Abstract
SET is elevated and mislocalized in the neuronal cytoplasm in brains of Alzheimer’s disease (AD) and Down syndrome (DS) patients. Cytoplasm SET leads to inhibition of protein phosphatase 2A and is involved in the tau pathology. However, the regulation of SET gene expression remains elusive. In the present study, we cloned a 1399-bp segment of the 5′ flanking region of the human SET gene and identified that the transcription start site (TSS) of SET transcript 1 is located at 123 bp upstream of the translation start site ATG in exon 1. Sequence analysis reveals several putative regulatory elements including NFkB, Sp1, and HSE. Luciferase assay and electrophoretic mobility shift assay (EMSA) identified a functional cis-acting NFkB-responsive element in the SET gene promoter. Overexpression and activation of NFkB upregulate transcription of SET isoform 1 but not isoform 2, indicating that the expression of these two isoforms is differentially regulated. The results demonstrate that NFkB plays an important role in regulation of the human SET gene expression. Our findings suggest that oxidative stress and inflammatory responses could result in abnormal SET gene expression, contributing to the tauopathy in AD pathogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disease that causes progressive memory loss and cognitive impairment. The key hallmark pathologies of AD are the extracellular neuritic plaques and intracellular neurofibrillary tangles (NFTs). NFTs mainly consist of abnormally hyperphosphorylated tau and neritic plaques are formed by amyloid β protein (Aβ) deposition.
SET gene, firstly identified as a fusion gene to oncogene CAN of the patient with acute undifferentiated leukemia, is located on chromosome 9q34 [1]. It contains 11 exons and 10 introns. Four isoforms differed in the first exon and identical in the last seven exons are generated from the SET gene through alternative splicing. SET gene has a tissue-specific expression pattern. Isoform 1, known as template-activating factor-I α (TAF-Iα), is absent from some early-stage hemopoietic cell lines. However, isoform 2 (TAF-Iβ) is ubiquitously present among different tissues and remains a relatively constant expression level [2]. These two isoforms consist of 290 and 277 amino acids, respectively. They have common structure with a blocked N-terminal region, an earmuff domain, and a long acidic tail in the C-terminal region. Both isoforms predominantly express in the nuclei and the cytosol and are associated with the endoplasmic reticulum [2–4]. The difference in the N-terminal region of isoforms 1 and 2 is responsible for their dimerization [5].
SET protein has multiple functions. The long acidic region is essential for the TAF-I activity involved in the cell-free adenovirus (Ad) core replication system and histone chaperone activity [6, 7]. As a subunit of inhibitor of acetyltransferases (INHAT), it helps to regulate both histone acetylation and transcription [8, 9]. Furthermore, SET is also identified as inhibitor 2 of protein phosphatase 2A, a family of major serine/threonine phosphatases involved in regulating cell proliferation and differentiation [10]. Thus, the crucial role is implied in renal development and the early ontogenesis of the nervous system [11, 12]. SET itself can work as a DNA binding protein and transcriptional activator of cytochrome P450c17, a key enzyme for the synthesis of dehydroepiandrosterone (DHEA) in the brain. Activation of P450c17 transcription leading an increased DHEA production could be an important signal for modulating neurotransmission to trigger formation of neuronal circuits [12]. SET can also specifically bind Jcasp, a pro-apoptotic domain of the amyloid β precursor protein (APP), leading to neuronal apoptosis [13]. Moreover, recent studies revealed that the abnormal localization of SET promotes abnormal tau hyperphosphorylation by affecting PP2A signaling [14–18]. Our recent report also found that overexpression beta-site APP cleaving enzyme 1 (BACE1) significantly upregulated the level of SET, resulting in neurodegeneration in Alzheimer-associated dementia in Down syndrome (DS) patients [19]. These studies strongly demonstrate that SET plays a critical role in AD pathogenesis, and abnormal regulation of SET may contribute to the development of AD. However, the regulatory mechanism of SET gene expression remains elusive.
In this paper, we cloned the promoter of SET and identified its transcription regulation. We mapped that the transcription start site (TSS) of SET transcript 1 and identified the NFkB cis-acting element in the SET promoter region as a positive regulator for SET gene transcription.
