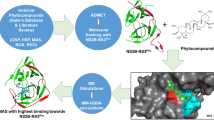
Abstract
Dengue fever (DF) is an endemic disease that has become a public health concern around the globe. The NS3 protease-helicase enzyme is an important target for the development of antiviral drugs against DENV (dengue virus) due to its impact on viral replication. Inhibition of the activity of the NS3 protease-helicase enzyme complex significantly inhibits the infection associated with DENV. Unfortunately, there are no scientifically approved antiviral drugs for its prevention. However, this study has been developed to find natural bioactive molecules that can block the activity of the NS3 protease-helicase enzyme complex associated with DENV infection through molecular docking, MM-GBSA (molecular mechanics-generalized born surface area), and molecular dynamics (MD) simulations. Three hundred forty-two (342) compounds selected from twenty traditional medicinal plants were retrieved and screened against the NS3 protease-helicase protein by molecular docking and MM-GBSA studies, where the top six phytochemicals have been identified based on binding affinities. The six compounds were then subjected to pharmacokinetics and toxicity analysis, and we conducted molecular dynamics simulations on three protein–ligand complexes to validate their stability. Through computational analysis, this study revealed the potential of the two selected natural bioactive inhibitors (CID-440015 and CID-7424) as novel anti-dengue agents.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dengue fever (DF) is a tropical and subtropical endemic infection that is caused by the dengue virus (DENV) belonging to the Flaviviridae family [1, 2]. It is estimated that there are approximately 100 countries worldwide that are affected by dengue. In Southeast Asia and the Americas, the prevalence of the disease is on the rise. This encompasses nations such as India, China, and Taiwan, among others [3, 4]. From 2000 to 2010, the number of dengue cases increased by over eightfold, reaching over 2.4 million, and from 2010 to 2019, it climbed to 5.2 million [5]. Each year, as many as 390 million people are affected by dengue globally, but only 96 million are clinically manifest, and approximately 20,000 die of the infection [6]. In 2080, dengue fever will affect about 60 percent of the world's population [7].
DENV is primarily transmitted by Aedes aegypti mosquitoes to humans through the bite, although it can also be transmitted by Aedes albopictus to a lesser extent [8]. Patients with dengue can either show no symptoms (about 75% of cases) or show symptoms ranging from a mild case of the flu to more severe symptoms after an incubation period of 4–10 days [9]. DF, the milder version of the illness, is characterized by a sudden, high fever that lasts 7–14 days and is followed by intense headaches, myalgias, arthralgias, and gastrointestinal distress. More severe instances, known as dengue hemorrhagic fever (DHF), exhibit increased vascular fragility and permeability, coagulopathy, spontaneous bleeding, and mild to moderate liver damage [10]. Dengue virus is composed of four distinct serotypes: DENV1, DENV2, DENV3, and DENV4, and they all belong to the family Flaviviridae, genus Flavivirus [1]. There is a genetic variation between each of the four serotypes, having 65–70% similar amino acid sequences [11], and among the four serotypes, DENV-2 is the cosmopolitan genotype [12]. The key component of the DENV genome is a single-stranded RNA (~ 11 kb), which can be translated directly into a long polypeptide. Therefore, it is regarded as positive-sense RNA. Ten distinct proteins are encoded by the polypeptide; these are classified as seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) and three structural proteins, namely the capsid (C), envelope (E), and membrane (M) [13]. The envelope protein is one of these structural proteins that allows the virus to enter the host. Since envelope proteins act as receptors for viral entry and are crucial targets for vaccine development for adaptive immune responses [14]. Anti-E antibody production is the primary way that cells respond to dengue virus by blocking virus binding and neutralizing viral infection [15]. The progression of the virus life cycle, such as replication, assembly, and other cellular functions in host cells, is accomplished by the seven nonstructural proteins [16]. The nonstructural 4B transmembrane protein inhibits the interferon (IFN) response. Consequently, it plays a key role in virus replication and proliferation [17]. Also, NS3 is responsible for viral replication [18]. Viral polyproteins and virus replication are entirely dependent on the NS2B-NS3 protease complex [18], therefore it is a desirable target for the development of antiviral drugs [19]. Many research studies have been done against DENV infection, but the development of natural inhibitors has yet to be explored.
Medicinal plants are utilized as an abundant supply of therapeutic compounds all over the world [20]. They are a valuable source of potential pharmaceutical candidates due to their diverse chemical composition. Herbal medicine can increase the therapeutic capacity of conventional antiviral medications [21] and produce many primary and secondary components in trace amounts that have pharmacological activity. Phytochemicals are used for drug development because of their harmless effects compared to chemical substances [4]. These have potential antiviral, antibacterial, antifungal, anticancer, and other properties [22]. According to reports, phytochemical substances such as flavonoids, phenolics, and terpenoids are important for treatments against DENV [23]. Therefore, the study examined the anti-DENV activity of natural bioactive compounds targeting the NS3 protease-helicase enzyme complex from twenty (20) medicinal plants (Acorus calamus, Andrographis paniculata, Annona reticulata, Azadirachta indica, Catharanthus roseus, Vitex negundo, Cissampelos pareira, Citrus limon, Cymbopogon citratus, Curcuma longa, Euphorbia hirta, Houttuynia cordata, Tinospora cordifolia, Mentha arvensis, Mikania cordata, Momordica charantia, Myristica fragrans, Phyllanthus urinaria, Spondias mombin, and Ricinus communis) selected based on traditional uses through an in silico approach.
At present, computer-aided drug design (CADD) has proven to be a useful and effective tool in the development of many therapeutics [24]. Through this approach, a lengthy and costly procedure that was previously required during medication research and development has been eliminated [25]. The use of in silico drug design has become increasingly important in the modern drug design process. To find effective and promising drug compounds against dengue, the study used a variety of in silico techniques, which include molecular docking, MM/GBSA, ADMET (adsorption, distribution, metabolism, excretion, and toxicity), and molecular dynamic (MD) simulation.
Methods and Materials
Chemical Library Preparation
Medicinal plants are a significant source of pharmacologically significant chemicals since they have historically played a significant role in medical systems. Around 342 phytochemicals were obtained from the IMPPAT (Indian Medicinal Plants, Phytochemistry, and Therapeutics) database in 3D SDF format from 20 medicinal plants.
Protein and Ligands Preparation
The 3D structure of the NS3 protease-helicase protein (PDB ID: 2VBC) was retrieved from the RCSB Protein Data Bank (PDB) (https://www.rcsb.org/) [26]. The dengue NS3 protease-helicase protein structure was identified with a resolution of 3.15 Å, consisting of 618 AA residues. To prepare the protein, a protein preparation wizard was deployed with default settings [27]. By using Maestro v-12.5 of Schrödinger Suite 2020–3, hydrogen atoms and omitted side chains were added, while water, metal ions, cofactors, and other molecules were removed. The protein's 3D crystal structure was stabilized by employing the OPLS-3e force field [28]. Additionally, the LigPrep module accessible in the Maestro Schrodinger Suite v11.5 was utilized to prepare ligands [29]. The chemical structures were optimized using the OPLS3e force field [30].
