Abstract
Pseudomonas, being the common inhabitant of colder environments, are suitable for the production of cold-active enzymes. In the present study, a newly isolated strain of Pseudomonas from cold desert site in Indian Himalayan Region, was investigated for the production of cold-active lipase. The bacteria were identified as Pseudomonas proteolytica by 16S rDNA sequencing. Lipase production by bacteria was confirmed by qualitative assay using tributyrin and rhodamine-B agar plate method. The bacterium produced maximum lipase at 25 °C followed by production at 15 °C while utilizing olive, corn, as well as soybean oil as substrate in lipase production broth. Enzyme produced by bacteria was partially purified using ammonium sulphate fractionation. GBPI_Hb61 showed aggregation behaviour which was confirmed using several techniques including gel filtration chromatography, dynamic light scattering, and native PAGE. Molecular weight determined by SDS-PAGE followed by in-gel activity suggested two lipases of nearly similar molecular weight of ~50 kDa. The enzyme showed stability in wide range of pH from 5 to 11 and temperature up to 50 °C. The enzyme from GBPI_Hb61 exhibited maximum activity toward p-nitrophenyldecanoate (C10). The stability of enzyme was not affected with methanol while it retained more than 75% activity when incubated with ethanol, acetone, and hexane. The bacterium is likely to be a potential source for production of cold-active lipase with efficient applicability under multiple conditions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Lipases (EC 3.1.1.3) are enzymes that catalyse hydrolysis of lipids and synthesis of esters at lipid water interface and water-insoluble substrate interface, respectively [1]. Their wide applications in many industries, including organic synthesis, paper manufacturing, food, dairy, detergent, biosensors, pharmaceuticals, and biodiesel production, make them extremely versatile biocatalyst of biotechnology industry [2, 3]. According to J. B. S. Haldane, a biochemist, who wrote in his monograph named as ‘Enzymes’, “The possible substrates for lipase are to be numbered in millions” [4]. With increasing trend for microbial lipases [5], which have the advantages including catalysis of diverse reactions, high yield, and less production costs over animal and plant based lipases, search for new source with novel characteristics is still continued. Cold-adapted lipases, with high rate of catalysis in low temperature, find its application in many areas unlike mesophilic and thermophilic enzymes which remain less active or inactive at low temperatures [3]. Cold-adapted lipases are mostly distributed among microbes which grow at low temperatures. Most microbes for exploration of cold-adapted lipase have been isolated from different cold habitats including Antarctic/polar region [6], and deep sea sediments/marine environments [7, 8]. Attempts have been made from time to time to isolate new microorganisms to produce cold-active lipases with higher catalytic activity in low-temperature conditions. These include several species of Acinetobacter [9–11], Achromobacter [12], Pseudoalteromonas [7, 13], Psychrobacter [14, 15], Pseudomonas [16–18], etc.
Himalayan mountains, which are gifted with valued source of biodiversity, still remain unexplored in several ways. Microbial diversity of Indian Himalayan region (IHR) with respect to bio-prospection for beneficial products has been reported in several studies [19–22]. Studies on microbiota of this region suggest the microorganisms to possess adaptations to various extremophilic conditions of temperatures, pH, and salt [20, 23]. These reports provide a fundamental base to explore novel enzymes which help microbes in these regions to survive under harsh conditions. Such enzymes can be overproduced in vitro for their applicability in biotechnological industries and lipase being one of them. Previous studies from high-altitude soils in IHR suggest dominance of psychrotolerant species of Pseudomonas [24]. Pseudomonas being the common inhabitant of cold environment is an important microorganism suitable for producing cold-active lipase with stability in wide range of temperature as well as pH. According to Yabuchi et al. [25], Pseudomonas lipases were probably the first to be studied; called as true lipases. Based on a recent study by Daiha et al. [2], use of lipases as biocatalysts is still a relevant topic to study for industrial sector. Therefore, present study focuses on production of a cold-active lipase from a newly isolated strain of Pseudomonas proteolytica from cold desert soil with emphasis on lipase aggregation studies and its characterization.
Methods
Bacterial Strain and Culture Conditions
The bacterium was isolated from soil collected from Mana (Distt. Chamoli) in Uttarakhand state under IHR (altitude of 3200 masl) following tenfold serial dilution. The pure bacterial culture was preserved in glycerol stocks at −20 °C while it was routinely cultured using tryptone yeast extract (TY) agar at 25 °C for 24 h.
Biochemical and Physiological Characterization and Identification of the Bacterium
The biochemical tests including oxidase, catalase, urea hydrolysis, indole production, nitrate reduction, utilization of carbohydrates, and enzyme production including protease, amylase, and lipase production were performed following standard methods. Physiological characterization of bacteria was carried out for temperature, pH, and salt tolerance through visual inspection for growth as described in Jain and Pandey [20].
For identification, DNA was isolated following the method of Chen and Kuo [26]. PCR amplification of 16SrRNA gene by using 27F and 1492R primers was performed followed by sequencing of amplified product (Courtesy: NCCS Pune, India). The nucleotide sequence was identified using EzTaxon database [27]. Phylogenetic tree was constructed employing maximum likelihood method using MEGA version 6 [28]. The type culture and its nucleotide sequence are deposited in Microbial Culture Collection, National Centre for Cell Science, Pune, India, and NCBI, respectively.
Lipase Production
Production of extracellular lipase by GBPI_Hb61 was first assayed qualitatively using tributyrin agar (HiMedia, India) and rhodamine B-olive oil agar plates [29].