Materials and Methods
Plasmids
The 5′ flanking region of the human SET gene exon 1 was firstly amplified by PCR from the genome DNA extracted from human embryonic kidney 293 (HEK293) cells. Nine fragments covering the 5′ flanking region of the SET from -1276 bp upstream to +123 bp downstream of the transcription start site at +1 (cytosine) were amplified by PCR and inserted upstream of the luciferase reporter gene (Luc) in the pGL3-basic expression vector (Promega). Primers were designed to include restriction enzyme digestion sequence at the 5′-end that is the same as the cloning sites of pGL3-basic. A series of deletion mutations (pSET-A, pSET-B, pSET-C, pSET-D, pSET-E, pSET-F, pSET-G, pSET-H, pSET-I) of SET promoter were constructed using primers listed as the following: -1276fXhoI: 5′-ccgacgcgtccaaaccctgtgcctact, -680fXhoI: 5′-ccgacgcgtcaagcgattctcctgcctc, -483fXhoI: 5′-ccgacgcgtttttaaagcaggcatcta, -399fXhoI: 5′-ccgacgcgtaagggaggtcaggaagga, -197fXhoI: 5′-ccgacgcgttcacgtgaaaccaggagg, +123rMluI: 5′-ccgctcgaggctgttagggaagtccca, +108rMluI: 5′-ccgctcgagccagaaccagaccacgag, +40rMluI: 5′-ccgctcgagcagcggcttaaatcgca, -48rMluI: 5′-ccgctcgagagcctcatcccacccaccct, -136rMluI: 5′-ccgctcgagggggcaagttcttccttg. A series of substitution mutations of pSET-C nuclear factor binding sites were constructed according to the protocol of KOD-Plus-Mutagenesis Kit (TOYOBO) with primers listed as the following: fMA: 5′-gcgaaaccaaatccaaaaggtgtggt, rMA: 5′-taactccttcctgacctcccttgtc, fMB: 5′-aaaaatttactgaggaatgttccaagctta, rMB: 5′-tttttactgcccggtttcgctaactccttc, fMC: 5′-aaaaaagcttaggtgaggcaacagcacgtg, rMC: 5′-gggggctcagtaaaccacacccccactgcc, fMD: 5′-tgaggcaaggctgaaacgttaaatc, rMD: 5′-cctaagcttggaacattcctcagta, fME: 5′-aaaaaaaagccaagagtgcccctgcagagcga, rME: 5′-ttttttcaagttcttccttgaggagaggat, fMF: 5′-aaaaactccagtgcagatttaagccgctggca, rMF: 5′-ttttttggaagggatatttacatgacagtggga, fMG: 5′-aaaaaactcgtggtctggttctgggacttcccta, rMG: 5′-tttttgtccggaagcaggctgaacactgaga, fMH: 5′-aaaaataacagcctcgagatctgcgatcta, rMH: 5′-tttttagaaccagaccacgagtctcctcgcccgt. Plasmid pSET-C was used as a template.
5′ RACE
Following the manufacturer’s instructions (FirstChoice® RLM-RACE Kit, Invitrogen), 10 μg total RNA extracted from HEK293 cells with the TRI reagent (Sigma) was used to generate the 5′ RACE complementary DNA (cDNA). With the cDNA as a template, two nested PCR reactions were carried out to generate specific 5′ RACE products. Forward primers are adapter primers provided in the kit. Specifically designed reverse primers are 5′-ggtttcttcttttgaggcgggagt (outer primer) and 5′-ccggaattcgaagtcccagaaccagaccacgagt (inner primer). After the inner PCR products were generated, they were digested and inserted into the pcDNA4/myc-HisA vector using the BamHI and EcoRI sites. The generated plasmids were sequenced to map the TSS.
Cell Culture, Transfection, and Luciferase Assays
HEK293 cells and SH-SY5Y cells were cultured in complete Dulbecco’s modified Eagle’s medium (DMEM, Gibco) containing 10 % fetal bovine serum (FBS, Gibco). All cells were maintained at 37 °C in an incubator containing 5 % CO2. HEK293 cells were transfected with 300 ng plasmid DNA per well of 48-well plate for luciferase assay, 10 μg plasmid DNA of 10-cm plate for nuclear extraction, and 2 μg plasmid DNA per well of 6-well plate for RNA extraction. All transfections were carried out with Lipofectamine™ 2000 transfection reagent (Invitrogen). The Renilla luciferase vector pRluc was cotransfected as an internal control to normalize the transfection efficiency in the luciferase assays. Cells were harvested at 24 h after transfection and lysed with 100 μl passive lysis buffer (Promega) per well. Firefly luciferase activities and Renilla luciferase activities were measured sequentially by the Dual-Luciferase Reporter Assay System (Promega). The firefly luciferase activity was normalized according to Renilla luciferase activity and expressed as relative luciferase units to reflect the promoter activity.