Receptor Grid Generation
Receptor grid generation is an important process to identify the target protein's active site and find compounds that will bind well to the catalytic site. It is insufficient to perform ligand-based molecular docking with the targeted protein without generating a receptor grid. For creating the receptor grid, the FTSite server (https://ftsite.bu.edu/) was used to generate the binding site [31]. A receptor grid using binding site residues determined a box range of X = 5.32, Y = 2.59, and Z = 40.43.
Molecular Docking
Molecular docking helps in determining the binding geometry and atomic-level interactions between a small molecule and a protein. This technique can screen large compound libraries, for which it is significantly considered an essential tool in drug design [32]. In this study, the Glide v-8.8 and Maestro v-12.5.139 packages from the Schrödinger Suite 2020–3 were used to analyze and visualize the best binding scores. To conduct docking calculations, a coordinate-defined grid box was used. System optimization was performed using the OPLS3e force field [30]. The Maestro viewer was used to visualize molecular-binding residues and chemical bonds.
MM-GBSA Analysis
An MM-GBSA study was conducted using the Prime MM-GBSA package to determine the binding-free energy of ligands and confirm the docking process between the NS3 protease-helicase and compounds [33]. The studies considered five of the most highly interacting ligands in terms of negative G binds (NS), Coulomb (Coulomb energy), hydrogen bond (hydrogen bond energy), lipophilicity (lipophilicity energy), and van der Waals interactions (VdW) [34]. In addition to providing useful insights into ligands, receptors, and complex structures, these features also provide insight into energy differences caused by strain and binding.
ADME and Toxicity Analysis
Pharmacokinetics and toxicity are important criteria for designing and developing drugs since they allow researchers to find potential compounds as drugs with effective features [35]. Essentially, this refers to the movement of drugs within, through, and out of the body, as well as the intensity and duration of their action. SwissADME (www.swissadme.ch) was used to assess the pharmacokinetic properties of our identified compounds at an early stage [36]. Subsequently, toxicity prediction is one of the essential steps in the drug development process. To evaluate the quality of a chemical compound that can cause organ damage to humans or animals, its toxicity must be predicted. Therefore, the evaluation procedure for the selected compounds' toxicity was conducted using the ProTox-II server (https://tox-new.charite.de/) [37].
MD Simulation Studies
MD simulation provides atomistic information about the movement of molecules and atoms in an artificial environment. In addition, protein–ligand complexes are assessed for structural stability [38]. An evaluation of the thermodynamic stability of the receptor-ligand complex was performed using the 'Desmond v3.6 Program' in Schrodinger (academic version) on a Linux platform [39]. The system was operated utilizing a preset TIP3P water model, where an orthorhombic periodic boundary box shape with a diameter of 10 × 10 × 10 Å3 was selected on both sides to ensure suitable volume and ions like Na+ and Cl− with a level of salt of 0.15 M throughout the whole system for electrical neutralization. The stabilization of the system was done using OPLS3e force field settings [38]. In the NPT (constant pressure-constant temperature) ensemble, the temperature was maintained at 300.0 K and the pressure at 1.01325 bar throughout the simulation. Through the computation of parameters including RMSD (root mean square deviation), RMSF (root mean square fluctuation), Rg (radius of gyration), and SASA (solvent-accessible surface area), the stability and dynamic properties of the complexes were assessed.
Simulation Trajectory Analysis
Every MD simulation snapshot was generated using Schrodinger's Maestro interface version 9.5. The simulation event has been analyzed using the Simulation Interaction Diagram (SID) of the Desmond module in the Schrodinger package. The RMSD, RMSF, protein–ligand interactions (P-L contact), and hydrogen bond interactions were all used to evaluate the stability of the complex structure based on the trajectory output.
RMSD Analysis
RMSD is used to calculate the dislocation distance averaged over a particular period in a protein–ligand complex system [40]. The RMSD of protein-suited molecules from all frames is fitted and calculated in comparison to the 100 ns simulation time. The RMSD should be calculated using the following formula (Eq. 1).
Here, N indicates the number of select atoms, tref stands for the reference or specified time, r indicates the specified atom's location in frame x following superimposition on the reference frame, and tx specifies the duration of the recording intervals.
RMSF Analysis
The RMSF describes the local conformational shift that occurs within a protein structure [41]. The following equation (Eq: 2) is assigned to get the RMSF value of a protein with the number of residues in the MD simulation.
Here, T primarily refers to the trajectory time, tref is the reference or given time, r is the selected atoms' position in frame i after superimposition on the reference frame, and (< >) expresses the average square distance traveled over residues.
Results
Plant Selection and Phytochemical Library Preparation
After a comprehensive review of the literature, a phytochemical library of 342 compounds was identified from twenty traditionally used medicinal plants, along with their chemical identifiers as presented in Table 1.
Molecular Docking Analysis
Molecular docking employs computational methods to forecast the binding interactions between target proteins and phytochemicals, facilitating the identification of potential drug candidates as well as the comprehension of their binding mechanisms. The resultant binding affinities obtained from the molecular docking of these phytochemical compounds have exhibited a diverse range between 4.21 and −7.16 kcal/mol, as shown in Supplementary Table 1.
The compounds selected for further analysis comprise the top two percentages (%) from a set of 342 phytochemicals (total 6) that have a binding score > −6.4 kcal/mol. The top six molecules that have the strongest binding affinities are displayed in Table 2.
Interpretations of Protein–Ligand Interactions
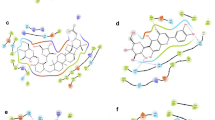
The interactions between the target protein and six selected ligands (CID 440015, CID 3033938, CID 11980943, CID 17750979, CID 7424, and CID 8018607) have been figured out by the Maestro module of the Schrodinger suite illustrated in Figs. 1 and 2. Various types of non-covalent interactions between receptors and ligands have been identified, including hydrogen bonds, electrostatic interactions, and hydrophobic interactions. It has been demonstrated that CID 440015 obtained a docking score of −7.16 kcal/mol and found two hydrogen bonds with residues GLU285 and ARG463 (Table 3). Moreover, it established interaction with the NS3 protease-helicase enzyme by forming three hydrophobic bonds with LEU193, PRO195, and ALA316 residues and two polar bonds with THR200 and ASN416, as shown in Table 3.