Lipase production medium (LPM) (composition in g/l: NaNO3 3 g, MgSO4·7H2O 0.5 g, KCl 0.5 g, K2HPO4 0.1 g, FeSO4·7H2O 0.01 g, Yeast extract 5 g, pH 7.0) amended with 1% olive oil [30] was used for quantitative production of lipase in 250-ml flask containing 50-ml sterilized media under static condition. 1% bacterial culture grown in LPM but in absence of olive oil (OD600 = 0.5) was used as inoculum. The production was carried out up to 10 days and the lipase activity was measured after every 2nd day of incubation. Bacterial culture was filtered through Whatman filter paper 1 and then centrifuged at 10,000 rpm for 10 min and the clear supernatant was used for lipase assay.
Lipase Assay
Lipase activity was measured using p-nitrophenyldodecanoate (pNPL) (Sigma, US) as substrate by the method described by Pinsirodom and Parkin [31]. Briefly, reaction mixture contained 1 ml of tris–Cl buffer (50 mM, pH 9.0), 1 ml substrate solution (420 µm pNPL), and 0.4 ml crude enzyme. The reaction was allowed to occur for 5 min at room temperature (20 °C). Release of pNP was recorded at 410 nm using a UV/Vis spectrophotometer (Ultrospec 2100 Pro, Amersham Biosciences). Enzyme activity was calculated using standard curve of pNP in tris–Cl buffer (pH 9.0). Enzyme activity was expressed in terms of enzyme unit which is defined as the enzyme required to release 1.0 µm of pNP per min under standard assay conditions.
Effect of Culture Conditions on Lipase Production
Lipase production was carried out in different culture conditions by altering temperature, pH and media component. For estimation of lipase production at different temperatures, the sterilized lipase production broth was inoculated with 1% inoculum and incubated at four different temperatures (5, 15, 25, and 35 °C). Effect of initial medium pH on lipase production was quantified by inoculating bacteria in lipase production medium of different pH (adjusted using 1 N NaOH/HCl) and incubation at 25 °C.
In all the experiments for media component optimization, original medium with 1% olive oil served as control. In carbon source, three oil source including soybean oil, corn oil, and tributyrin were screened for lipase production. Nitrogen source included organic as well as inorganic nitrogen source. In case of organic nitrogen, yeast extract in original medium was replaced with same concentration of peptone, malt extract, casein, urea, or no organic nitrogen source. Similarly, for inorganic N-source, sodium nitrate in original medium was replaced with ammonium nitrate, ammonium chloride, ammonium sulphate, or no inorganic nitrogen source added. Effect of detergents (1%) including tweens 20, 40, 60, 80, and triton X-100 was also investigated on lipase production by the bacterial strain.
Partial Purification of Enzyme
Ammonium sulphate fractionation (0–50% and 50–80%) of cell free crude supernatant after enzyme production for 10 days was performed using solid ammonium sulphate at 4 °C. Resulting precipitates were collected after centrifugation at 4 °C for 20 min at 15,000 rpm and dissolved in minimal volume of tris–Cl (50 mM, pH 8.0) buffer. Both the fractions were dialysed overnight against same buffer at 4 °C with two change of same buffer. Lipase activity was measured in both the fractions. Active fraction was used for further studies. Protein was estimated spectrophotometrically by Bradford method [32] using bovine serum albumin as standard. Specific activity in culture supernatant and active fraction obtained after ammonium sulphate fractionation was calculated.
Aggregation Studies on Lipase
Lipase produced by the bacterium showed very high degree of aggregation. Various techniques were used to confirm the formation of protein aggregates. All the experiments related to aggregation studies were conducted using partially purified lipase obtained after ammonium sulphate precipitation.
Gel Filtration Chromatography
Gel filtration chromatography using Sephadex G-100 was performed to purify the lipase in initial steps. The column was equilibrated with 50 mM tris–Cl buffer (pH 8.0). 500 µl of concentrated lipase solution was loaded on to the column and elution was carried out with same buffer. Column was run at a flow rate of 0.5 ml/min and a total of 25 fractions of 1 ml each were collected.
Dynamic Light Scattering
Experiment based on dynamic light scattering (DLS) using Zetasizer Nano (Malvern Instruments) was also performed [33]. Different treatments including several detergents and reagents were tested to break aggregate of proteins in the sample. Protein sample diluted (1:4) in tris–Cl buffer (50 mM, pH 8.0) was used for DLS measurements at 25 °C. Data were analysed using Zetasizer software (v 7.11) to compare reduction in molecular size (d.nm) of proteins in the samples after several treatments.
Native PAGE
To study aggregation behaviour of lipase, non-denaturing polyacrylamide gel electrophoresis was performed. Native PAGE of ammonium sulphate precipitate was performed using gel system of Laemmli [34] with resolving gel of 10% and stacking gel of 4%. Sample was treated with different detergents including CHAPS, Triton-X100, NP-40, Brij-35, Tween-20, SDS, l-arginine, glycerol, and PEG. After electrophoretic separation, zymogram was developed following the method of Prim et al. [35] using 4-methyl umbelliferyl butyrate (Sigma, US) as substrate. Same gel was also stained with coomassie brilliantblue R250 (CBB).
Determination of Molecular Weight by SDS-PAGE
SDS-PAGE (12.5%) (non-reducing as well as reducing) was performed using gel system of Laemmli [34] for determination of molecular weight of the lipase. After electrophoresis was complete, gel was washed in 2.5% triton-X (only in case of non-reducing SDS-PAGE) in tris–Cl (50 mM, pH 8.0) buffer to remove SDS from gel. Zymogram was developed as described above and the same gel after activity staining was also stained with CBB. Reducing gel was directly stained with CBB after electrophoresis. Molecular weight was determined by comparing active bands with standard pre-stained molecular weight markers (Puregene, Genetix Biotech) and protein bands in CBB stained gels.