Electrophoretic Mobility Shift Assay
Electrophoretic mobility shift assay (EMSA) was performed as previously described [20]. HEK293 cells were transfected with the pMTF-P65 expression vector for 24 h to obtain NFkB-enriched nuclear extract [21]. Nuclear extraction was collected using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermo Scientific) according the manufacturer’s instructions. Two oligonucleotides probes (wild-type NFkB consensus-forward and SET 3XNFkB-forward) were labeled with IR700 dye (Bioneer Corporation) and annealed with corresponding anti-sense oligos to generate double-stranded probes at a final concentration of 0.01 pmol/μl. For competition experiments, 2 μl of nuclear extract was incubated with the 0.01 pmol of labeled probe and 100× (1 pmol) unlabeled competition probe for 20 min at room temperature. For supershifting assay, monoclonal anti-p65 (Cell Signaling) was added. The reaction mixtures were separated on a 4 % Tris-glycine-EDTA gel for 70 min at 70 V in darkness. The gel was scanned using the LI-COR Odyssey (LI-COR Biosciences) at a wavelength of 700 nm. The sequences of the oligonucleotides were as follows: wild-type NFkB consensus oligonucleotides (forward: agttgaggggactttcccaggc, reverse: gcctgggaaagtcccctcaact), mutant NFkB consensus oligonucleotides (forward: agttgaggccactttcccaggc, reverse: gcctgggaaagtggcctcaact), SET 3XNFkB oligonucleotides (forward: gttctgggacttccctactgggacttccctactgggacttccctaaca, reverse: tgttagggaagtcccagtagggaagtcccagtagggaagtcccagaac), SET NFkB oligonucleotides (forward: gttctgggacttccctaaca, reverse: tgttagggaagtcccagaac), mutant SET NFkB oligonucleotides (forward: gttctgccacttccctaaca, reverse: tgttagggaagtggcagaac).
Quantitative RT-PCR
Cells transfected with empty vector (pMTF or pCGN) or nuclear factor plasmids (pMTF-P65 or pCGN-Sp1) were harvested 24 h after the transfection, and total RNA was extracted using TRI reagent (Sigma). ThermoScript™ RT-PCR system kit (Invitrogen) was used to synthesize the first-strand cDNA using 1 μg of total RNA as a template. The human SET isoform 1 mRNA-specific primers (forward: 5′-gactcgtggtctggttctg, reverse: 5′-aatcgcttcttgctgttct) were used to amplify a 170 bp fragment. The human SET isoform 2 mRNA specific primers (forward: 5′-ggcggccaaagtcagtaaa, reverse: 5′-caatcgcttcttgctgttcttt) were used to amplify an 86 bp fragment. GAPDH gene primers (forward: 5′-aggtccaccactgacacgtt, reverse: 5′-gcctcaagatcatcagcaat) were used to amplify a 307-bp fragment as an internal control. Quantitative real-time PCR was performed with SYBR® Premix Ex Taq™ II (TaKaRa) and the reaction included one initial denaturation step at 94 °C for 3 min, then 40 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s (Bio-Rad CFX96). The level of cDNA was calculated based on standard curve and data were normalized by the level of GAPDH.
Statistical Analysis
Three or more independent experiments were performed. All results are presented as mean ± SEM and were analyzed by ANOVA or two-tailed Student’s t test. p < 0.05 was considered as statistically significant.
Results
Cloning the Human SET Promoter and Mapping Its Transcription Initiation Site
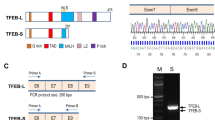
The human SET gene spans a region of 12,964 bp on chromosome 9q34. The human SET gene has four transcript variants which are distinct in the first exon and share the identical exons 5 to 11 (Fig. 1a). The human SET gene transcript 1 (NCBI Reference Sequence: NM_001122821) is 2863 bp long and composed of eight exons. Human genomic DNA samples were extracted from HEK293 cells, and a 1399-bp 5′ flanking region of the SET exon 1 was amplified and cloned. The DNA fragment was sequenced (Fig. 1b). To identify the transcription initiation site of the human SET gene, 5′ RACE assay was performed. The PCR product amplified with the outer primer pairs is approximately 260 bp in length (Fig. 1c). The DNA fragment amplified by the inner primers and inserted in vector was sequenced. DNA sequencing mapped the transcription start site at 123 bp upstream of the translation start site ATG (Fig. 1d). This transcription start site is designated as +1 and begins with cytosine. Further sequence analysis and a computer-based transcription factor binding site search using TRANSFAC, TF2 SEARCH, and Promoter Scan revealed that the human SET gene promoter lacks typical TATA and CAAT boxes, but contains several putative regulatory elements, such as HSE, Sp1, and NFkB (Fig. 1b).