CID 3033938 demonstrated a docking score of −7.00 kcal/mol when interacting with the NS3 protease-helicase enzyme. It formed four hydrogen bonds with LYS199, LYS201, ASN416, and ASN464 residues, along with six hydrophobic bonds involving LEU193, PRO195, ALA234, VAL227, TYR394, and PHE417. Additionally, there are three polar bonds with THR200, ASN416, and ASN464. CID 11980943 showed a docking score of −6.99 kcal/mol with the target protein. The analysis of the interaction revealed the presence of ten hydrogen bonds that bound with GLU66, LYS199, ALA229, GLU233, ASH284, GLU285, LYS398, ASN416, PHE417, and ASN464. In addition, there were ten hydrophobic bonds with LEU193, PRO195, ALA229, MET231, ALA234, VAL226, VAL227, TYR394, PRO395, and PHE417. Furthermore, there were three polar bonds with THR200, ASN416, and ASN464, similar to CID 3033938.
For the compound CID 17750979, it has been found that the binding affinity is −6.68 kcal/mol. CID 17750979 engages in six hydrogen bonds at the positions of GLU66, ASH284, LYS201, GLU233, GLU285, and LYS398. Five hydrophobic bonds with LEU193, ALA234, VAL227, TYR394, and PHE417 and three polar bonds with THR200, ASN416, and ASN464 have also been observed during the interaction.
In the case of CID 7424, it exhibited a docking score of −6.45 kcal/mol and showed the ability to form three hydrogen bonds with ALA316, ASN416, and GLN456 residues, and four hydrocarbon bonds with LEU193, PRO195, MET314, and ALA316 residues. Notably, three polar bonds have been noticed at the positions of HIE287, THR315, and GLN456.
The docking score for CID 8018607 against the target protein was −6.45 kcal/mol. The interaction study of the CID 8018607 found seven hydrogen bonds at the positions of LYS201, GLU230, GLU233, ASN284, GLU285, ASN416, ARN460, and two hydrocarbon bonds at LEU193, PRO195, VAL227, MET231, ALA234, MET314, ALA316, and TYR394 residual positions. In addition, there were six polar bonds with THR200, HIE287, THR315, THR317, ASN416, and GLN456.
MM-GBSA Analysis
In MM-GBSA, binding energy can be estimated to identify ligands that bind efficiently with receptors, helping to detect strong molecular interactions. The compounds or ligands' validity identified through docking underwent additional validation by performing MM-GBSA binding free energy estimation calculations. For the protein, the highest ΔG bind was seen for the compound CID 11980943 at ΔG = −63.95 kcal/mol, which provides a strong interaction between this ligand and the receptor. In addition, compounds CID 8018607, CID 3033938, and CID 17750979 showed a high ΔG bind at −31.97 kcal/mol −29.82 kcal/mol, and −17.47 kcal/mol, respectively, while compounds CID 440015 and CID 7424 displayed the least binding affinity, with binding energy values of −16.11 kcal/mol and −8.49 kcal/mol, respectively. The analysis of 2VBC-ligand complexes following post-docking and MD simulations showed diverse interaction energies. These included Coulomb energy (ΔG bind Coulomb), hydrogen bond energy (ΔG bind H bond), electrostatic interactions (ΔG bind covalent), and lipophilicity energy (ΔG bind lipo). These various energies collectively contribute to gaining a comprehensive insight into the overall binding stability within these complexes. In conclusion, our results suggest that these compounds exhibit a strong binding affinity, demonstrating their suitability as novel candidates worthy of further development.
Pharmacokinetic Properties and Toxicity Analysis
To evaluate the drug-likeliness properties, the top six compounds have been subjected to ADME analysis; among them, only three compounds passed our screening criteria. A pharmacokinetic profile of each of these three compounds is presented in Table 4. The ProTox-II web server was utilized to conduct an in silico toxicity investigation of the top six phytochemicals and provided good toxicological effects of the selected three compounds, as presented in Table 4.
MD Simulation Analysis
MD simulation is used in CADD to investigate the stability and interactions between molecules within a protein–ligand complex over a specified time frame. It also facilitates the identification of the conformational alterations occurring in the complex system within a simulated environment. MD simulation was performed over a duration of 100 ns to investigate the structural dynamics, binding interactions, and flexibility of the protein–ligand complexes. The results led to six specific conclusions, addressing parameters such as RMSD, RMSF, Rg, SASA, and intra-molecular hydrogen bond.
RMSD Analysis
The RMSD of the protein backbone atoms is recognized as an important factor for evaluating the equilibrium and stabilization of MD trajectories. In our study, we investigated the interactions of three compounds, CID 7424, CID 440015, and CID 8018607, in complex with the protein (2VBC) shown in Fig. 3. The average shift in values between the 2VBC backbone and the compounds CID 7424, CID 440015, and CID 8018607 falls within the 0.5 to 3 Å range. The CID 8018607 has the lowest overall RMSD among the three compounds, suggesting it has the most stable conformation during the simulation. CID 8018607 showed a higher resemblance to the apo protein than other compounds, but unlike the apo protein at 1.5 ns, there was a fluctuation observed. However, this fluctuation eventually stabilized after 12 ns of MD simulation time. CID 8018607 exhibited stability throughout the simulation until 72 ns, after which fluctuations were observed up to 85 ns, indicating a degree of flexibility in its structure. During the initial 9 to 29 ns of MD simulation, noticeable fluctuations were observed in the behavior of the CID 7424 compound, which has shown a stabilized or equilibrated state after 30 ns of MD simulation. The average distance between the three compounds consistently falls within the 0.5 to 3 Å range, so as a result, fluctuations are expected to be optimized over an extended duration of the simulation.
RMSD of Ligand in Complex With Protein
The stability of the selected compounds was determined by calculating the RMSD. An acceptable average variation in RMSD for the protein–ligand complex falls within the range of 1–3 Å or 0.1–0.3 nm. If the RMSD value exceeds this limit, it indicates a significant conformational shift in the protein structure. The RMSD trajectories of the CID 7424 complex displayed values reaching a peak of 0.381 Å at 93 ns and averaging at 0.173 Å, as shown in Fig. 4. Similarly, the ligand CID 440015 exhibited stability with a few minor fluctuations. The complex showed a maximum RMSD of 0.67 Å and an average RMSD of 0.25 Å. The RMSD trajectories of the CID 8018607 complex displayed an abrupt shift initially, followed by a phase of stable fluctuations between 1.5 and 50 ns. The maximum trajectory value of this complex was 0.554 Å at 51 ns, while the average RMSD was found to be 0.84 Å, which is within the acceptable range. The RMSD trajectories demonstrate that both CID 440015 and CID 7424 maintained their structural integrity throughout the simulation without any substantial alterations from their initial configurations. Conversely, CID 8018607 underwent slight structural changes during the simulation, in contrast to the stability observed in the other two compounds.
