Abstract
Streptococcus thermophilus is a lactic acid bacterium and used as starter culture in the dairy industry, mainly in the manufacture of yoghurt, with Lactobacillus delbrueckii subsp. bulgaricus. It produces lactic acid as a major fermentation end product and some carbonyl compounds through sugar metabolism. The level of metabolites could be improved using molecular biotechnology. The genes of als, encoding α-acetolactate synthase (Als), the pflA, encoding pyruvate-formate lyase activating enzyme (PflA), and the adhB which encodes alcohol dehydrogenase (AdhB) of S. thermophilus NCFB2393 strain were amplified by polymerase chain reaction and separately cloned into the overexpression vector pNZ276 under the control of the lacA promoter. The strains were transformed individually with the constructed plasmids. Their abilities to generate important metabolites such as pyruvate, lactate, formate, acetaldehyde, acetoin, ethanol, and 2,3-butanediol in LM17 medium were analyzed using high-performance liquid chromatography. High level of 2,3-butanediol was obtained by overexpressing the als gene. The level of formate increased slightly by overexpressing the pflA gene. The overexpression of the adhB gene, on the other hand, resulted in a significant increase in the ethanol level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Streptococcus thermophilus is an important lactic acid bacterium in the dairy industry and has long been used in combination with Lactobacillus delbrueckii subsp. bulgaricus as a starter culture for yoghurt production. It has also been used in the manufacture of certain types of cheese. The main role of S. thermophilus in fermentation is to acidify the environment through the conversion of lactose into lactic acid and to produce a variety of compounds that contribute to flavor and texture of the final product. S. thermophilus is a “generally recognized as safe” (GRAS) species and it is the second important industrial strain following Lactococcus lactis. S. thermophilus belongs to the group of the thermophilic lactic acid bacteria and it is used not only by itself but also in combination with other species in dairy fermentations [16]. S. thermophilus can metabolize five different sugars such as lactose, sucrose, glucose, galactose, and fructose [34] and can degrade all amino acids [27]. Therefore, S. thermophilus strains show better growth than L. bulgaricus strains when grown in pure culture. However, the S. thermophilus—L. bulgaricus cooperation stimulates the growth of S. thermophilus strain [11].
Ten S. thermophilus strain genomes have been sequenced and published until 2015. The genomes and their four completed direct submission sequences are present in the NCBI Genome database [7, 18, 20, 21, 31–33, 36]. Most of the transport proteins and the proteins that have a role in energy metabolism related to carbohydrate degradation, uptake, and fermentation are pseudogenes. S. thermophilus is an obligate homolactic bacterium and produces l-lactate as main end products. The production of low-level additional fermentation end products, such as formate, acetoin, diacetyl, acetaldehyde, α-acetolactate, and acetate by S. thermophilus has also been reported [26, 28]. l-lactate dehydrogenase (Ldh), pyruvate-formate lyase (Pfl), pyruvate-formate lyase activating enzyme (PflA), and the α-acetolactate (Als) enzyme are known to be active in S. thermophilus pyruvate pathways. S. thermophilus has alternative routes other than lactate dehydrogenase for the production of certain metabolites; the phosphotransacetylase (EutD) and acetate kinase (AckA) pathways for acetate production, the α-acetolactate decarboxylase (AldB) route for acetoin production, and alcohol dehydrogenase (AdhB) for ethanol production [16]. Alcohol/acetaldehyde dehydrogenase genes (ψadhA and ψadhE) and the diacetyl/acetoin reductase genes (ψbutA, Ψstr0909, Ψstr0910) were determined as pseudogenes in completed genomes sequences (Fig. 1).