Sequence features of the human SET gene promoter. a The genomic organization of human SET gene on chromosome 9. E represents exon. SET gene consists of 11 exons. ATG is the translation start codon and TAA is the stop codon. b The nucleotide sequence of the human SET gene from -1276 to +150 bp. The cytosine +1 represents the transcription start site. The putative transcription factor binding sites are underlined in bold face. c 5′ RACE was performed to map the SET transcription start site. The PCR product was run on a 2 % agarose gel. d The PCR product was cloned into pcDNA4/myc-His A vector and sequenced to identify the transcription start site. The arrow points to the 5′ end of the 5′ RACE product insert in the plasmid, which corresponds to the transcription start site
Functional Analysis of the Human SET Gene Promoter
To investigate the transcriptional regulation of the human SET gene, a series of deletions of the 5′ flanking fragments were cloned into a promoterless luciferase reporter plasmid pGL3-basic. Expression of luciferase in cells transfected with pGL3-basic relies on insertion and proper orientation of a functional promoter upstream of the luciferase gene. The pGL3-basic vector, which lacks eukaryotic promoter and enhancer sequences upstream of the luciferase reporter gene, has little luciferase expression. Eight fragments covering various lengths from -1276 to +123 bp of the 5′ flanking region of the human SET gene were amplified by PCR and cloned into pGL3-basic vector according to restriction enzyme cutting sites as described in the “Materials and Methods” section (Fig. 2a). Plasmid DNA was transfected into HEK293 cells along with plasmid pRLuc, and luciferase activity was measured by a luminometer to reflect promoter activity (Fig. 2b).
Deletion analysis of the human SET gene promoter. a Schematic diagram of the SET promoter deletion constructs in pGL3-basic vector. The arrow shows the direction of transcription. The numbers represents the end points of the human SET inserts relative to the TSS site. b The deletion plasmids were cotransfected with pRLuc into HEK293 cells. Twenty-four hours after the transfection, the luciferase activity was measured and expressed in relative luciferase units (RLU). The pRLuc was used to normalize for transfection efficiency. The values represent means ± SEM. n = 3, *p < 0.01 vs. pGL3-basic by one-way ANOVA followed by post hoc Tukey’s multiple comparisons test
Compared with cells transfected with an empty vector pGL3-basic, pSET-A-transfected cells showed a significant luciferase activity (61.42 ± 5.82 RLU). This result indicated that the 1399-bp fragment contains the functional promoter region of the human SET gene. A deletion from -1276 to -680 shows little impact on promoter activity, while a further deletion from -680 to -483 (pSET-C) resulted in robust increase of promoter activity (131.75 ± 9.87 RLU). These results suggest that the 197-bp fragment may contain negative regulatory cis-acting elements. In addition, different deletions from both 5′ and 3′ (pSET-D, pSET-E, pSET-F, and pSET-G) showed reduced promoter activity compared with pSET-C. The sharp decrease caused by deletion from -483 to -399 and +123 to +108 indicates potential positive regulatory cis-acting elements in the region. Deletions that lack transcription initiation site (pSET-H and pSET-J) had no promoter activity as was expected. These data showed that the region from -483 to +123 has the highest promoter activity and potential positive regulatory elements.
NFkB Upregulates the Human SET Gene Promoter Activity
Sequence analysis revealed several putative cis-acting binding elements from -483 to +123 (Fig. 1b). To determine whether these putative sites are functional, mutation analysis is performed (Fig. 3a). A mutation in the last two Sp1 (pSET-C MF and pSET-C MG) and the NFkB binding sites (pSET-C MH) significantly reduced SET gene promoter activity in HEK293 cells (p < 0.05), whereas a mutation in other putative cis-acting elements had no significant effect on the SET gene promoter activity (p > 0.05) (Fig. 3b). In order to further investigate the effects of transcriptional factor Sp1 and NFkB on SET promoter, we cotransfected pMTF-P65 or pCGN-Sp1 with SET promoter plasmids in HEK293 cells and SH-SY5Y cells. Results showed that in HEK293 cells, NFkB and Sp1 overexpression significantly increased SET promoter activity to 1.92-fold (p < 0.0001) and 1.47-fold (p < 0.05), respectively (Fig. 3c, d). In SH-SY5Y cells, NFkB upregulates the human SET gene promoter activity to 1.37-fold (p < 0.05) while Sp1 does not (p > 0.05) (Fig. 3e, f). These results indicated that NFkB plays a more vital role in regulating SET gene promoter activity in neuronal-like cells, compared with Sp1. Moreover, SET promoter which lacks the NFkB binding sites (pSET-F and pSET-G) or contains the mutant NFkB binding sites (pSET-C MH) could not be upregulated by cotransfection with pMTF-P65 in SH-SY5Y cells (Fig. 3g). These data further confirmed that NFkB activates SET gene transcription.