RMSF Analysis
RMSF measures the flexibility and variability of individual atoms or groups of atoms within a biomolecule over time in computational biology and structural bioinformatics [69]. It quantifies the extent to which atomic positions deviate from their average positions during simulations or other dynamic analyses. In our 100 ns MD simulation, we calculated the RMSF values for each residue analysis. The RMSF analysis revealed regions of increased flexibility, such as LEU30, THR62, THR120, LYS157, THR252, GLY347, GLU512, GLU585 residues at 12, 44, 101, 140, 234, 329, 449, and 567 positions, which are known to be involved in ligand binding, as represented in Fig. 5. The reduced RMSF values of 2VBC in complex with CID 8018607 indicate a trend of decreased atomic fluctuations in the biomolecule as compared to Apo protein (2VBC). In contrast to the Apo protein, the fluctuation of residues in the complex structure is notably limited, pointing towards a high level of rigidity in the protein structure. Fluctuations in the protein are highest at the N- and C-terminal domains, suggesting a low probability of atom displacement in real-life environments for the identified compounds.
RMSF of the amino acid residues over a 100 ns simulation timescale. The RMSF values were derived from the protein Cα atoms of the protein–ligand complex. Apo protein is shown in orange colors, and the three complexes bound to the compounds CID 7424, CID 440015, and CID 8018607 were indicated by yellow, green, and magenta colors, respectively
Rg Analysis
The Rg characterizes how atoms are distributed around the axis in a protein–ligand complex system. The calculation of Rg is a crucial indicator for the structural activity of a macromolecule, providing insights into changes in the compactness of the complex. It has been shown that a protein in a consistently folded state retains a stable Rg, and a lower Rg signifies a higher level of compactness. In the process of protein unfolding, its Rg value fluctuates over time. So, the analysis of the stability of CID 7424, CID 440015, and CID 8018607 in complex with the 2VBC protein involved studying their Rg over a 100 ns simulation time shown in Fig. 6. The compounds CID 7424, and CID 440015 showed average Rg values of 2.56 Å, 2.20 Å, respectively, which is an acceptable range and exhibited no significant conformational changes in the protein's active site after binding to the specified compounds, while the CID 8018607 ligand displayed an average Rg value of 5.24 Å, indicating a considerable difference compared to the other two ligands. This substantial difference in Rg values for CID 8018607 suggests that its binding may induce a distinct configuration of the protein.
SASA Analysis
SASA analysis was performed to evaluate the protein surface area that is readily accessible to the solvent, which is shown in Fig. 7. This assessment involved estimating the hydrophilic and hydrophobic residues of the protein, with a higher SASA value indicating a relatively expanded surface area. The average SASA values of CID 7424 and CID 440015 in complex with 2VBC were 10.78 Å2 and 14.66 Å2, respectively, while CID 8018607 showed a SASA value of 76.16 Å2 when in complex with 2VBC. Among the three compounds, CID 7424 and CID 440015 have the most compact structure, as indicated by their lower SASA values. On the other hand, the CID 8018607 has the highest SASA throughout the simulation, which suggests that it has the most extensive surface area exposed to the solvent. The interpretation of effective exposure of amino acid residues to the identified compound within the complex systems requires further analysis beyond SASA values alone.
P-L Contact Analysis
The protein's complex structure, along with identified ligands and their intermolecular interactions, has been assessed through a 100 ns simulation using the SID. Various factors, including hydrogen bonds, non-covalent bonds (hydrophobic bonds), ionic bonds, and water bridge bonds, were taken into consideration. The interactions between the protein and the designated compounds with CID 7424, CID 440015, and CID 8018607 were analyzed and visually represented in Fig. 8. The stacked bar charts representing compound CID 440015 indicate an interaction fraction value (IFV) of 2.0 at residue ARG 460. This suggests that the interaction at this specific residue, facilitated by hydrogen bonds, has been consistently maintained throughout the entire simulation, accounting for over 100% of the simulation time. Significant hydrogen bonding and water bridges are found at the GLU 285, GLY 414, and ARG 463 sites. Additionally, there were some observed interactions involving ionic bonds. In the case of CID 7424, significant contacts were established through hydrogen bonds and water bridges. Residues LYS 199, GLU 285, GLN 456, ARG 460, and ARG 463 were significant sites where these interactions occurred. Additionally, a hydrophobic interaction was also observed at ALA 316. For CID 8018607, all four types of interactions have been found, controlled by H-bonds and water bridges. Significant interactions were found at THR 200, LYS 200, GLU 285, ANS 416, ARG 460, ARG 463, and ANS 464. This suggests that there are a variety of forces at play that stabilize the complex.
Discussion
The NS3 protein represents an attractive target for the development of antiviral drugs [70]. Due to NS3's dual functions as an RNA helicase and a protease, it plays an essential role in the life cycle of DV [71]. RNA helicase activity facilitates the unwinding of viral RNA duplexes, which is crucial for replication, while protease activity is responsible for the cleavage of viral polyproteins, which is critical for the generation of functional viral proteins [72]. Furthermore, NS3 has been implicated in modulating the immune response of the host. The virus may be able to evade host defenses and establish infection if it interferes with the host's innate immune signaling pathways [73]. The development of novel therapeutics against dengue fever may be achieved by inhibiting NS3 enzymatic activity or disrupting its interactions with host factors [74]. For rational drug design and optimization, it is imperative to understand the structural dynamics of NS3-inhibitor complexes [75]. Based on the results of our in silico study, we have gained valuable insight into the mechanism of NS3 inhibition in dengue virus infection.
CADD is a useful tool for identifying new compounds against an identified protein since it contains a variety of advanced characteristics and techniques. It has reduced the time and costs associated with the entire drug development process, which includes molecular docking, molecular MM-GBSA, MD simulation, and ADMET [76]. Molecular docking is a crucial technique in CADD, enabling the virtual screening of compound libraries to identify potential drug candidates [77]. It assesses binding affinity and intermolecular interactions at the atomic level between a target protein and bioactive compounds [78]. We identified six compounds (CID 440015, CID 3033938, CID 11980943, CID 17750979, CID 7424, and CID 8018607) with good binding affinity (> −6.4 kcal/mol, Table 2) for the targeted protein NS3 protease-helicase through molecular docking.
In the MM-GBSA study, the lowest ∆G-Bind score (the most negative score) was regarded as the best score [79]. In this study, CID 11980943 and CID 7424 demonstrated lower ∆G Bind scores compared to other docked compounds for NS3 protease-helicase, indicating the best ∆G-Bind score (Fig. 9). ADME and toxicity prediction play a major role in the preclinical phase of drug research and development [76]. In the drug design process, toxicity analysis is indispensable for identifying possible harmful effects of chemical substances on humans, animals, and the environment [40]. In our studies, three compounds revealed satisfactory outputs in terms of pharmacokinetics and toxicity profiles depicted in Table 4.