Pyruvate metabolic pathway of Streptococcus thermophilus. Existence of the genes in all completed genomes was searched. ldh l-lactate dehydrogenase, pfl/pflA pyruvate-formate lyase/pyruvate-formate lyase activating enzyme, als acetolactate synthase, aldB alpha-acetolactate decarboxylase, eutD phosphotransacetylase, ackA acetate kinase, gapN NADP-dependent glyceraldehyde-3-phosphate, butA, str0909 and str0910 acetoin reductase, nox NADH oxidase, acoABL acetoin dehydrogenase complex, glyA serine hydroxymethyltransferase, adhB, str0880, str0881, str0882 alcohol dehydrogenase, Ψstr1879-1884 aldehyde dehydrogenase, Str_1034-1035 pyruvate dehydrogenase, and pdc pyruvate decarboxylase are present in Z. mobilis. ψ: pseudogenes
Inactivation of the ldh gene was attempted to change the functionality of the pyruvate dissipating pathways and for the implication of its role in the formation of aromatic products. However, inactivation of ldh gene was unsuccessful due to the absence of an alternative route for NAD+ regeneration from pyruvate as shown by the reconstruction of pyruvate metabolism in S. thermophilus [16]. If the genes coding an enzyme involved in conversion of pyruvate to aromatic compounds can be overexpressed, the production of desired metabolites will increase. In this study, we describe a metabolic engineering approach to achieve high levels of enzyme production. For this purpose, we amplified the als, pflA, and adhB genes of S. thermophilus NCFB 2393 strain by PCR, cloned them into overexpression vector pNZ276, and created three new transformant plasmids. Effects of constructed plasmids on metabolite production were detected in aerobic conditions.
Materials and Methods
Bacterial Strains and Growth Conditions
The bacterial strains and plasmids used in this study are listed in Table 1. S. thermophilus NCFB 2393 [2] strains were routinely cultured in M17 medium supplemented with 1 % (w/v) lactose (LM17 medium) at 42 °C. Escherichia coli strains were grown on L broth or L agar [35] in a shaking incubator at 37 °C. Agar plate media contained 1.5 % (w/v) Bacto agar and antibiotic selection was used when appropriate for E. coli, 100 μg/mL ampicillin, 15 μg/mL of chloramphenicol and 200 μg/mL of streptomycin, and for S. thermophilus NCFB 2393 5 μg/mL of chloramphenicol.
Molecular Cloning Techniques
Plasmid DNA isolation, manipulation of DNA, and transformation of E. coli strains with recombinant plasmids were carried out as described previously [1, 19]. Restriction enzymes and other DNA-modifying enzymes from various sources were used according to the suppliers’ recommendations. Constructed plasmids were introduced into E. coli by transformation using calcium chloride method, and introduced into S. thermophilus by electroporation according to Mollet et al. method [24]. Transformant colonies were selected on LB and LM17 agar plates containing appropriate antibiotics, respectively. Chromosomal DNA was isolated from S. thermophilus NCFB 2393 [2] strain according to the procedure reported by Vos et al. [35].
The als, pflA, and adhB genes were amplified by PCR. In order to clone the als, pflA, and adhB genes, the published S. thermophilus LMG18311 complete genome sequence (Accession no: NC006448) [7] was used to design the primers. Primers were designed for amplification of the entire als, pflA, and adh genes including ribosome binding sites. Oligonucleotides were designed and obtained from Iontek (Istanbul, Turkey). Appropriate restriction sequence was added to each primer to insert into the overexpression vector pNZ276. The als gene fragment primers were engineered to contain AccIII and XhoI restriction sites in AlsF3 (5′-gcgaaaatccGGAAAGTT GGCTCTACCATTTTA-3′) and AlsR2 (5′-tatccgctcgagTAGTAAAATTCATCT GGCAA-3′) primers, respectively. The als amplification gave 1743 bp length DNA, including open reading frame and ribosome binding site regions. AccIII and XhoI restriction endonuclease sites containing PflAF (5′-tgcttccggaTGTAAAAATCGTTTTTCAAG-3′) and PflAR (5′-gcgaaactcgagTTAATCCAA GGTTTTAACC-3′) primers were used to amplify 841 bp length pflA gene. The adhB gene fragment (1078 bp length) was amplified using PstI and XhoI restriction sites added AdhBF (5′-gcgaaaactgcagACTATAACTAAAGAAATAG-3′) and AdhBR (5′-tatccgctcgagCTAAT CTAAAACAATCA-3′) primers.