Biochemical Characterization of Lipase
Effect of Temperature and pH on Lipase Activity and Stability
Effect of temperature on lipase activity was determined after incubating enzyme reaction mixture at different temperatures from 10 to 70 °C in tris–Cl buffer (50 mM, pH 8). Thermal stability of partially purified lipase was determined by incubating the enzyme at different temperatures for 1 h. Residual activity was then measured by assaying the enzyme reaction in tris–Cl buffer (pH 8.0) as described earlier.
Optimal pH for lipase activity was determined by performing enzyme assay in buffers of different pH values ranging from 4 to 12 at 20 °C. Similarly, for pH stability, enzyme was incubated in different pH buffers for 2 h and residual activity was measured after performing standard enzyme assay. Buffers used for different pH values include 50 mM citrate phosphate buffer (pH 4, 5), 50 mM sodium phosphate buffer (6, 7), 50 mM tris–Cl (pH 8, 9), 50 mM glycine–NaOH buffer (pH 10, 11), and 50 mM Na2HPO4–NaOH buffer (pH 12).
Effect of Organic Solvents on Stability
Nine different organic solvents including methanol, ethanol, n-propanol, n-butanol, acetone, dichloromethane, acetonitrile, toluene, and hexane were used for determining effect of these solvents on lipase stability. Each solvent (25% v/v) was incubated with enzyme for 30 min and residual activity (%) against control without any solvent was calculated.
Substrate Specificity of Lipase
4-Nitrophenyl esters including pNP-octanoate (C8), pNP-decanoate (C10), pNP-dodecanoate (C12), pNP-myristate (C14), and pNP-palmitate (C16) were used for determination of substrate specificity of partially purified lipase. 10 mM of each substrate was dissolved in ethanol and 25 µl of substrate mixed with 475 µl of 1% triton X-100 in water was added to 480 µl of tris–Cl buffer (50 mM, pH 8.0). Enzyme assay was performed at 20 °C using 20 µl of enzyme.
Statistical Analysis
One-way ANOVA following Duncan’s test was performed for measuring statistical difference (p < 0.05) in enzyme production between different treatments. Mean and standard error of three replicates were calculated using Microsoft Excel 2013 software.
Results
Biochemical Characteristics and Identification of the Bacterium
GBPI_Hb61 produced fluorescent light yellow coloured colonies on TY agar at 25 °C following 24 h of incubation. The bacterial cells were observed as tiny rods which were Gram negative and scattered under oil immersion. The bacterium could only utilize dextrose sugar among all the tested carbohydrates. Temperature tolerance by GBPI_Hb61 was in the range of 5–35 °C suggesting it as a psychrotolerant bacterium with a wide range of pH tolerance (pH 2–14). In extreme acidic (pH 2) and alkaline pH (12–14), the bacterium showed restricted growth. Salt tolerance was recorded up to 10% with no restriction in cell biomass. All characteristics of GBPI_Hb61 are summarized in Table 1.
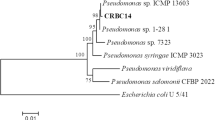
Following 16S rRNA sequencing and BLAST analysis using EzTaxon, the bacterium was identified as P. proteolytica (99.54% similarity). A phylogenetic tree to understand the evolutionary relation with closely matched bacteria was also re-constructed, which is shown in Fig. 1. The tree indicates its close relation with other psychrophilic Pseudomonas spp. isolated from Antarctic/polar regions.
Lipase Production by the Bacterium
Bacteria showed lipase production in tributyrin agar indicated by zone of clearance while orange fluorescence was observed following olive oil utilization in rhodamine-B agar around the bacterial colony. Prolonged production of lipase by the bacterium i.e., up to 10 days of incubation (at 25 °C) was investigated during quantitative estimation of lipase.
Effect of Temperature and pH on Lipase Production
Temperature played a significant role to facilitate enzyme production by the bacterial isolate (Fig. 2A). Out of the four temperatures screened, the bacterium showed maximum lipase production at 25 °C followed by production at 14 °C. Production of lipase enzyme initiated from 2nd day of incubation at 25 °C which continued till 10th day after which enzyme production reached a plateau. Production at 35 °C was completely seized while at 4 °C low enzyme production was recorded. Enzyme production at 14 °C as well as 4 °C was found increasing with further incubation. Maximum enzyme activity on 10th day of incubation at 25, 14 and 4 °C was recorded as 33.29, 21.94, and 6.6 U/ml, respectively.
The pH did not affect enzyme production in initial stages of incubation while in later stages acidic pH (pH 5) aided higher enzyme production. Maximum production (39.81 U/ml) was achieved at pH 5 after 10 days of incubation. Alkaline pH (pH 11) inhibited the enzyme production by GBPI_Hb61 (Fig. 2B).
Effect of Oil and Nitrogen Source and Surfactant on Enzyme Production
The bacterium was able to equally utilize all tested oils as substrate for lipase production, except tributyrin oil. No significant difference (p < 0.05) between enzyme activities was observed when corn, soybean, and olive oil were used as sole source of carbon for lipase induction on day 10 of incubation (Fig. 3A). When screening various organic nitrogen and inorganic nitrogen sources, yeast extract as organic nitrogen source was found to be necessary for lipase production, while other tested N-source inhibited the production of enzyme. A significant difference (p < 0.05) in enzyme production was observed when control was compared with different treatments. Sodium nitrate did not affect the enzyme production in initial incubation stages in comparison to when no inorganic nitrogen source was added in the production media (Fig. 3B). Yeast extract (34.94 U/ml) as organic nitrogen source followed by casein (28.19 U/ml) and peptone (27.32 U/ml) were found to be the best supplement for lipase production (Fig. 3C).