NFkB upregulates the human SET gene promoter activity. a The putative cis-acting element binding mutant plasmids were constructed by site mutagenesis from -483 to +123. b The mutant plasmids were cotransfected with pRLuc into HEK293 cells. Twenty-four hours after the transfection, the luciferase activity was measured and expressed in relative luciferase units (RLU). The values represent means ± SEM, n = 3, *p < 0.05 vs. pSET-C by one-way ANOVA followed by post hoc Tukey’s multiple comparisons test. c–f The effects of NFkB and Sp1 on the human SET promoter in HEK293 cells (c, d) and SH-SY5Y cells (e, f) were analyzed by luciferase reporter assays. The SET promoter reporter plasmid pSET-C was cotransfected into cells with NFkB expression plasmid pMTF-P65 or the empty vector pMTF (b, d), Sp1 expression plasmid pCGN-Sp1, or the empty vector pCGN (c, e). g SET promoter which lacks the NFkB binding sites (pSET-F and pSET-G) or contains the mutant NFkB binding sites (pSET-C MH) was cotransfected into SH-SY5Y cells with pMTF-P65 or pMTF. Values indicate means ± SEM. n = 3, *p < 0.05 by Student’s t test
The SET Gene Promoter Contains a Functional NFkB Binding Site
The putative NFkB cis-acting binding elements is located at +107 to +116 bp in the 5′ untranslated region of the SET gene promoter. To investigate whether NFkB signaling regulates SET gene transcription by interacting with the putative NFkB cis-acting elements, we performed EMSA. First, HEK293 cells were transfected with plasmid pMTF-P65 to obtain NFkB-enriched nuclear extracts. Wild-type NFkB consensus oligonucleotides and repetitive SET NFkB oligonucleotides were synthesized and labeled with IR700 dye to generate six probes: NFkBwt-IR700, NFkBwt, NFkBmu, SET3XNFkB-IR700, SETNFkB, and SETNFkBmu. The NFkB binding probe NFkBwt served as a positive binding probe and the mutant NFkB binding probe NFkBmu which loses the NFkB binding ability served as a negative binding probe [22]. A shifted DNA–protein complex band was observed after incubating the NFkBwt-IR700 with nuclear extract (Fig. 4a, lane 2). This shifted band disappeared by an addition of the 100× unlabeled consensus competition probe NFkBwt (Fig. 4a, lane 3), while the mutant one NFkBmu had no impact (Fig. 4a, lane 4). Similarly, the probe SETNFkB containing the putative NFkB cis-acting site on SET promoter sharply reduced the intensity of the shifted band while probe SETNFkBmu had no effect (Fig. 4a, lanes 5 and 6). An addition of anti-p65 antibody resulted in a slower-migrating supershifted band, which confirmed the binding between the NFkB and the probe (Fig. 4a, lane 7).
The SET gene promoter contains a cis-acting NFkB element. Nuclear extract was isolated from HEK293 cells transfected with NFkB expression plasmid. a EMSA with NFkB p65 consensus probe. Lane 1 is labeled human consensus NFkB probe only. Lane 2 shows a shifted DNA–protein complex formed between the labeled NFkB with nuclear extracts. Competition assays were performed by further adding different competition oligonucleotides including consensus wild-type NFkB (lane 3), mutant NFkB (lane 4), putative SET NFkB, and mutant SET NFkB (lanes 5 and 6). Lane 7 shows supershifted band with the anti-p65 antibody. b EMSA with SET NFkB probe. Lane 1 is labeled SET3XNFkB-IR700 probe only. Lanes 3 to 6 shows the competitor homologous SET NFkB (lane 3), mutant SET NFkB (lane 4), wild-type NFkB (lane 5), and mutant NFkB (lane 6) were added at 100-fold. Lane 7 showed supershifted band with the anti-p65 antibody
To further investigate the binding between SETNFkB in the human SET promoter region and NFkB, we performed EMSA with IR700 dye-labeled SETNFkB. An IR700 dye-labeled probe SET3XNFkB-IR700 contained three repeats of putative NFkB cis-acting site of SET promoter. Consistently, 100× competition probes SETNFkB and NFkBwt weaken the shifted band but mutant probes made no effect (Fig. 4b, lanes 2–6). A super-shifted band was also detected (Fig. 4b, lane 7). Taken together, these data clearly demonstrate that the human SET promoter contains a functional NFkB binding site in the region of +107 to +116 bp.
NFkB Increases Human SET Isoform 1 Transcription in SH-SY5Y Cells
After testing the effect of NFkB and Sp1 on SET gene promoter activity, we performed a quantitative RT-PCR to measure the mRNA level of endogenous SET isoform 1 and isoform 2. NFkB overexpression has significantly increased the human SET isoform 1 mRNA level by 56.94 % in SH-SY5Y cells (Fig. 5a). Sp1 overexpression had no effect in SH-SY5Y cells (Fig. 5a). NFkB overexpression has no impact on SET isoform 2 mRNA level in both SH-SY5Y cells (Fig. 5a). These data clearly demonstrated SET isoform 1 as a downstream target of the NFkB signaling, and NFkB overexpression results in a significant upregulation of the human SET isoform 1 transcription in neuronal-like cells.