The polymerase chain reaction (PCR) was performed in a DNA thermal cycler with the following thermal cycling program: initial cycle, 2 min at 94 °C; next 30 cycles, 1 min at 94 °C; 1 min at annealing temperature; and 2 min at 72 °C. Different annealing temperatures (60, 68, and 55 °C) were applied for the amplification of the als, pflA, and adhB gene fragments, respectively. Colony PCR for routine screening of recombinant clones was performed after 5 min at 95 °C pre-denaturation step. PCR was done in 40 μL of reaction mixture containing 1 μL (20 pmol) forward primer, 1 μL (20 pmol) reverse primer, 1 μL dNTP (1 mM), 4 μL buffer (NH4)2SO4 (10X), 1,6 μL MgCl2, 1 μL DNA polymerase (5 μ/μL) [25 mM Tris–HCl (pH 7.5), 0.1 mM EDTA, 1 mM DTT ve %50 (v/v) glycerol], 1 μL template DNA (~400 ng/mL), abd 29.4 μL dH2O.
Fragments generated for the construction of vectors were amplified with Fermantas proof-reading DNA polymerase (Thermo Scientific, Turkey) and cloned into pJET1.2/blunt Cloning vector using the original CloneJET™ PCR Cloning kit (Thermo Scientific, Turkey) before nucleotide sequence confirmation. For routine PCR screening of recombinant clones, Taq DNA polymerase (Thermo Scientific, Turkey) was used. Ligation reactions were performed according to Sambrook et al. [29]. The nucleotide sequences of PCR-generated fragments were confirmed on purified plasmid DNA with an Applied Biosystems DNA sequencer (model 3130xl). DNA sequencing reactions were conducted using the DNA sequencing kit (ABI BigDye®) supplied by Applied Biosystems. pJET1.2 forward and pJET1.2 reverse primers were used with the DNA sequencing kit. The results of sequencing were analyzed using ChromasPro and Clone Manager 9 program.
One mL overnight culture was centrifuged at 14,000×g for 5 min and then resulting supernatant was filtered and transferred into a fresh eppendorf tube. The sample was then analyzed by HPLC or frozen in a dry ice/ethanol bath, and stored at −20 °C until analysis. HPLC analysis was carried out by diluting the sample onefold in 0.5 % meta-phosphoric acid, and injecting 20 μL of it onto the HPLC system. The separation was performed on a Capcell Pak® 5 μm C18 MG column (150 × 4.6 mm) at 30 °C. A Shimadzu Prominence HPLC apparatus (Shimadzu, Kyoto, Japan) equipped with a PD-M20A diode array detector and two binary gradient pumps (Shimadzu LC-10AT), autosampler (SIL 20AC), column oven (CTO-20AC), and a communication bus module (CBM-20A) with valve unit FCV-11AL was used. Standards of all metabolites were purchased from Sigma-Aldrich (Munich, Germany). The mobile phase consisted of meta-phosphoric acid and HPLC grade water. The HPLC system was calibrated using standards for sodium pyruvate (10 mg/mL), sodium lactate (10 mg/mL), sodium formate (10 mg/mL), acetoin (10 mg/mL), acetaldehyde (10 mg/mL), ethanol (10 mg/mL), and 2,3-butanediol (10 mg/mL) prepared in LM17 broth background. Quantification of the peak height was performed using a PC1000 chromatographic data analysis system (Thermo Separation Products). Average of each metabolite was calculated from the results of three independent analyses.
Results
Construction of Overexpression Vectors Containing the als, pflA, and adhB Genes Under Control of the lacA Promoter
The genome sequence showed that the als, pflA, and adhB genes were transcribed mRNA in S. thermophilus [7]. The als gene encodes acetolactate synthase enzyme (ALS) containing 560 amino acid residues. The PFLA, pyruvate-formate lyase activating enzyme (267 a.a.), and ADHB, alcohol dehydrogenase enzyme (345 a.a.), were encoded by pflA and adhB genes, respectively. The genes containing ribosome binding sites were amplified with appropriate primers (see Fig. 2a–c), ligated into pJET1.2 vector, and transformed into E. coli MC1022.
