = 676.19, P = 1.35×10–8), indicating the presence of horizontal pleiotropy. However, the MR-Egger intercept test (P = 0.99) indicated no directional pleiotropy, suggesting that the horizontal pleiotropy was unlikely to bias the IVW estimate. Furthermore, the LOO analysis and forest plot indicated that no single SNP drove the association (Supplementary Figure 1-2). After removing 5 potential pleiotropic SNPs (Supplementary Table 3), the IVW estimate was not significantly changed (IVW OR, 0.924; 95% CI, 0.857–0.997; P = 0.042). The MR Steiger test for directionality indicated that using ALS as the outcome was the correct causal direction for sCr (P < 0.001).
sCr and ALS in the East Asian Population
We further evaluated the casual association between sCr and ALS in the East Asian population. The casual-effect estimate of sCr on ALS is not statistically significant in East Asian population (OR, 0.92; 95% CI, 0.84–1.01; P = 0.084; Figs. 1 and 2), although it was similar with European population in direction and magnitude, which may be due to limited power. Similar effects estimates were observed in other MR estimates (Figs. 1 and 2). The LOO analysis and forest plot indicated that no single SNP can influence the casual-effect estimate (Supplementary Figure 3-4). Furthermore, there was no evidence of substantial heterogeneity (Corchan’s Q = 74.52, P = 0.132) and directional pleiotropy (MR-Egger intercept, P = 0.780).
Discussion
In this study, we investigated the causal relationship between sCr and ALS in both European and East Asian populations using a two-sample MR approach. Our findings showed that genetically predicted sCr was inversely associated with the risk of ALS in European population but not in East Asian population.
Previous observational studies have shown that sCr are decreased in ALS patients compared to controls in European population [4, 5]. Our MR study found similar inversely association between sCr and ALS in European population. These findings suggest that sCr can protect against the risk of ALS in European population. In East Asian population, some observational studies also have reported ALS cases have lower sCr compare to the controls [7, 8]. However, we did not find a statistically significant association between sCr and ALS (OR, 0.92; 95% CI, 0.84–1.01; P = 0.084), although it was similar with European population in direction and magnitude, suggesting that it is may be due to limited power. Given that the type 1 error is 5%, the sample size is 4084, the proportion of variance explained by SNPs is 2.4%, and the true casual OR of sCr on ALS is 0.90 (or equivalently 1.1), the statistical power of the two-sample MR to detect such causal effect is only 8% in the East Asian population. Therefore, whether a causal relationship between sCr and ALS in East Asian population is still needed to further investigate in larger sample.
The exact underlying mechanism linking sCr with ALS still remains unclear. Two potential mechanisms involve oxidative stress and mitochondrial dysfunction. Oxidative stress and mitochondrial dysfunction have been proved to contribute to the pathogenesis of ALS [30,31,32,33,34,35]. sCr, the end product of creatine and creatine phosphate, can reflect the change of creatine pool, which plays a key role in mitochondrial function and has neuro-protective function in vivo and in vitro [36]. Several in vivo [37, 38] and in vitro [39,40,41] studies have demonstrated that creatine has antioxidant capacity. Further studies are needed to elucidate the exact mechanism linking sCr with ALS.
Our study has some strengths. First, MR study is less prone to reverse causation and confounding, providing relatively robust estimate of casual relationship between sCr and ALS. Second, we used large-scale GWASs in the European population. Our study has several limitations. First, a major limitation of MR study is pleiotropy, which may bias the IVW estimates. Hence, several approaches were performed to assess the pleiotropy. For example, MR-RAPS, weighted median, and MR-Egger estimates that are robust to pleiotropy were calculated to compare with IVW estimate. In addition, we performed the LOO analyses to evaluate whether a single SNP drove the result. All these analyses have similar results, indicating that pleiotropy is unlikely to bias the IVW estimate. Second, another limitation is that we cannot assess the correlation between sCr and other parameters of ALS (e.g., clinical phenotype, genetic background, cognitive impairment, survival, sit of onset, and age), because these parameters are not available. More studies are needed to investigate this correlation. Third, the two-sample MR assumes a linear association between exposure and outcome. Hence, we did not assess a potential nonlinear association between sCr and ALS. Fourth, the statistical power was low in the analyses of ALS in East Asian population (8%). Hence, we cannot rule out the causal relationship between sCr and ALS in East Asian population. Finally, we cannot assess sex-specific effects of sCr on ALS, because no sex-specific GWASs are available.