Based on pharmacokinetics and toxicity analysis, the best three compounds were subjected to MD simulation analysis to observe the structural stability of protein–ligand complexes. An MD simulation determines a protein's stability when complexed with its ligand. The RMSD values indicate the stability of the protein–ligand complex, while the RMSF values indicate the fluctuation of the residues upon ligand binding [80]. The protein complexed with the compounds CID 440015 and CID 7424 showed lower RMSD and RMSF values over the 100 ns simulation time, indicating the stability of the complexes (Figs. 3, 4, and 5). The lower Rg value indicates a high level of compactness, and the larger value indicates a disassociation of the compounds from the protein [81]. Protein-compound CID 440015 complex and protein-compound CID 7424 complexes demonstrated better Rg values compared to the protein-compound CID 8018607 complex (Fig. 6). Higher SASA values indicate less stable structures, whereas lower values indicate tightly contracted complexes of water molecules and amino acids [81]. In this study, the protein-CID 440015 complexes and the protein-CID 7424 complexes demonstrated lower mean SASA values compared to the protein-CID 8018607 (Fig. 7). Using the SID, we observed the organization of proteins bound to the specified ligands as well as their molecular interactions over a 100 ns simulation. Moreover, CID 440015 and CID 7424 formed higher hydrogen bonds, hydrophobic contacts, and water bridge bonds with NS3 protease-helicase compared to the ligand CID 8018607.
Based on the aforementioned results, it can be said that CID 440015 and CID 7424 could be used as potential lead compounds to treat DENV infection. However, a range of laboratory trial methods are necessary to demonstrate the anti-viral properties of these compounds, which may provide alternatives for the treatment of the DENV outbreak.
Conclusion
The NS3 protease-helicase enzyme is the key responsible protein for the replication and infection of the DENV virus and serves as a very important target for the development of antiviral drugs. Consequently, the study aims to identify effective natural phytochemicals that can block the activity of the NS3 protease-helicase enzyme. The findings of our study suggest that CID 440015 and CID 7424 are potential inhibitors that can selectively block DENV replication by inhibiting the activity of the NS3 protease-helicase protein. Further in vitro and in vivo studies are required to verify the activity of these inhibitors against the NS3 protease-helicase protein.
Data Availability
Data will be made available on request.
Abbreviations
- DF:
-
Dengue fever
- DENV:
-
Dengue virus
- IMPPAT:
-
Indian Medicinal Plants, Phytochemistry, and Therapeutics
- MM-GBSA:
-
Molecular mechanics-generalized born surface area
- ADME:
-
Absorption, Distribution, Metabolism, and Excretion
- SID:
-
Simulation Interaction Diagram
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuation
- Rg:
-
Radius of gyration
- SASA:
-
Solvent-accessible surface area
References
Adawara, S. N., Shallangwa, G. A., Mamza, P. A., & Ibrahim, A. (2021). In-silico approaches towards the profiling of some anti-dengue virus as potent inhibitors against dengue NS-5 receptor. Scientific African, 13, e00907. https://doi.org/10.1016/j.sciaf.2021.e00907
Tantawichien, T. (2012). Dengue fever and dengue haemorrhagic fever in adolescents and adults. Paediatrics and International Child Health, 32(s1), 22–27. https://doi.org/10.1179/2046904712Z.00000000049
Hasan, S., Jamdar, S. F., Alalowi, M., Beaiji, A. A. A., & S. M. (2016). Dengue virus: A global human threat: Review of literature. Journal of International Society of Preventive & Community Dentistry, 6(1), 1–6. https://doi.org/10.4103/2231-0762.175416
Halder, S. K., Ahmad, I., Shathi, J. F., Mim, M. M., Hassan, M. R., Jewel, M. J. I., & Hossen, M. S. (2022). A Comprehensive Study to Unleash the Putative Inhibitors of Serotype2 of Dengue Virus: Insights from an In Silico Structure-Based Drug Discovery. Molecular Biotechnology, 1, 1–14. https://doi.org/10.1007/s12033-022-00582-1
Abutaha, N., & Almutairi, B. O. (2023). Exploring the therapeutic potential of GC–MS separated compounds from Dracaena cinnabari against dengue virus and Aedes aegypti using in silico tools. Journal of King Saud University - Science, 35(2), 102478. https://doi.org/10.1016/j.jksus.2022.102478
Ononamadu, C. J., Abdalla, M., Ihegboro, G. O., Li, J., Owolarafe, T. A., John, T. D., & Tian, Q. (2021). In silico identification and study of potential anti-mosquito juvenile hormone binding protein (MJHBP) compounds as candidates for dengue virus - Vector insecticides. Biochemistry and biophysics reports, 28, 101178. https://doi.org/10.1016/j.bbrep.2021.101178
Messina, J. P., Brady, O. J., Golding, N., Kraemer, M. U. G., Wint, G. R. W., Ray, S. E., & Hay, S. I. (2019). The current and future global distribution and population at risk of dengue. Nature Microbiology, 4(9), 1508–1515. https://doi.org/10.1038/s41564-019-0476-8
Otu, A., Ebenso, B., Etokidem, A., & Chukwuekezie, O. (2019). Dengue fever - an update review and implications for Nigeria, and similar countries. African Health Sciences, 19(2), 2000–2007. https://doi.org/10.4314/ahs.v19i2.23
Guzman, M. G., Gubler, D. J., Izquierdo, A., Martinez, E., & Halstead, S. B. (2016). Dengue infection. Nature reviews. Disease primers, 2, 16055. https://doi.org/10.1038/nrdp.2016.55
Halstead, S. B., Mahalingam, S., Marovich, M. A., Ubol, S., & Mosser, D. M. (2010). Intrinsic antibody-dependent enhancement of microbial infection in macrophages: Disease regulation by immune complexes. The Lancet. Infectious diseases, 10(10), 712–722. https://doi.org/10.1016/S1473-3099(10)70166-3
Azhar, E. I., Hashem, A. M., El-Kafrawy, S. A., Abol-Ela, S., Abd-Alla, A. M. M., Sohrab, S. S., & Jamjoom, G. (2015). Complete genome sequencing and phylogenetic analysis of dengue type 1 virus isolated from Jeddah Saudi Arabia. Virology Journal, 12, 1. https://doi.org/10.1186/s12985-014-0235-7
Ernst, T., McCarthy, S., Chidlow, G., Luang-Suarkia, D., Holmes, E. C., Smith, D. W., & Imrie, A. (2015). Emergence of a new lineage of dengue virus type 2 identified in travelers entering Western Australia from Indonesia, 2010–2012. PLoS Neglected Tropical Diseases, 9(1), e0003442. https://doi.org/10.1371/journal.pntd.0003442
Gebhard, L. G., Filomatori, C. V., & Gamarnik, A. V. (2011). Functional RNA elements in the dengue virus genome. Viruses, 3(9), 1739–1756. https://doi.org/10.3390/v3091739
Chakraborty, S., Chakravorty, R., Ahmed, M., Rahman, A., Waise, T. M. Z., Hassan, F., & Shamsuzzaman, S. (2010). A computational approach for identification of epitopes in dengue virus envelope protein: a step towards designing a universal dengue vaccine targeting endemic regions. In silico Biology, 10(5–6), 235–246. https://doi.org/10.3233/ISB-2010-0435
J.S, S., J.M, C., C.O, N., N.M, O., K.A, F., S, I., … J.E, R. (2010). Neutralizing and non-neutralizing monoclonal antibodies against dengue virus e protein derived from a naturally infected patient. Virology Journal, 7(M), 1–11.