NFkB increases the transcription of human SET gene isoform 1 but not isoform 2 in SH-SY5Y cells. Cells were transfected with either empty vector (pMTF or pCGN) or the nuclear factor expression plasmids (pMTF-P65 or pCGN-Sp1) for 24 h. a NFkB overexpression facilitated SET isoform 1 mRNA level in SH-SY5Y cells while not in Sp1 overexpression. b NFkB expression has no effect on SET isoform 2 mRNA level. RT-PCRs were performed using either primers specific to the human SET isoform 1 or isoform 2 coding sequence or the human GAPDH coding sequence. Values indicate means ± SEM. n = 3, *p < 0.05 by Student’s t test
Discussion
SET was first discovered in 1992 as part of a fusion protein with nucleoporin Nup214 (CAN) in a patient with acute undifferentiated leukemia [1]. Interestingly, the upregulation and abnormal location of SET were also observed in the brain of AD and DS patients [15, 19, 23, 24]. Neurofibrillary tangles are formed by the hyperphosphorylated tau at over 30 serine/threonine residues in AD brains [25]. PP2A is the major enzyme that accounts for 71 % of the total tau phosphatase activity [26], and the activity of PP2A is tightly regulated by the inhibitor protein SET/I2PP2A whose C-terminal region is responsible for the binding to PP2A sub-unit PP2Ac [16]. Inactivation of nuclear localization signal (NLS) in the C-terminal region or cleavage lead to N- and C-terminal peptides causes cytoplasm retention of SET/I2PP2A and leads to abnormal tau pathology and neuronal death [18, 27]. Thus, studying regulation of SET promoter activity could provide new insights into the disease pathogenesis.
We cloned the human SET gene promoter and mapped the transcription initiation site at 123 bp 5′ upstream of the translation start site. Our deletion analysis revealed that the fragment from -483 to +123 has the highest promoter activity. Further deletion from either 5′ end or 3′ end significantly decreases the promoter activity. We found that NFkB overexpression significantly increased SET promoter activity in SH-SY5Y cell. Consistent with the results of SET promoter assay in SH-SY5Y cell, NFkB overexpression also increases the mRNA level of endogenous SET isoform 1 in SH-SY5Y cell. Our study showed that the regulation of SET gene expression by NFkB activation is mediated by the cis-acting NFkB binding elements in the SET gene promoter. All these results demonstrate that NFkB plays an important role in the regulation of human SET gene transcription of isoform 1. NFkB overexpression has no effect on SET isoform 2 expression. The data showing distinct transcriptional regulation between SET gene isoform 1 and isoform 2 is consistent with previous study of isoform 2 being driving from a distinct promoter [28].
NFkB is a family of transcription factors and consists of five members including p50, p52, p65 (RelA), RelB, and C-Rel [29]. NFkB-activating stimuli include oxidative stress, mitogens, cytokines (TNF-α), and bacterial products (LPS) [30]. In the clinical stage of mild cognitive impairment (MCI) due to AD, significant oxidative damage occurs without the remarkable Aβ plaque and NFTs [31]. Oxidative stress causes lipid peroxidation [32, 33], protein oxidation [34], DNA/RNA oxidation [35, 36], and downregulation of antioxidants [37] in AD patients. Oxidative stress could trigger the pathological process of AD. Recently, two functional NFkB binding elements were identified in the human BACE1 promoter region [38]. We also reported that glycogen synthase kinase-3β (GSK-3β) regulates BACE1 gene expression via NFkB signaling pathway [39]. BACE1 is essential for Aβ generation in the AD amyloidogenic process. Our results here extend the mechanism linking NFkB with expression of SET, a molecule associated with tau pathology. Oxidative stress could activate NFkB signaling pathway, leading to upregulation of the human SET gene expression in neural cells. Such activation may contribute to abnormal tau phosphorylation in AD pathogenesis. Further studies should explore the potential impact of SET different isoforms on AD pathogenesis.