A previous population-based study has demonstrated that sCr may serve as a prognostic biomarker for ALS [42]. This study assessed the correlation of sCr evaluated at diagnosis with ALS outcome, and found that patients with lower sCr are significantly associated with worse clinical function at diagnosis of ALS and shorter survival [42]. In line with this study, our study found that patients with higher sCr was associated with lower risk of ALS in European population. These findings suggested that sCr is possibly involved in the pathogenesis of ALS and may play a protective role in the development of ALS. Therefore, sCr may serve as a reliable and easily accessible blood markers of the risk and outcome for ALS. Large multi-center prospective studies are still needed to investigate the association of sCr with the risk and outcome of ALS. Besides, further functional studies are warranted to investigate the mechanisms linking the association of sCr with ALS.
In conclusions, using a two-sample MR approach, our study provides evidence to support that sCr protects against the risk of ALS in European but not in East Asian population.
Data Availability
The data used to perform the analyses in this study were obtained from public genome-wide association studies summary statistics.
References
Brown RH, Al-Chalabi A (2017) Amyotrophic lateral sclerosis. New England Journal of Medicine 377(2):162–172. https://doi.org/10.1056/NEJMra1603471
Chiò A, Logroscino G, Traynor BJ, Collins J, Simeone JC, Goldstein LA, White LA (2013) Global epidemiology of amyotrophic lateral sclerosis: a systematic review of the published literature. Neuroepidemiology 41(2):118–130. https://doi.org/10.1159/000351153
Arthur KC, Calvo A, Price TR, Geiger JT, Chiò A, Traynor BJ (2016) Projected increase in amyotrophic lateral sclerosis from 2015 to 2040. Nat Commun 7:12408. https://doi.org/10.1038/ncomms12408
Zoccolella S, Simone IL, Capozzo R, Tortelli R, Leo A, D’Errico E, Logroscino G (2011) An exploratory study of serum urate levels in patients with amyotrophic lateral sclerosis. J Neurol 258(2):238–243. https://doi.org/10.1007/s00415-010-5735-9
Lawton KA, Cudkowicz ME, Brown MV, Alexander D, Caffrey R, Wulff JE, Bowser R, Lawson R et al (2012) Biochemical alterations associated with ALS. Amyotroph Lateral Scler 13(1):110–118. https://doi.org/10.3109/17482968.2011.619197
Lawton KA, Brown MV, Alexander D, Li Z, Wulff JE, Lawson R, Jaffa M, Milburn MV et al (2014) Plasma metabolomic biomarker panel to distinguish patients with amyotrophic lateral sclerosis from disease mimics. Amyotroph Lateral Scler Frontotemporal Degener 15(5-6):362–370. https://doi.org/10.3109/21678421.2014.908311
Chen X, Guo X, Huang R, Zheng Z, Chen Y, Shang H-F (2014) An exploratory study of serum creatinine levels in patients with amyotrophic lateral sclerosis. Neurol Sci 35(10):1591–1597. https://doi.org/10.1007/s10072-014-1807-4
Chen X, Wei Q-Q, Chen Y, Cao B, Ou R, Hou Y, Yuan X, Zhang L et al (2019) Clinical disease stage related changes of serological factors in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener 20(1-2):53–60. https://doi.org/10.1080/21678421.2018.1550516
Smith GD, Ebrahim S (2003) ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease. Int J Epidemiol 32(1)
Burgess S, Thompson SG (2015) Mendelian randomization: methods for using genetic variants in causal estimation. Chapman & Hall/CRC Press, London
Sinnott-Armstrong N, Tanigawa Y, Amar D, Mars NJ, Aguirre M, Venkataraman GR, Wainberg M, Ollila HM, Pirruccello JP, Qian J, Shcherbina A, Rodriguez F, Assimes TL, Agarwala V, Tibshirani R, Hastie T, Ripatti S, Pritchard JK, Daly MJ, Rivas MA (2019) Genetics of 38 blood and urine biomarkers in the UK Biobank. bioRxiv:660506. doi:https://doi.org/10.1101/660506
Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, Iwata N, Ikegawa S et al (2018) Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet 50(3):390–400. https://doi.org/10.1038/s41588-018-0047-6
Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R, Dominov JA, Kenna BJ et al (2018) Genome-wide analyses identify KIF5A as a novel ALS gene. Neuron 97(6):1268–1283.e6. https://doi.org/10.1016/j.neuron.2018.02.027
Benyamin B, He J, Zhao Q, Gratten J, Garton F, Leo PJ, Liu Z, Mangelsdorf M et al (2017) Cross-ethnic meta-analysis identifies association of the GPX3-TNIP1 locus with amyotrophic lateral sclerosis. Nat Commun 8(1):611. https://doi.org/10.1038/s41467-017-00471-1
Shim H, Chasman DI, Smith JD, Mora S, Ridker PM, Nickerson DA, Krauss RM, Stephens M (2015) A multivariate genome-wide association analysis of 10 LDL subfractions, and their response to statin treatment, in 1868 Caucasians. PLoS ONE 10(4):e0120758. https://doi.org/10.1371/journal.pone.0120758
Pierce BL, Ahsan H, Vanderweele TJ (2011) Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol 40(3):740–752. https://doi.org/10.1093/ije/dyq151
Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S et al (2018) The MR-Base platform supports systematic causal inference across the human phenome. Elife 7. https://doi.org/10.7554/eLife.34408
Verbanck M, Chen C-Y, Neale B, Do R (2018) Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 50(5):693–698. https://doi.org/10.1038/s41588-018-0099-7
Zhao Q, Wang J, Hemani G, Bowden J, Small DS (2020) Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann Statist 48(3):1742–1769. https://doi.org/10.1214/19-AOS1866
Bowden J, Davey Smith G, Haycock PC, Burgess S (2016) Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 40(4):304–314. https://doi.org/10.1002/gepi.21965
Bowden J, Davey Smith G, Burgess S (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44(2):512–525. https://doi.org/10.1093/ije/dyv080
Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR (2016) Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol 45(6):1961–1974. https://doi.org/10.1093/ije/dyw220
Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, Datta G, Davila-Velderrain J et al (2019) Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 51(2):237–244. https://doi.org/10.1038/s41588-018-0307-5
Zhan Y, Fang F (2019) Smoking and amyotrophic lateral sclerosis: a Mendelian randomization study. Ann Neurol 85(4):482–484. https://doi.org/10.1002/ana.25443
Yu X, Wang T, Chen Y, Shen Z, Gao Y, Xiao L, Zheng J, Zeng P (2020) Alcohol drinking and amyotrophic lateral sclerosis: an instrumental variable causal inference. Ann Neurol 88(1):195–198. https://doi.org/10.1002/ana.25721
Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J et al (2013) Discovery and refinement of loci associated with lipid levels. Nat Genet 45(11):1274–1283. https://doi.org/10.1038/ng.2797
Zeng P, Zhou X (2019) Causal effects of blood lipids on amyotrophic lateral sclerosis: a Mendelian randomization study. Hum Mol Genet 28(4):688–697. https://doi.org/10.1093/hmg/ddy384
Hemani G, Tilling K, Davey Smith G (2017) Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet 13(11):e1007081. https://doi.org/10.1371/journal.pgen.1007081
Brion M-JA, Shakhbazov K, Visscher PM (2013) Calculating statistical power in Mendelian randomization studies. Int J Epidemiol 42(5):1497–1501. https://doi.org/10.1093/ije/dyt179
Barber SC, Shaw PJ (2010) Oxidative stress in ALS: key role in motor neuron injury and therapeutic target. Free Radic Biol Med 48(5):629–641. https://doi.org/10.1016/j.freeradbiomed.2009.11.018
Bond L, Bernhardt K, Madria P, Sorrentino K, Scelsi H, Mitchell CS (2018) A metadata analysis of oxidative stress etiology in preclinical amyotrophic lateral sclerosis: benefits of antioxidant therapy. Front Neurosci 12:10. https://doi.org/10.3389/fnins.2018.00010