Effect of A oil source, B inorganic nitrogen, C organic nitrogen source, and D non-ionic detergent on lipase production. AN ammonium nitrogen, AC ammonium chloride, AS ammonium sulphate, Pep peptone, ME malt extract, Cas casein, U urea, N 0 no nitrogen source (inorganic/organic, respectively), TX-100 triton X-100, T tween. Different alphabets in a bar group indicate significant difference (p < 0.05) calculated by Duncan’s test
In another experiment, where effect of different non-ionic detergents was tested, no detergent was able to enhance lipase production in comparison to control. Although triton X-100 showed no significant difference (p < 0.05) in lipase production in comparison to control, tweens significantly (p < 0.05) inhibited lipase production (Fig. 3D).
Partial Purification of Lipase
Among the two fractions (0–50% and 50–80%), obtained after ammonium sulphate fractionation, 0–50% fraction contained active lipase which was confirmed by performing activity assay for both the fractions. No activity was observed in 50–80% fraction. Therefore, active fraction containing lipase was utilized for further studies. Specific activity of active fraction was calculated to be 262.93 U/mg of protein in comparison to 79.08 U/mg of protein in case of crude supernatant.
Aggregation Studies
Gel Filtration Chromatography
During gel filtration of ammonium sulphate precipitate, active lipase fraction was collected in void volume due to high molecular weight aggregates of lipase or formation of oligomers. Elution profile of gel filtration performed using Sephadex G-100 is shown in Fig. 4, which suggest size of native protein to be more than 250 kDa.
Dynamic Light Scattering
DLS in the presence of different compounds showed no effect on lipase aggregation except NP-40. NP-40 was able to disaggregate lipase but only to a little extent reflected by shifting of curve towards lower size (Z-average decreased to 26.17 d.nm in comparison to 80.70 d.nm for control). DLS profile of protein samples for molecular size distribution obtained in the presence of various detergents and compounds is shown in Fig. 5.
Native PAGE
On the basis of native PAGE followed by in-gel enzyme activity analysis, high molecular weight aggregates of >250 kDa were observed. When same gel was stained with CBB, dark protein bands of large molecular weight were visible near interface of stacking and resolving gel. NP-40, as suggested by DLS analysis also, was able to break the large aggregates to some extent and therefore active bands were visible near 200 kDa. Also, SDS to some scope was able to break the large aggregates of lipase indicated by the absence of dark band near gel interface but activity could not be detected in gel may be due to denaturation of lipase in the presence of SDS (Fig. 6). Rest of the treatments were ineffective in breaking the protein aggregates.
Native PAGE analysis followed by in-gel enzyme activity after sample treatment with various detergents and reagents showing large molecular weight aggregates of lipase (indicated by arrows). Lane 1 0.5% NP-40; 2 0.1% Brij-35; 3 50 mM imidazole; 4 0.5% SDS; 5 10 mM CHAPS; 6 5% PEG; 7 0.5 M arginine; 8 10% glycerol; 9 control
Molecular Weight Determination by SDS-PAGE
SDS-PAGE was first performed in non-reducing condition followed by removal of SDS from gel and development of zymogram. Two active bands very close to each other, with minor difference in their molecular weight, were visible. This was further confirmed by staining same gel with CBB. Two protein bands which could not be resolved from each other in gel showed molecular weight of approximately 50 kDa. Similar protein bands were visible when SDS-PAGE was performed under reducing condition (Fig. 7).
Characterization Studies on Lipase
Effect of Temperature and pH on Lipase Activity and Stability
GBPI_Hb61 lipase showed maximum activity at 40 °C (relative activity of 136.22%) while it retained 88.85% activity at low temperature (10 °C) in comparison to the activity recorded at 20 °C. Similarly, lipase produced by bacterial isolate was almost equally stable from 10 to 50 °C, while sharp decrease in stability was recorded at 60 °C (Fig. 8A). Optimum pH for lipase activity was observed to be pH 8 with no significant variation at pH 9 as well as pH 10. In contrast, lipase was equally stable in pH ranging from 6 to 11 even after 24 h. This indicates wide pH stability of lipase for its use in acid as well as alkaline conditions (Fig. 8B).
Effect of Organic Solvents
Stability of lipase produced by GBPI_Hb61 decreased when incubated with solvents of various polarities except methanol where small increase (103.45% residual activity) in activity was recorded. Decrease in stability along with decrease in solvent polarity was observed. Residual activity of lipase, when incubated with different organic solvents, is given in Table 2. More than 75% residual activity was observed only in the presence of hexane, methanol, and acetone.
Substrate Specificity of Lipase
Lipase caused maximum hydrolysis (relative activity of 227.65%) of pNPdecanoate (C10) while pNPoctanoate and pNPdodecanoate were equally utilized as substrates. This showed substrate specificity if GBPI_Hb61 lipase towards medium chain esters. Specificity for higher carbon chain (C14 and C16) ester was recorded very less (Fig. 9).
Discussion
The present study explains isolation of lipase from a newly isolated strain of P. proteolytica which has been isolated from high altitude in IHR. The bacterium itself was found to possess several extremophilic traits including tolerance to low temperature (psychrotolerant), wide range of pH as well as salt concentration equivalent to 1.7 M (halotolerant). Soil in Himalayan region are reported to inhabit microbes with several extremophilic characteristics [20, 36]. These studies explain the richness of Himalayan soil with novel microbiota which still remains unmapped. Answer to their adaptation capabilities in harsh condition might be explored after digging into their genome and finding out their evolutionary relation with their ancestors. In the present study, phylogenetic tree explains closeness of GBPI_Hb61 with that of several psychrophilic microorganisms including P. antarctica, P. extremaustralis, and P. yamanorum, which has been originally isolated from extreme colder environments [37, 38].