Construction of the als Overexpression Vector, p131
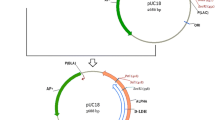
The pJET1.2 plasmid (p121) containing the als fragment was digested with restriction enzymes (Fig. 3a), and the als DNA fragment purified from the agarose gel. The plasmid pNZ276 was also digested (Fig. 3b), and the approximate 4261 bps fragment was purified from the gel. The AccIII/XhoI-digested als fragment was ligated into AccIII and XhoI sites of pNZ276, and transformed into E. coli MC1022. Colonies were screened by PCR using AlsF3 and AlsR2 primers, and then confirmed with AccIII and XhoI digestion. The 1743 bp fragment containing the als gene cloned into pNZ276 vector under control of lacA promoter. After the confirmation with the DNA sequencing, the resulting plasmid was named as p131 (Fig. 4; Table 1). E. coli strain harboring this plasmid was designated as E-MMG131 (Table 1).
a. Agarose gel electrophoresis of AccIII- and XhoI-digested pJET1.2 vector (p121) containing als fragment, Lane 1 is uncut plasmid DNA; Lanes 2 is the plasmid p121 digested with AccIII and XhoI b Agarose gel electrophoresis of the digestion of pNZ276 plasmids with AccIII and XhoI. Lane 1 is uncut pNZ276 plasmid DNA; Lane 2 is the plasmid pNZ276 digested with AccIII and XhoI. Lane M DNA marker
Construction of the pflA Overexpression Vector, p132
The 801-bp length pflA fragment was isolated by digestion with AccIII and XhoI from transformant pJET1.2 plasmid (p122; Table 1) and ligated into AccIII- and XhoI-digested pNZ276 plasmid and transformed into E. coli MC1022. Following colony screening by PCR with PflAF and PflAR primers and sequencing analysis, the plasmid containing pflA fragment was named as p132 (Fig. 4; Table 1). E. coli strain harboring this plasmid was designated as E-MMG132 (Table 1).
Construction of the adhB Overexpression Vector, p134
The 1078-bp length PCR amplified adhB fragment (Fig. 2c) was ligated into pJET1.2 plasmid forming p124 (Table 1). The adhB gene was then excised with PstI and XhoI restriction enzymes from transformant pJET1.2 plasmid. Purified fragment from agarose gel was cloned into PstI- and XhoI-digested pNZ276 plasmid and transformed into E. coli MC1022. After PCR, restriction and sequence analyses, the plasmid containing adhB fragment was named as p134. E. coli strain harboring this plasmid was designated as E-MMG134 (Table 1).
The overexpression plasmids p131, p132, and p134 (Fig. 4) were electroporated separately into S. thermophilus NCFB 2393 strain, and resulting strains named as S-MMG131, S-MMG132, and S-MMG134, respectively (Table 1).
Analysis of Metabolites Production
Metabolic engineering strategies aimed to enhance desired metabolite production by the over production of relevant enzymes. The end of the molecular construction, als ++, pflA ++, and adhB ++ overexpressed S. thermophilus strains were generated and their end product diversion behaviors were analyzed. In order to evaluate the desired metabolite production, the small-scale cell cultures (10 mL) of the reference and the constructed strains carrying the overexpression vector were grown with the lactose substrate. The levels of end products produced from NCFB 2393 strain were used as a reference. Depending on the overexpressed gene, the conversion of the end metabolites may be redirected (Table 2). The supernatants after overnight growth were analyzed by HPLC to measure the effect of the overexpression of the als, pflA, or adhB genes on the metabolic activity. The overexpression of the als gene increases the level of ALS enzyme and this enzyme is regulated by the increasing amount of α-acetolactate, diacetyl, and 2,3-butanediol. The result of metabolite screening showed that the highest amount of end product was produced through 2,3-butanediol conversion. The amount of acetoin, the intermediate metabolite produced from α-acetolactate and diacetyl, slightly increased with the overexpression of the als gene (Table 2).
The overexpression of pflA gene resulted in increased amount of pyruvate diversion to formate. The pathway between pyruvate and formate directed to acetyl-coenzyme, and pyruvate is not the only route to convert formate. As expected, the overexpression of pflA gene increased formate accumulation slightly, and pflA ++ strain showed similarities with the reference strain (Table 2).