Yusuf M, Khan M, Robaian MA, Khan RA (2018) Biomechanistic insights into the roles of oxidative stress in generating complex neurological disorders. Biol Chem 399(4):305–319. https://doi.org/10.1515/hsz-2017-0250
Cozzolino M, Carrì MT (2012) Mitochondrial dysfunction in ALS. Prog Neurobiol 97(2):54–66. https://doi.org/10.1016/j.pneurobio.2011.06.003
Joshi AU, Mochly-Rosen D (2018) Mortal engines: Mitochondrial bioenergetics and dysfunction in neurodegenerative diseases. Pharmacol Res 138:2–15. https://doi.org/10.1016/j.phrs.2018.08.010
Calió ML, Henriques E, Siena A, Bertoncini CRA, Gil-Mohapel J, Rosenstock TR (2020) Mitochondrial dysfunction, neurogenesis, and epigenetics: putative implications for amyotrophic lateral sclerosis neurodegeneration and treatment. Front Neurosci 14:679. https://doi.org/10.3389/fnins.2020.00679
Beal MF (2011) Neuroprotective effects of creatine. Amino Acids 40(5):1305–1313. https://doi.org/10.1007/s00726-011-0851-0
Deminice R, Portari GV, Vannucchi H, Jordao AA (2009) Effects of creatine supplementation on homocysteine levels and lipid peroxidation in rats. Br J Nutr 102(1):110–116. https://doi.org/10.1017/S0007114508162985
Guimarães-Ferreira L, Pinheiro CHJ, Gerlinger-Romero F, Vitzel KF, Nachbar RT, Curi R, Nunes MT (2012) Short-term creatine supplementation decreases reactive oxygen species content with no changes in expression and activity of antioxidant enzymes in skeletal muscle. Eur J Appl Physiol 112(11):3905–3911. https://doi.org/10.1007/s00421-012-2378-9
Lenz H, Schmidt M, Welge V, Schlattner U, Wallimann T, Elsässer H-P, Wittern K-P, Wenck H et al (2005) The creatine kinase system in human skin: protective effects of creatine against oxidative and UV damage in vitro and in vivo. J Invest Dermatol 124(2):443–452
Fimognari C, Sestili P, Lenzi M, Cantelli-Forti G, Hrelia P (2009) Protective effect of creatine against RNA damage. Mutat Res 670(1-2):59–67. https://doi.org/10.1016/j.mrfmmm.2009.07.005
Sestili P, Martinelli C, Bravi G, Piccoli G, Curci R, Battistelli M, Falcieri E, Agostini D et al (2006) Creatine supplementation affords cytoprotection in oxidatively injured cultured mammalian cells via direct antioxidant activity. Free Radic Biol Med 40(5):837–849
Chiò A, Calvo A, Bovio G, Canosa A, Bertuzzo D, Galmozzi F, Cugnasco P, Clerico M et al (2014) Amyotrophic lateral sclerosis outcome measures and the role of albumin and creatinine: a population-based study. JAMA Neurol 71(9):1134–1142. https://doi.org/10.1001/jamaneurol.2014.1129
Acknowledgements
We thank all the GWAS consortiums for providing summary statistics. In addition, Mengmeng Wang wants to thank, in particular, the support from Dandan Liu over the years. Will you marry me?
Funding
This study was supported by the Science and Technology Bureau of Changzhou (No. CE20195042), Major Science and Technology Projects of Changzhou Health Commission (No. ZD201904), and Applied Basic Research Project of Changzhou Science and Technology Bureau (No. CJ20190085).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics Approval
All relevant ethics approval are from original genome-wide association studies.
Consent to Participate
This study only used publicly available summary statistics from published genome-wide association studies. No individual-level data were involved, and no additional informed consent are needed in this study.
Consent for Publication
No individual-level data were involved, and no consent for publication is needed for this study.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, M., Liu, D., Zhang, Z. et al. Serum Creatinine Protects Against Amyotrophic Lateral Sclerosis: a Mendelian Randomization Study. Mol Neurobiol 58, 2910–2915 (2021). https://doi.org/10.1007/s12035-021-02309-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-021-02309-w