Extremophiles are the source for several enzymes for their multiple functionality in several industrial sectors. Pseudomonas, being an ecologically successful microorganism [39], has been a choice for the production of several active metabolites including cold-active lipases. This study also explains the production and properties of a cold-active lipase from a psychrotolerant Pseudomonas species. It was interesting to find that the bacterium produced lipase in the production medium only when incubated at or below 25 °C. Production was recorded maximum at 25 °C with slow but continuous enhancement in lipase production at low temperatures. Rashid et al. [40] reported production of a cold-active lipase from Pseudomonas sp. strain KB700A when grown below 25 °C, while no activity in culture supernatant was observed at 30 °C. Maharana and Ray [18] also reported minimum lipase (CALip5) production by a psychrotolerant Pseudomonas sp. AKM-L5 at 35 °C. The production of lipase at higher pH (9 and above) was significantly inhibited while at pH 5 and 7 not much variation in lipase production was observed. The pH of culture supernatant checked (data not included) after lipase production lied between pH 7.5 and 8.0 and is suggested to be the ideal medium pH for lipase production by GBPI_Hb61. In a similar study, optimum production of a lipase from Aeromonas caviae AU04 has been observed between pH 7 and 8 [41].
The bacterium, in the present study, utilized all tested oil sources as sole carbon source for the production of lipase. As lipase is an inducible enzyme and require lipid source for its production, oil in medium is necessary to achieve lipase production. While tributyrin being a short chain fatty acid ester could not facilitate lipase production at par to other oil source indicating production of lipolytic enzyme in the production medium as true lipase. In a recent study on Botryococcus sudeticus, Yong et al. [42] observed maximum lipase production while using olive oil as an inducer. In contrast, Bisht et al. [43] and Kulkarni and Garde [44] found castor oil as suitable oil source for the production of lipase by P. aeruginosa and P. fluorescens, respectively. Difference in fatty acid composition of different oils might account for the variation in lipase production.
Apart from the oil source as lipase inducer, nitrogen source in the medium played a critical role in lipase production by GBPI_Hb61. Through this study, it was conferred that organic nitrogen source was a limiting nutrient for lipase production by GBPI_Hb61 as no lipase production was observed in its absence while it was not true for inorganic nitrogen. It has been observed that, in general, microorganisms give high yield of lipase when organic nitrogen is used in the production media [43, 45]. Contrary to this, Kiran et al. [46] indicated that the supply of nitrogen is not an absolute requirement for lipase production from Pseudomonas MS1057. While examining the effect of detergents on lipase production, no significant enhancement was observed using triton X-100. The results are in contrast to the study by Lin et al. [47], where they observed increased lipase from P. pseudoalcaligenes by 50-fold on addition of triton X-100 in the medium but in support where they observed inhibitory effect of tween.
Similar to several earlier studies [11, 41, 48–50] on lipases, lipase aggregation in the production medium as well as while purifying lipase making it difficult to purify to homogeneity was also noticed in the present study. Various attempts to purify lipase using chromatographic techniques including gel filtration, anion exchange, and hydrophobic interaction chromatography (data not shown) were unsuccessful. It was considered that the aggregation of lipase is responsible for this failure; hence, the aggregation behaviour of lipase was studied and confirmed using several techniques including DLS (Fig. 5) and native PAGE (Fig. 6). Aggregated lipase in native PAGE was not able to move beyond the gel interface even in 7.5% polyacrylamide gel (data not shown) due to its high molecular weight. Native PAGE followed by in-gel activity confirmed formation of these aggregates. Data based on DLS analysis and native PAGE in the presence of several detergents and reagents indicate their inability to disaggregate lipase to full extent. Further work is needed to disaggregate the lipase from GBPI_Hb61.
Non-reducing SDS-PAGE showed the presence of two lipase bands in gel having minor difference in their molecular weight. The one with high molecular weight as well as with higher activity was not visible in CBB stained gel due to smear formation in the gel (Fig. 7). This could be due to the formation of micelle structure by interaction with SDS in gel. Molecular weight of both the lipases was detected to be approximately 50 kDa after SDS-PAGE while native PAGE as well as gel filtration chromatography showed the molecular weight of lipase aggregates to be >250 kDa. Rashid et al. [40] and Chung et al. [51] reported molecular weight of lipases from Pseudomonas (KB700A) and P. fluorescens (SIK W1) to be 49.9 and 50 kDa, respectively. Maharana and Ray [18] recently reported a lipase of 57 kDa from psychrotolerant Pseudomonas sp. AKM-L5. The lipA gene, a structural gene encoding for lipase of molecular mass 48 kDa from P. aeruginosa B2264, has been expressed by Madan and Mishra [52]. On the other side, large molecular weight of aggregates of lipase has been described by various researchers. Ugras and Uzmez [11] reported that lipase from Acinetobacter sp. (SU15) forms very large, active aggregates (>250 kDa) during native-PAGE. Similarly, thermophilic alkaline lipases from Thermosyntropha lipolytica also form high molecular mass aggregates (≥280 kDa) on a gradient native-PAGE [53].