Welsch, S., Miller, S., Romero-Brey, I., Merz, A., Bleck, C. K. E., Walther, P., & Bartenschlager, R. (2009). Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host & Microbe, 5(4), 365–375. https://doi.org/10.1016/j.chom.2009.03.007
Umareddy, I., Chao, A., Sampath, A., Gu, F., & Vasudevan, S. G. (2006). Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. The Journal of General Virology, 87(Pt 9), 2605–2614. https://doi.org/10.1099/vir.0.81844-0
Yusof, R., Clum, S., Wetzel, M., Murthy, H. M., & Padmanabhan, R. (2000). Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. The Journal of Biological Chemistry, 275(14), 9963–9969. https://doi.org/10.1074/jbc.275.14.9963
Assenberg, R., Mastrangelo, E., Walter, T. S., Verma, A., Milani, M., Owens, R. J., & Mancini, E. J. (2009). Crystal structure of a novel conformational state of the flavivirus NS3 protein: implications for polyprotein processing and viral replication. Journal of Virology, 83(24), 12895–12906. https://doi.org/10.1128/JVI.00942-09
Zandi, K., Teoh, B. T., Sam, S. S., Wong, P. F., Mustafa, M. R., & AbuBakar, S. (2012). Novel antiviral activity of baicalein against dengue virus. BMC Complementary and Alternative Medicine, 12, 1. https://doi.org/10.1186/1472-6882-12-214
Silva-Trujillo, L., Quintero-Rueda, E., Stashenko, E. E., Conde-Ocazionez, S., Rondón-Villarreal, P., & Ocazionez, R. E. (2022). Essential Oils from Colombian Plants: Antiviral Potential against Dengue Virus Based on Chemical Composition, In Vitro and In Silico Analyses. Molecules (Basel, Switzerland), 27, 20. https://doi.org/10.3390/molecules27206844
Briskin, D. P. (2000). Medicinal plants and phytomedicines Linking plant biochemistry and physiology to human health. Plant Physiology, 124(2), 507–514. https://doi.org/10.1104/pp.124.2.507
Rosmalena, R., Elya, B., Dewi, B. E., Fithriyah, F., Desti, H., Angelina, M., & Seto, D. (2019). The Antiviral Effect of Indonesian Medicinal Plant Extracts Against Dengue Virus In Vitro and In Silico. Pathogens (Basel, Switzerland), 8, 2. https://doi.org/10.3390/pathogens8020085
Hasan, M. R., Alsaiari, A. A., Fakhurji, B. Z., Molla, M. H. R., Asseri, A. H., Sumon, M. A. A., & Kim, B. (2022). Application of Mathematical Modeling and Computational Tools in the Modern Drug Design and Development Process. Molecules (Basel, Switzerland), 27, 13. https://doi.org/10.3390/molecules27134169
Aljahdali, M. O., Molla, M. H. R., & Ahammad, F. (2021). Compounds identified from marine mangrove plant (Avicennia alba) as potential antiviral drug candidates against WDSV, an in-silico approach. Marine Drugs, 19, 5. https://doi.org/10.3390/md19050253
Luo, D., Xu, T., Hunke, C., Grüber, G., Vasudevan, S. G., & Lescar, J. (2008). Crystal structure of the NS3 protease-helicase from dengue virus. Journal of Virology, 82(1), 173–183. https://doi.org/10.1128/JVI.01788-07
Orgován, Z., Ferenczy, G. G., & Keserű, G. M. (2019). The role of water and protein flexibility in the structure-based virtual screening of allosteric GPCR modulators: An mGlu 5 receptor case study. Journal of Computer-Aided Molecular Design, 33, 787–797.
Roos, K., Wu, C., Damm, W., Reboul, M., Stevenson, J. M., Lu, C., & Harder, E. D. (2019). OPLS3e: Extending Force Field Coverage for Drug-Like Small Molecules. Journal of Chemical Theory and Computation, 15(3), 1863–1874. https://doi.org/10.1021/acs.jctc.8b01026
Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R., & Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design, 27(3), 221–234. https://doi.org/10.1007/s10822-013-9644-8
Harder, E., Damm, W., Maple, J., Wu, C., Reboul, M., Xiang, J. Y., & Friesner, R. A. (2016). OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. Journal of Chemical Theory and Computation, 12(1), 281–296. https://doi.org/10.1021/acs.jctc.5b00864
Kozakov, D., Grove, L. E., Hall, D. R., Bohnuud, T., Mottarella, S. E., Luo, L., & Vajda, S. (2015). The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins. Nature Protocols, 10(5), 733–755. https://doi.org/10.1038/nprot.2015.043
Adyani, F., Fuad, A., & Ahammad, F. (2020). Virtual Screening and Molecular Docking Approaches for Identification of Natural Lead Compounds for Improved Anti-Dengue Therapeutics. Researchgate.Net, . https://doi.org/10.13140/RG.2.2.27630.43849
Jacobson, M. P., Pincus, D. L., Rapp, C. S., Day, T. J. F., Honig, B., Shaw, D. E., & Friesner, R. A. (2004). A hierarchical approach to all-atom protein loop prediction. Proteins, 55(2), 351–367. https://doi.org/10.1002/prot.10613
Nobel, F. A., Islam, S., Babu, G., Akter, S., Jebin, R. A., Sarker, T. C., & Islam, M. J. (2022). Isolation of multidrug resistance bacteria from the patients with wound infection and their antibiotics susceptibility patterns: A cross-sectional study. Annals of Medicine and Surgery, 84, 104895. https://doi.org/10.1016/j.amsu.2022.104895
Boezio, B., Audouze, K., Ducrot, P., & Taboureau, O. (2017). Network-based Approaches in Pharmacology. Molecular Informatics, 36(10), 1–10. https://doi.org/10.1002/minf.201700048
Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7(1), 42717. https://doi.org/10.1038/srep42717
Veber, D. F., Johnson, S. R., Cheng, H. Y., Smith, B. R., Ward, K. W., & Kopple, K. D. (2002). Molecular properties that influence the oral bioavailability of drug candidates. Journal of Medicinal Chemistry, 45(12), 2615–2623. https://doi.org/10.1021/jm020017n
Opo, F. A. D. M., Rahman, M. M., Ahammad, F., Ahmed, I., Bhuiyan, M. A., & Asiri, A. M. (2021). Structure based pharmacophore modeling, virtual screening, molecular docking and ADMET approaches for identification of natural anti-cancer agents targeting XIAP protein. Scientific Reports, 11(1), 1–18. https://doi.org/10.1038/s41598-021-83626-x
Bharadwaj, S., Dubey, A., Yadava, U., Mishra, S. K., Kang, S. G., & Dwivedi, V. D. (2021). Exploration of natural compounds with anti-SARS-CoV-2 activity via inhibition of SARS-CoV-2 Mpro. Briefings in Bioinformatics, 22(2), 1361–1377. https://doi.org/10.1093/BIB/BBAA382
Samad, A., Alam, R., Hossen, S., & Al-ghamdi, K. (2021). Calculations , and Molecular Dynamics Simulation Approaches, (Md).