References
von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G (1992) CAN, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3’ half to different genes: characterization of the set gene. Mol Cell Biol 12(8):3346–3355
Nagata K, Saito S, Okuwaki M, Kawase H, Furuya A, Kusano A, Hanai N, Okuda A, Kikuchi A (1998) Cellular localization and expression of template-activating factor I in different cell types. Exp Cell Res 240(2):274–281
Kim EG, Choi ME, Ballermann BJ (1994) Spatially restricted expression of set mRNA in developing rat kidney. Am J Physiol 266(1 Pt 2):F155–161
Fan Z, Beresford PJ, Zhang D, Xu Z, Novina CD, Yoshida A, Pommier Y, Lieberman J (2003) Cleaving the oxidative repair protein Ape1 enhances cell death mediated by granzyme A. Nat Immunol 4(2):145–153
Muto S, Senda M, Akai Y, Sato L, Suzuki T, Nagai R, Senda T, Horikoshi M (2007) Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc Natl Acad Sci U S A 104(11):4285–4290
Nagata K, Kawase H, Handa H, Yano K, Yamasaki M, Ishimi Y, Okuda A, Kikuchi A, Matsumoto K (1995) Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc Natl Acad Sci U S A 92(10):4279–4283
Kato K, Miyaji-Yamaguchi M, Okuwaki M, Nagata K (2007) Histone acetylation-independent transcription stimulation by a histone chaperone. Nucleic Acids Res 35(3):705–715
Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D (2001) Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104(1):119–130
Miyamoto S, Suzuki T, Muto S, Aizawa K, Kimura A, Mizuno Y, Nagino T, Imai Y, Adachi N, Horikoshi M, Nagai R (2003) Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol Cell Biol 23(23):8528–8541
Li M, Makkinje A, Damuni Z (1996) The myeloid leukemia-associated protein SET is a potent inhibitor of protein phosphatase 2A. J Biol Chem 271(19):11059–11062
Carlson SG, Eng E, Kim EG, Perlman EJ, Copeland TD, Ballermann BJ (1998) Expression of SET, an inhibitor of protein phosphatase 2A, in renal development and Wilms’ tumor. JASN 9(10):1873–1880
Compagnone NA, Zhang P, Vigne JL, Mellon SH (2000) Novel role for the nuclear phosphoprotein SET in transcriptional activation of P450c17 and initiation of neurosteroidogenesis. Mol Endocrinol 14(6):875–888
Madeira A, Pommet JM, Prochiantz A, Allinquant B (2005) SET protein (TAF1beta, I2PP2A) is involved in neuronal apoptosis induced by an amyloid precursor protein cytoplasmic subdomain. FASEB J 19(13):1905–1907
Qu D, Zhang Y, Ma J, Guo K, Li R, Yin Y, Cao X, Park DS (2007) The nuclear localization of SET mediated by impalpha3/impbeta attenuates its cytosolic toxicity in neurons. J Neurochem 103(1):408–422
Wang X, Blanchard J, Kohlbrenner E, Clement N, Linden RM, Radu A, Grundke-Iqbal I, Iqbal K (2010) The carboxy-terminal fragment of inhibitor-2 of protein phosphatase-2A induces Alzheimer disease pathology and cognitive impairment. FASEB J 24(11):4420–4432
Arnaud L, Chen S, Liu F, Li B, Khatoon S, Grundke-Iqbal I, Iqbal K (2011) Mechanism of inhibition of PP2A activity and abnormal hyperphosphorylation of tau by I2(PP2A)/SET. FEBS Lett 585(17):2653–2659
Yu G, Yan T, Feng Y, Liu X, Xia Y, Luo H, Wang JZ, Wang X (2013) Ser9 phosphorylation causes cytoplasmic detention of I2PP2A/SET in Alzheimer disease. Neurobiol Aging 34(7):1748–1758
Arif M, Wei J, Zhang Q, Liu F, Basurto-Islas G, Grundke-Iqbal I, Iqbal K (2014) Cytoplasmic retention of protein phosphatase 2A inhibitor 2 (I2PP2A) induces Alzheimer-like abnormal hyperphosphorylation of tau. J Biol Chem 289(40):27677–27691
Zhang X, Wu Y, Duan X, Chen W, Zou H, Zhang M, Zhang S, Cai F, Song W (2015) Upregulation of SET expression by BACE1 and its implications in Down syndrome. Mol Neurobiol 51(2):781–790
Duan X, Tong J, Xu Q, Wu Y, Cai F, Li T, Song W (2014) Upregulation of human PINK1 gene expression by NFkappaB signalling. Mol Brain 7:57
Wang K, Liu S, Wang J, Wu Y, Cai F, Song W (2014) Transcriptional regulation of human USP24 gene expression by NF-kappa B. J Neurochem 128(6):818–828
Wang R, Zhang M, Zhou W, Ly PT, Cai F, Song W (2011) NF-kappaB signaling inhibits ubiquitin carboxyl-terminal hydrolase L1 gene expression. J Neurochem 116(6):1160–1170
Tanimukai H, Grundke-Iqbal I, Iqbal K (2005) Up-regulation of inhibitors of protein phosphatase-2A in Alzheimer’s disease. Am J Pathol 166(6):1761–1771