Reduction of acetaldehyde to ethanol terminates the fermentative pathways of various ethanol producing microorganisms. This reaction was catalyzed by NAD(P)H-dependent alcohol dehydrogenases enzyme (see Fig. 1). There genes (adhA, adhE, and adhB) were described in acetaldehyde to ethanol pathway. The adhA and adhE genes are pseudogenes and adhB gene overexpressed in adhB ++ strain. When compared to the reference strain, the ethanol production was increased 5.4-fold but acetaldehyde, formate, pyruvate, and lactate production were decreased with the overexpression of the adhB gene (Table 2).
Discussion
Streptococcus thermophilus is a thermophilic species of lactic acid bacteria that can be used as a dairy starter culture for the production of the yoghurt with the combination of Lactobacillus delbrueckii subsp. bulgaricus and a variety of cheese. Enormous gene inactivation and the loss of the most virulence determinants support the ‘generally recognized as safe’ status of S. thermophilus [7]. The aroma and the flavor of yoghurt are fundamentally related to the production of non-volatile and volatile acids and carbonyl compounds [6]. Yoghurt starter cultures generate lactate, acetaldehyde, diacetyl, ethanol, propanone, and 2-butanone metabolites via the pyruvate pathway. Acetaldehyde is responsible for a typical yoghurt aroma and it is the most prominent compound with the concentrations in the range of 5–21 mg/L in the final product [25, 26]. The metabolite formation abilities of starter cultures are important to determine the properties of the fermented product. To improve the metabolite formation abilities of starter cultures by redirecting the metabolic pathways, metabolic engineering strategies are widely used. Redirection of metabolic pathways is important for industrial compound production.
In this study, we have overexpressed the als, pflA, and adhB genes individually in S. thermophilus NCFB 2393. The metabolites were analyzed by HPLC from the cultures grown in LM17 broth medium. The improving level of 2,3-butanediol was obtained by overexpressing the als gene. The level of formate increased slightly by overexpressing the pflA gene. The overexpression of the adhB gene resulted in remarkable increase in the ethanol level. The overexpression of adhB gene in wild-type strain made a clear metabolic shift in the production of metabolic end product.
Completed genome sequence of S. thermophilus showed that butA gene (Fig. 1) taking role in acetoin and 2,3-butanediol productions [16]. However, our results showed that the overexpression of the als gene increased 2,3-butanediol production in S-MMG131 strain. This strain is also able to produce detectable amount of acetoin compared to other strains (Table 2). Therefore, the als gene overexpression with the overexpression of the aldB gene could increase 2,3-butanediol production. In the other study, the overexpression of the als ve ar (acetoin reductase) genes in Klebsiella pneumonia resulted in the high level of 2,3-butanediol production for industrial use [15]. α-ALS, α-acetolactate decarboxylase (ALDC), and acetoin reductase (AR) enzymes catalyze the production of acetolactate from pyruvate, acetoin from acetolactate, and 2,3-butanediol from acetoin, respectively.
In S. thermophilus, threonine is converted into acetaldehyde and glycine by threonine aldolase activity [10]. The serine hydroxymethyltransferase enzyme, which has threonine aldolase activity, was encoded by the glyA gene and the inactivation of the glyA gene resulted in complete loss in acetaldehyde formation during fermentation. When the growth medium was supplemented with l-threonine, acetaldehyde production increased [10]. We previously observed an increase in acetaldehyde production when glyA gene was overexpressed (Akyol et al. unpublished result). In Zymomonas mobilis, the pyruvate decarboxylase (Pdc) enzyme catalyzes the conversion of pyruvate to acetaldehyde (see Fig. 1), which is finally reduced to ethanol by alcohol dehydrogenase [37]. The NADH oxidase (Nox) is known to decrease lactate production by NADH-dependent Ldh under aerobic conditions [12], leading to increased pyruvate availability for alternative reactions. Both pdc and nox genes were overexpressed in L. lactis and Pdc overproduction rerouted the pyruvate metabolism towards acetaldehyde and ethanol. In the reference L. lactis strain, lactate and acetate were main fermentation products under aerobic condition while a clear decrease of lactate production and increased acetoin production were resulted in nox overexpressed strains [8].