Han, Y., Wang, Z., Ren, J., Wei, Z., & Li, J. (2021). Potential inhibitors for the novel coronavirus (SARS-CoV-2). Briefings in Bioinformatics, 22(2), 1225–1231. https://doi.org/10.1093/bib/bbaa209
Rajput, S. B., Tonge, M. B., & Karuppayil, S. M. (2014). An overview on traditional uses and pharmacological profile of Acorus calamus Linn (Sweet flag) and other Acorus species. Phytomedicine, 21(3), 268–276. https://doi.org/10.1016/j.phymed.2013.09.020
Hossain, M. S., Urbi, Z., Sule, A., & Rahman, K. M. H. (2014). A Review of Ethnobotany, Phytochemistry, and Pharmacology. The Scientific World Journal, 2014, 1–28.
Jamkhande, P. G., & Wattamwar, A. S. (2015). Annona reticulata Linn (Bullock’s heart): Plant profile, phytochemistry and pharmacological properties. Journal of Traditional and Complementary Medicine, 5(3), 144–152. https://doi.org/10.1016/j.jtcme.2015.04.001
Alzohairy, M. A. (2016). Therapeutics Role of Azadirachta indica (Neem) and Their Active Constituents in Diseases Prevention and Treatment. Evidence-based complementary and alternative medicine : ECAM, 2016, 7382506. https://doi.org/10.1155/2016/7382506
Kumar, S., Malhotra, R., & Kumar, D. (2010). Euphorbia hirta: Its chemistry, traditional and medicinal uses, and pharmacological activities. Pharmacognosy Reviews, 4(7), 58–61. https://doi.org/10.4103/0973-7847.65327
Venkateswarlu, K. (2012). Vitex negundo: Medicinal values, biological activities, toxicity studies and phytopharmacological actions. International Journal of Pharmaceutical and Phytopharmacological Research, 2, 126–133.
Amresh, G., Singh, P. N., & Rao, C. V. (2007). Antinociceptive and antiarthritic activity of Cissampelos pareira roots. Journal of Ethnopharmacology, 111(3), 531–536. https://doi.org/10.1016/j.jep.2006.12.026
Brügger, B. P., Martínez, L. C., Plata-Rueda, A., de Castro, B. M., & C. E., Soares, M. A., Wilcken, C. F., … Zanuncio, J. C. (2019). Bioactivity of the Cymbopogon citratus (Poaceae) essential oil and its terpenoid constituents on the predatory bug, Podisus nigrispinus (Heteroptera: Pentatomidae). Scientific Reports, 9(1), 8358. https://doi.org/10.1038/s41598-019-44709-y
Omosa, L. K., Midiwo, J. O., & Kuete, V. (2017). Chapter 19 - Curcuma longa. In V. Kuete (Ed), Medicinal Spices and Vegetables from Africa (pp. 425–435). Academic Press. https://doi.org/10.1016/B978-0-12-809286-6.00019-4
Yang, L., & Jiang, J.-G. (2009). Bioactive components and functional properties of Hottuynia cordata and its applications. Pharmaceutical Biology, 47(12), 1154–1161. https://doi.org/10.3109/13880200903019200
Devi, P. (2021). A review on Tinospora cordifolia: as an Immunomodulating agent. Himalayan Journal of Health Sciences, 10(10), 6–14. https://doi.org/10.22270/hjhs.v6i1.88
Nazim, M., Sadiq, Q.-U.-A., Nawaz, A., Anjum, S., Ali, M., & Maryam, H. (2020). Mentha arvensis, a medicinal and aromatic plant, has high nutritional value and several-uses: A review. Buletin Agroteknologi, 1(2), 37. https://doi.org/10.32663/ba.v1i2.1180
Al Nayeem, A., Khatun, A., Rahman, M. S., & Rahman, M. (2011). Evaluation of phytochemical and pharmacological properties of Mikania cordata (Asteraceae) leaves. Journal of Pharmacognosy and Phytotherapy, 3(8), 118–123.
Anilakumar, K. R., Kumar, G. P., & Ilaiyaraja, N. (2015). Nutritional, Pharmacological and Medicinal Properties of Momordica Charantia. International Journal of Nutrition and Food Sciences, 4(1), 75–83. https://doi.org/10.11648/j.ijnfs.20150401.21
Jinous, A. (2012). Phytochemistry and pharmacologic properties of Myristica fragrans Hoyutt: A review. African Journal of Biotechnology, 11(65), 12787–12793. https://doi.org/10.5897/ajb12.1043
Geethangili, M., & Ding, S. T. (2018). A review of the phytochemistry and pharmacology of Phyllanthus urinaria L. Frontiers in Pharmacology, 9, 1–20. https://doi.org/10.3389/fphar.2018.01109
Maria, A. C. B., Simões, T. R., de Ramos, A., & S., Haddad de Almeida, M. M., Mpalantinos da Silva, M. A., Diocesano da Cruz, J., … Amaral, A. C. F. (2022). Spondias mombin L: An Updated Monograph. Pharmacognosy Reviews, 16(31), 45–61. https://doi.org/10.5530/phrev.2022.16.8
Chouhan, N. S., Mohan, K., & Ghose, A. (2017). <Chouhan et al 2016.pdf>, 12(7), 3632–3642. https://doi.org/10.13040/IJPSR.0975-8232.12(7).3632-42
Wu, G. (2020). Important roles of dietary taurine , creatine , carnosine , anserine and 4 ‑ hydroxyproline in human nutrition and health. Amino Acids. Springer Vienna. https://doi.org/10.1007/s00726-020-02823-6
Lee, H., Kim, C. E., Ahn, B., & Yang, J. (2017). Anti-inflammatory effect of in desiccation stress-induced experimental dry eye mouse. Scientific Reports, 1, 1–12. https://doi.org/10.1038/s41598-017-07965-4
Chao, X., & Ding, W. (2019). Role and mechanisms of autophagy in alcohol-induced liver injury. Drug-induced Liver Injury (1st ed., Vol. 85). Elsevier Inc. https://doi.org/10.1016/bs.apha.2019.01.008
Hippocampus, C. (1989). The protease inhibitor leupeptin interferes with the development of LTP in hippocampal slices, 505, 233–238.