Interestingly, the lipase produced by GBPI_Hb61 was found to be stable in wide range of temperature as well as pH. While the enzyme showed maximum activity at 40 °C in pH 8, it was active as well as stable in low temperature allowing use of GBPI_Hb61 lipase for low temperature catalysis such as cold washing, bioremediation in cold environments, etc. In the present study, a decline in enzyme activity and stability above 50 °C was recorded. According to Joseph et al. [3], most of the cold-active lipases are unstable above 65 °C. Unlike other cold-active lipases which easily get deactivated when subjected to extreme pH range [11, 54, 55], stability of GBPI_Hb61 lipase over wide pH was observed. More than 80% activity was retained between pH 5.0 and 12.0. A cold-active lipase from Pseudomonas sp. also showed stability at wide range of pH from pH 2 to 11 [18]. In the present study, cold-active lipase produced by GBPI_Hb61 utilized substrate with mid acyl chain esters (C8 to C12). Maximum activity was recorded against pNP ester with C10, which was more than twofold higher in comparison to C8 and C12 acyl esters (Fig. 9). Similar results were reported by Rashid et al. [40], who showed maximum activity of KB-Lip towards pNPcaprate (C10). Lee et al. [12] and Velu et al. [41] also reported that for lipase of Aeromonas sp. (LPB 4) and A. caviae (AU04), medium chain acyl group pNP esters appear to be good substrates. While several lipases have been reported to be active in the presence of organic solvents [56, 57], residual activity of GBPI_Hb61 was found to be decreased in the presence of solvents of different polarities. Overall, GBPI_Hb61 is a potential isolate for cold-active lipase production for use in detergent industry for formulating detergents active in low temperatures. The production cost of enzyme can be reduced by using low-cost oil source as GBPI_Hb61 is able to use several oils for lipase production. The lipase produced by GBPI_Hb61 was found to be functional and stable under various conditions of temperature and pH making it useful for several biotechnological applications. Further investigations on enzyme purification are needed to unleash its full potential.
Conclusion
Research on microorganisms from low-temperature environments extent opportunity to harness their potential in biotechnological applications. To the best of our knowledge, this appears to be the first report on the lipolytic potential of a psychrotolerant strain of P. proteolytica. The bacterium can be utilized for lipase production using low-cost substrates to reduce the cost of production. Cold and pH activity as well as stability may be useful for harnessing the enzyme in wide range of industrial as well as environmental uses including cold washing, bioremediation in cold environments, etc. Further, advance studies, including, genomics and proteomics, will be important in understanding the adaptation strategies associated with these organisms.
Abbreviations
- pNP:
-
p-Nitrophenol
- CHAPS:
-
3-[(3-Cholamidopropyl)dimethylammonio]-1-propane sulfonate
- NP-40:
-
Nonylphenoxy polyethoxyethanol
- Brij-35:
-
Polyethylene glycol dodecyl ether
- SDS:
-
Sodium dodecyl sulphate
- PEG:
-
Poly ethylene glycol
References
Gupta, R., Hupta, N., & Rathi, P. (2004). Bacterial lipases: an overview of production, purification and biochemical properties. Applied Microbiology and Biotechnology, 64, 763–781.
de Godoy Daiha, K., Angeli, R., de Oliveira, S. D., & Almeida, R. V. (2015). Are lipases still important biocatalysts? A study of scientific publications and patents for technological forecasting. PLoS ONE, 10(6), e0131624. doi:10.1371/journal.pone.0131624.
Joseph, B., Ramteke, P. W., & Thomas, G. (2008). Cold active microbial lipases: Some hot issues and recent developments. Biotechnology Advances, 26, 457–470.
Haldane, J. B. S. (1930). Especificity of the lipases. In: R.H.A. Plimmer & F.G. Hopkins (Eds.), Monographs on biochemistry–enzymes (pp. 102), 1st ed. Logmans: Green and Co.
Tan, C. H., Show, P. L., Ooi, C. W., Ng, E. P., Lan, J. C., et al. (2015). Novel lipase purification methods—a review of the latest developments. Biotechnology Journal, 10, 31–44. doi:10.1002/biot.201400301.
Kulakovaa, L., Galkina, A., Nakayamab, T., Nishinob, T., & Esakia, N. (2004). Cold-active esterase from Psychrobacter sp. Ant300: gene cloning, characterization, and the effects of Gly Pro substitution near the active site on its catalytic activity and stability. Biochimica et Biophysica Acta, 1696, 59–65.
Zeng, X., Xiao, X., Wang, P., & Wang, F. (2004). Screening and characterization of psychrotrophic lipolytic bacteria from deep sea sediments. Journal of Microbiology and Biotechnology, 14, 952–958.
Ryu, H. S., Kim, H. K., Choi, W. C., Kim, M. H., Park, S. Y., et al. (2006). New cold-adapted lipase from Photobacterium lipolyticum sp. nov. that is closely related to filamentous fungal lipases. Applied Microbiology and Biotechnology, 70, 321–326.
Suzuki, T., Nakayama, T., Kurihara, T., Nishino, T., & Esaki, N. (2001). Cold-active lipolytic activity of psychrotrophic Acinetobacter sp. strain no. 6. Journal of Bioscience and Bioengineering, 92, 144–148.
Kasan, R. C., Kaur, B., & Yadav, S. K. (2008). Isolation and identification of a psychrotrophic Acinetobacter sp. CR9 and characterization of its alkaline lipase. Journal of Basic Microbiology, 48, 207–212. doi:10.1002/jobm.200700160.
Ugras, S., & Uzmez, S. (2016). Characterization of a newly identified lipase from a lipase-producing bacterium. Frontiers in Biology, 11(4), 323–330.
Lee, H. K., Min, J. A., Sung, H. K., Won, H. S., & Byeong, C. J. (2003). Purification and characterization of cold active lipase from psychrotrophic Aeromonas sp. LPB4. Journal of Microbiology, 41, 22–27.
Giudice, A. L., Michaud, L., de Pascale, D., Domenico, M. D., & di Prisco, G. (2006). Lipolytic activity of Antarctic cold adapted marine bacteria. Journal of Applied Microbiology, 101, 1039–1048.
Novototskaya-Vlasova, K. A., Petrovskaya, L. E., Kryukova, E., Rivkina, E. M., Dolgikh, D., et al. (2013). Expression and chaperone-assisted refolding of a new cold-active lipase from Psychrobacter cryohalolentis K5T. Protein Expression and Purification, 91, 96–103. doi:10.1016/j.pep.2013.07.011.