Facchinetti P, Dorard E, Contremoulins V, Gaillard MC, Deglon N, Sazdovitch V, Guihenneuc-Jouyaux C, Brouillet E, Duyckaerts C, Allinquant B (2014) SET translocation is associated with increase in caspase cleaved amyloid precursor protein in CA1 of Alzheimer and Down syndrome patients. Neurobiol Aging 35(5):958–968
Hanger DP, Betts JC, Loviny TL, Blackstock WP, Anderton BH (1998) New phosphorylation sites identified in hyperphosphorylated tau (paired helical filament-tau) from Alzheimer’s disease brain using nanoelectrospray mass spectrometry. J Neurochem 71(6):2465–2476
Liu F, Grundke-Iqbal I, Iqbal K, Gong CX (2005) Contributions of protein phosphatases PP1, PP2A, PP2B and PP5 to the regulation of tau phosphorylation. Eur J Neurosci 22(8):1942–1950
Trakhtenberg EF, Morkin MI, Patel KH, Fernandez SG, Sang A, Shaw P, Liu X, Wang Y, Mlacker GM, Gao H, Velmeshev D, Dombrowski SM, Vitek MP, Goldberg JL (2015) The N-terminal SET-beta protein isoform induces neuronal death. J Biol Chem 290(21):13417–13426
Asaka MN, Murano K, Nagata K (2008) Sp1-mediated transcription regulation of TAF-Ialpha gene encoding a histone chaperone. Biochem Biophys Res Commun 376(4):665–670
Baldwin AS Jr (1996) The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol 14:649–683
Gapuzan ME, Schmah O, Pollock AD, Hoffmann A, Gilmore TD (2005) Immortalized fibroblasts from NF-kappaB RelA knockout mice show phenotypic heterogeneity and maintain increased sensitivity to tumor necrosis factor alpha after transformation by v-Ras. Oncogene 24(43):6574–6583
Price JL, McKeel DW Jr, Buckles VD, Roe CM, Xiong C, Grundman M, Hansen LA, Petersen RC, Parisi JE, Dickson DW, Smith CD, Davis DG, Schmitt FA, Markesbery WR, Kaye J, Kurlan R, Hulette C, Kurland BF, Higdon R, Kukull W, Morris JC (2009) Neuropathology of nondemented aging: presumptive evidence for preclinical Alzheimer disease. Neurobiol Aging 30(7):1026–1036
Lovell MA, Ehmann WD, Butler SM, Markesbery WR (1995) Elevated thiobarbituric acid-reactive substances and antioxidant enzyme activity in the brain in Alzheimer’s disease. Neurology 45(8):1594–1601
Markesbery WR, Lovell MA (1998) Four-hydroxynonenal, a product of lipid peroxidation, is increased in the brain in Alzheimer’s disease. Neurobiol Aging 19(1):33–36
Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR (2001) Protein oxidation in the brain in Alzheimer’s disease. Neuroscience 103(2):373–383
Shan X, Tashiro H, Lin CL (2003) The identification and characterization of oxidized RNAs in Alzheimer’s disease. J Neurosci 23(12):4913–4921
Anderson AJ, Su JH, Cotman CW (1996) DNA damage and apoptosis in Alzheimer’s disease: colocalization with c-Jun immunoreactivity, relationship to brain area, and effect of postmortem delay. J Neurosci 16(5):1710–1719
Venkateshappa C, Harish G, Mahadevan A, Srinivas Bharath MM, Shankar SK (2012) Elevated oxidative stress and decreased antioxidant function in the human hippocampus and frontal cortex with increasing age: implications for neurodegeneration in Alzheimer’s disease. Neurochem Res 37(8):1601–1614
Chen CH, Zhou W, Liu S, Deng Y, Cai F, Tone M, Tone Y, Tong Y, Song W (2012) Increased NF-kappaB signalling up-regulates BACE1 expression and its therapeutic potential in Alzheimer’s disease. Int J Neuropsychopharmacol 15(01):77–90
Ly PT, Wu Y, Zou H, Wang R, Zhou W, Kinoshita A, Zhang M, Yang Y, Cai F, Woodgett J, Song W (2013) Inhibition of GSK3beta-mediated BACE1 expression reduces Alzheimer-associated phenotypes. J Clin Invest 123(1):224–235
Acknowledgments
We thank Shasha Meng and Mingjing Liu for providing valuable comments and technique support. This work was supported by the Graduate Student Research Innovation Project of Chongqing, National Natural Science Foundation of China (NSFC) grant 81161120498 (T. L.), and Canadian Institutes of Health Research (CIHR) Grant TAD-117948 (W.S.). W.S. is the holder of the Tier 1 Canada Research Chair in Alzheimer’s disease.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no competing financial interests.
Additional information
Yi Feng, Xiaoyong Li and Weitao Zhou contributed equally to this work.
Rights and permissions
About this article
Cite this article
Feng, Y., Li, X., Zhou, W. et al. Regulation of SET Gene Expression by NFkB. Mol Neurobiol 54, 4477–4485 (2017). https://doi.org/10.1007/s12035-016-9967-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-9967-2