End, A. G., Assessment, P. I., Docking, M., Abdel-naim, A. B., Koshak, A. E., Proksch, P., & Elhady, S. S. (2022). Phenolics from Chrozophora oblongifolia Aerial Parts as.
He, H., & Silva, A. (2022). Recognition of Gallotannins and the Physiological Activities : From Chemical View, 9(June), 1–9. https://doi.org/10.3389/fnut.2022.888892
Pandey, A., Bani, S., & Kumar, N. (2012). Anti-arthritic activity of agnuside mediated through the down-regulation of inflammatory mediators and cytokines, 293–304. https://doi.org/10.1007/s00011-011-0410-x
Activity, C., Lines, M.-C., Sylwia, M., Swiderski, G., Pietryczuk, A., Cudowski, A., … Swisłocka, R. (2021). Comprehensive Study of Structural , Anti- / Pro-Oxidant ,.
Brzobohaty, B., Id, H. T. H., & Martin, Č. (2020). trans-Zeatin-N-glucosides have biological activity in Arabidopsis thaliana, 1–20. https://doi.org/10.1371/journal.pone.0232762
Fatriansyah, J. F., Rizqillah, R. K., Yandi, M. Y., & Fadilah, & Sahlan, M. (2022). Molecular docking and dynamics studies on propolis sulabiroin-A as a potential inhibitor of SARS-CoV-2. Journal of King Saud University - Science, 34(1), 101707. https://doi.org/10.1016/j.jksus.2021.101707
Luo, D., Vasudevan, S. G., & Lescar, J. (2015). The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development. Antiviral research, 118, 148–158. https://doi.org/10.1016/j.antiviral.2015.03.014
Swarbrick, C. M. D., Basavannacharya, C., Chan, K. W. K., Chan, S.-A., Singh, D., Wei, N., & Vasudevan, S. G. (2017). NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication. Nucleic Acids Research, 45(22), 12904–12920. https://doi.org/10.1093/nar/gkx1127
Shi, R.-Z., Pan, Y.-Q., & Xing, L. (2021). RNA Helicase A Regulates the Replication of RNA Viruses. Viruses, 13, 3. https://doi.org/10.3390/v13030361
Lee, M. F., Voon, G. Z., Lim, H. X., Chua, M. L., & Poh, C. L. (2022). Innate and adaptive immune evasion by dengue virus. Frontiers in Cellular and Infection Microbiology, 12, 1004608. https://doi.org/10.3389/fcimb.2022.1004608
Krishnan, M. N., & Garcia-Blanco, M. A. (2014). Targeting host factors to treat West Nile and dengue viral infections. Viruses, 6(2), 683–708. https://doi.org/10.3390/v6020683
Ozen, A., Sherman, W., & Schiffer, C. A. (2013). Improving the Resistance Profile of Hepatitis C NS3/4A Inhibitors: Dynamic Substrate Envelope Guided Design. Journal of Chemical Theory and Computation, 9(12), 5693–5705. https://doi.org/10.1021/ct400603p
Sabe, V. T., Ntombela, T., Jhamba, L. A., Maguire, G. E. M., Govender, T., Naicker, T., & Kruger, H. G. (2021). Current trends in computer aided drug design and a highlight of drugs discovered via computational techniques: A review. European Journal of Medicinal Chemistry, 224, 113705. https://doi.org/10.1016/j.ejmech.2021.113705
Pinzi, L., & Rastelli, G. (2019). Molecular Docking : Shifting Paradigms in Drug Discovery.
Biswas, B., Golder, M., Abid, M. A., Mazumder, K., & Sadhu, S. K. (2021). Terpenoids enriched ethanol extracts of aerial roots of Ceriops decandra (Griff.) and Ceriops tagal (Perr.) promote diuresis in mice. Heliyon, 7(7), e07580. https://doi.org/10.1016/j.heliyon.2021.e07580
Zhang, X., Perez-sanchez, H., & Lightstone, F. C. (2017). Current Topics in Medicinal Chemistry, 1631–1639. https://doi.org/10.2174/15680266166661611171
Pavan, M., Menin, S., Bassani, D., Sturlese, M., & Moro, S. (2022). Qualitative Estimation of Protein − Ligand Complex Stability through Thermal Titration Molecular Dynamics Simulations. https://doi.org/10.1021/acs.jcim.2c00995
Samad, A., Huq, A., & Rahman, S. (2022). Bioinformatics approaches identified dasatinib and bortezomib inhibit the activity of MCM7 protein as a potential treatment against human cancer. Scientific Reports. https://doi.org/10.1038/s41598-022-05621-0
Acknowledgements
We are thankful to the researchers supporting project number (RSPD2024R740), King Saud University, Riyadh, Saudi Arabia.
Funding
Researchers supporting project number (RSPD2024R740), King Saud University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
Md. Tarikul Islam: Project Design, Data generation, Formal analysis, Writing. Md. Aktaruzzaman: Supervision, Formal analysis, Writing. Ahmed Saif: Writing. Aeysha Akter: Writing. Mashood Ahmed Bhat, PhD: Fund Acquisition, Writing and Review. Mirza Mahfuz Hossain: Visualization. Dr. S.M. Nur Alam: Critically reviewed the manuscript, Writing and Review. Rifat Rayhan: Data Curation. Saira Rehman: Data Curation, Writing and Review. Muhammad Yaseen, PhD: Resources, Writing and Review. Md. Obayed Raihan, PhD: Validation, Editing manuscript, Supervision.
Corresponding authors
Ethics declarations
Conflicts of Interests
The authors have disclosed no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Islam, M.T., Aktaruzzaman, M., Saif, A. et al. In Silico-Based Identification of Natural Inhibitors from Traditionally Used Medicinal Plants that can Inhibit Dengue Infection. Mol Biotechnol (2024). https://doi.org/10.1007/s12033-024-01204-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12033-024-01204-8