Zhang, J., Lin, S., & Zeng, R. (2007). Cloning, expression, and characterization of a cold-adapted lipase gene from an antarctic deep-sea psychrotrophic bacterium, Psychrobacter sp. 7195. Journal of Microbiology and Biotechnology, 17(4), 604–610.
Ali, M. S. M., Fuzi, S. F. M., Ganasen, M., Rahman, R. N. Z. R. A., Basri, M., et al. (2013). Structural adaptation of cold-active RTX lipase from Pseudomonas sp. strain AMS8 revealed via homology and molecular dynamics simulation approaches. BioMed Research International. doi:10.1155/2013/925373.
Alquati, C., de Gioia, L., Santarossa, G., Alberghina, L., Fantucci, P., et al. (2002). The cold-active lipase of Pseudomonas fragi: heterologous expression, biochemical characterization and molecular modelling. European Journal of Biochemistry, 269, 3321–3328.
Maharana, A. K., & Pratima, R. (2015). A novel cold-active lipase from psychrotolerant Pseudomonas sp. AKM-L5 showed organic solvent resistant and suitable for detergent formulation. Journal of Molecular Catalysis B Enzymatic, 120, 173–178. doi:10.1016/j.molcatb.2015.07.005.
Pandey, N., Dhakar, K., Jain, R., & Pandey, A. (2016). Temperature dependent lipase production from cold and pH tolerant species of Penicillium. Mycosphere. doi:10.5943/mycosphere/si/3b/5.
Jain, R., & Pandey, A. (2016). A phenazine-1-carboxylic acid producing polyextremophilic Pseudomonas chlororaphis (MCC2693) strain, isolated from mountain ecosystem, possesses biocontrol and plant growth promotion abilities. Microbiological Research, 190, 63–71. doi:10.1016/j.micres.2016.04.017.
Dhakar, K., Jain, R., Tamta, S., & Pandey, A. (2014). Prolonged laccase production by a cold and pH tolerant strain of Penicillium pinophilum (MCC 1049) isolated from a low temperature environment. Enzyme Research. doi:10.1155/2014/120708.
Pandey, A., Trivedi, P., Kumar, B., & Palni, L. M. S. (2006). Characterization of a phosphate solubilizing and antagonistic strain of Pseudomonas putida (B0) isolated from a sub-alpine location in the Indian Central Himalaya. Current Microbiology, 53, 102–107.
Dhakar, K., & Pandey, A. (2016). Wide pH range tolerance in extremophiles: towards understanding an important phenomenon for future biotechnology. Applied Microbiology Biotechnology, 100, 2499–2510.
Pandey, A., & Palni, L. M. S. (1998). Microbes in Himalayan soils: Biodiversity and potential applications. Journal of Scientific & Industrial Research, 57, 668–673.
Yabuchi, E., Kosako, Y., Oyaizu, H., Yano, I., Hotta, H., et al. (1992). Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiology and Immunology, 36(12), 1251–1275.
Chen, W., & Kuo, T. (1993). A simple rapid method for the preparation of gram negative bacterial genomic DNA. Nucleic Acids Research, 21(9), 2260.
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., et al. (2012). Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. International Journal of Systematic Evolutionary Microbiology, 62, 716–721. doi:10.1099/ijs.0.038075-0.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Shu, Z., Lin, R., Jiang, H., Zhang, Y., Wang, M., et al. (2009). A rapid and efficient method for directed screening of lipase-producing Burkholderia cepacia complex strains with organic solvent tolerance from rhizosphere. Journal of Bioscience and Bioengineering, 107(6), 658–661.
Kanwar, S. S., Kaushal, R. K., Jawed, A., Gupta, R., & Chimni, S. S. (2005). Methods of inhibition of residual lipase activity in colorimetric assay: A comparative study. Indian Journal of Biochemistry and Biophysics, 42, 233–237.
Pinsirodom, P., & Parkin, K. L. (2001). Current protocols in food analytical chemistry. New York: Wiley.
Bradford, M. M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein dye binding. Annals of Biochemistry, 72, 248–254.
Acharya, P., & Rao, N. M. (2003). Stability studies on a lipase from Bacillus subtilis in guanidinium chloride. Journal of Protein Chemistry, 22, 51–60.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227(5259), 680–685.
Prim, N., Sánchez, M., Ruiz, C., Pastor, F. I. J., & Díaz, P. (2003). Use of methylumbeliferyl-derivative substrates for lipase activity characterization. Journal of Molecular Catalysis B Enzymatic, 22, 339–346.
Pandey, A., Dhakar, K., Sharma, A., Priti, P., Sati, P., et al. (2015). Thermophilic bacteria, that tolerate wide temperature and pH range, colonize the Soldhar (95 °C) and Ringigad (80 °C) hot springs of Uttarakhand, India. Annals of Microbiology, 65, 809–816.
Arnau, V. C., Sánchez, L. A., & Delgado, O. D. (2015). Pseudomonas yamanorum sp. nov., a new psychrotolerant bacterium isolated from Sub-Antarctic environment (Tierra del Fuego, Ushuaia). International Journal of Systematic Evolutionary Microbiology, 65, 424–431. doi:10.1099/ijs.0.065201-0.
López, N. L., Pettinari, M. J., Stackebrandt, E., Tribelli, P. M., Põtter, M., et al. (2009). Pseudomonas extremaustralis sp. nov., a poly (3-hydroxybutyrate) producer isolated from an antarctic environment. Current Microbiology, 59, 514–519. doi:10.1007/s00284-009-9469-9.
Moreno, R., & Rojo, F. (2014). Features of Pseudomonas growing at low temperatures: another facet of their versatility. Environmental Microbiology Reports. doi:10.1111/1758-2229.12150.
Rashid, N., Shimada, Y., Ezaki, S., Atomi, H., & Imanaka, T. (2001). Low-temperature lipase from psychrotrophic Pseudomonas sp. strain KB700A. Applied and Environmental Microbiology, 67, 4064–4069.
Velu, N., Divakar, K., Nandhinidevi, G., & Gautam, P. (2012). Lipase from Aeromonas caviae AU04: Isolation, purification and protein aggregation. Biocatalysis and Agricultural Biotechnology. doi:10.1016/j.bcab.2011.08.004.
Yong, S. K., Lim, B. H., Said, S. & Tey, L.-H. (2016). Optimisation, purification and characterisation of extracellular lipase from Botryococcus sudeticus (UTEX 2629). Journal of Molecular Catalysis B Enzymatic. doi:10.1016/j.molcatb.2016.02.004.
Bisht, D., Yadav, S. K., Gautam, P., & Darmwal, N. D. (2012). Simultaneous production of alkaline lipase and protease by antibiotic and heavy metal tolerant Pseudomonas aeruginosa. Journal of Basic Microbiology, 52, 1–8. doi:10.1002/jobm.201200157.
Kulkarni, N., & Garde, R. V. (2002). Production and properties of an alkaline thermophilic lipase from Pseudomonas fluorescens NS2W. Journal of Industrial Microbiology and Biotechnology, 28, 344–348.
Sharma, R., Soni, S. K., Vohra, R. M., Jolly, R. S., Gupta, L. K., et al. (2002). Production of extracellular lipase from Bacillus sp. RSJ1 and its application in ester hydrolysis. Indian Journal of Microbiology, 42, 49–54.
Kiran, G. S., Shanmughapriya, S., Jayalakshimi, J., Selvin, J., Gandhimathi, R., et al. (2008). Optimization of extracellular psychrophilic alkaline lipase produced by marine Pseudomonas sp. (MSI057). Bioprocess and Biosystems Engineering, 31, 483–492. doi:10.1007/s00449-007-0186-0.
Lin, S., Chiou, C., & Tsai, Y. (1995). Effect of triton X-100 on alkaline lipase production by Pseudomonas pseudoalcaligenes F-111. Biotechnology Letters, 17, 959. doi:10.1007/BF00127434.
Snellman, E. A., Sullivan, E. R., & Colwell, R. R. (2002). Purification and properties of the extracellular lipase, LipA, of Acinetobacter sp. RAG-1. European Journal of Biochemistry, 269, 5771–5779. doi:10.1046/j.1432-1033.2002.03235.x.
Dandavate, V., Jinjala, J., Keharia, H., & Madamwar, D. (2009). Production, partial purification and characterization of organic solvent tolerant lipase from Burkholderia multivorans V2 and its application for ester synthesis. Bioresource technology, 100, 3374–3381. doi:10.1016/j.biortech.2009.02.011.
de Lima, L. N., Aragon, C. C., Mateo, C., Palomo, J. M., Giordano, R. L. V., et al. (2013). Immobilization and stabilization of a bimolecular aggregate of the lipase from Pseudomonas fluorescens by multipoint covalent attachment. Process Biochemistry, 48, 118–123. doi:10.1016/j.procbio.2012.11.008.
Chung, G. H., Lee, Y. P., Jeohn, G. H., Yoo, O. J., & Rhee, J. S. (1991). Cloning and nucleotide sequence of thermostable lipase gene from Pseudomonas fluorescens SIK W1. Agricultural and Biological Chemistry, 55, 2359–2365.
Madan, B., & Mishra, P. (2010). Co-expression of the lipase and foldase of Pseudomonas aeruginosa to a functional lipase in Escherichia coli. Applied Microbiology and Biotechnology, 85, 597. doi:10.1007/s00253-009-2131-4.
Salameh, M. A., & Wiegel, J. (2010). Effects of detergents on activity, thermostability and aggregation of two alkali thermophilic lipases from Thermosyntropha lipolytica. Open Biochemistry Journal, 4, 22–28.
Matsumoto, M., Kida, K., & Kondo, K. (2001). Enhanced activities of lipase pretreated with organic solvents. Journal of Chemical Technology and Biotechnology, 76, 1070–1073.
Noel, M., & Combes, D. (2003). Effects of temperature and pressure on Rhizomucor miehei lipase stability. Journal of Biotechnology, 102, 23.
Suen, W. C., Zhang, N., Xiao, L., Madison, V., & Zaks, A. (2004). Improved activity and thermostability of Candida antarctica lipase B by DNA family shuffling. Protein Engineering Design and Selection, 17, 133–140.
Li, X.-L., Shi, Y., Zhang, W.-H., Dai, Y.-J., Zhang, H.-T., et al. (2014). A high-detergent-performance, cold-adapted lipase from Pseudomonas stutzeri PS59 suitable for detergent formulation. Journal of Molecular Catalysis B Enzymatic, 102, 17–24. doi:10.1016/j.molcatb.2014.01.006.
Acknowledgements
The authors are thankful to Dr. P.P. Dhyani (Director, G.B. Pant National Institute of Himalayan Environment and Sustainable Development, India) for extending the facilities. Financial support from Ministry of Environment, Forest and Climate Change is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jain, R., Pandey, A., Pasupuleti, M. et al. Prolonged Production and Aggregation Complexity of Cold-Active Lipase from Pseudomonas proteolytica (GBPI_Hb61) Isolated from Cold Desert Himalaya. Mol Biotechnol 59, 34–45 (2017). https://doi.org/10.1007/s12033-016-9989-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-016-9989-z