The pyruvate-formate lyase (Pfl) enzyme catalyzes the non-oxidative transformation of pyruvate into acetyl-coenzyme A and formate. The regulation of the pfl expression and Pfl activity has been intensively studied in S. bovis [4, 5] and L. lactis [3, 22]. Under anaerobic conditions and in the presence of slowly fermentable sugars, Pfl is responsible for the shift from homolactic to mixed-acid fermentation in L. lactis. Formate production was not detected, which suggests that Pfl is irreversibly inactivated by the exposure to oxygen during preparation of the incubation mixtures [3, 17, 23]. In S. bovis, it was shown that the pfl expression is also dependent on the nature of the fermentable carbon source via a CcpA-dependent mechanism [5]. In the strain S. thermophilus LMG18311, a catabolite-responsive element (cre box) was detected the upstream of the pfl (sequence TGTAAGCGGTTACT, at −92 bp, the upstream of the putative ATG start codon), suggesting that the nature of the carbon source would also modulate the pfl expression in S. thermophilus. Proteomics and transcriptional studies of the pfl gene were made and the Pfl enzyme was just at the detectable level in M17 grown S. thermophilus. When S. thermophilus cultures grown exponentially in LM17 medium were shifted to milk medium for 15 min, a strong increase in the abundance of the pfl mRNA was observed [13]. The pfl transcript abundance was maximal during the exponential phase but drastically decreased at the start of stationary phase. The proteome analysis showed that the same trend was observed at the protein level so Pfl was the strongest overexpressed protein of S. thermophilus LMG18311 during exponential growth in milk [13]. In a mixed-culture transcriptome analysis of S. thermophilus and L. bulgaricus, it was found that the pfl ve pflA were higher expressed in mixed culture, especially in the first exponential phase (3.0- and 4.1-fold, respectively) compared to monocultures. In the pure S. thermophilus culture, the gene pflA and the pathway for purine were upregulated in the transition phase compared to the first exponential phase [30]. S. thermophilus provide L. bulgaricus with crucial components for purine nucleotide biosynthesis, including the precursor formic acid and the cofactor folic acid.
In conclusion, we have successfully cloned the als, pflA, and adhB genes of a S. thermophilus strain. The high level of the enzymes increased the production of 2,3-butanediol and ethanol metabolites. These metabolites can be purified for the industrial use. Additionally, some wild S. thermophilus strains [14] can be transformed with these created constructs in order to see the effects of overexpression on metabolite formation.
References
Akyol, I., Serdaroglu, K., Gezginc, Y., Dayisoylu, K. S., Ekinci, M. S., & Ozkose, E. (2009). Redirection of pyruvate pathway of lactic acid bacteria to improve cheese quality. Food Biotechnology, 23, 200–213.
Almiron-Roig, E., Mulholland, F., Gasson, M. J., & Griffin, A. M. (2000). The complete cps gene cluster from Streptococcus thermophilus NCFB 2393 involved in the biosynthesis of a new exopolysaccharide. Microbiol-SGM, 146, 2793–2802.
Arnau, J., Jorgensen, F., Madsen, S. M., Vrang, A., & Israelsen, H. (1997). Cloning, expression, and characterization of the Lactococcus lactis pfl gene, encoding pyruvate formate lyase. Journal of Bacteriology, 179, 5884–5891.
Asanuma, N., Iwamoto, M., & Hino, T. (1999). Structure and transcriptional regulation of the gene encoding pyruvate formate-lyase of a ruminal bacterium, Streptococcus bovis. Microbiology-UK, 145, 151–157.
Asanuma, N., Yoshii, T., & Hino, T. (2004). Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate lyase in the ruminal bacterium Streptococcus bovis. Applied and Environmental Microbiology, 70, 5244–5251.
Beshkova, D. M., Simova, E. D., Frengova, G. I., Simov, Z. I., & Dimitrov, Z. P. (2003). Production of volatile aroma compounds by kefir starter cultures. International Dairy Journal, 13, 529–535.
Bolotin, A., Quinquis, B., Renault, P., Sorokin, A., Ehrlich, S. D., Kulakauskas, S., et al. (2004). Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nature Biotechnology, 22, 1554–1558.
Bongers, R. S., Hoefnagel, M. H. N., & Kleerebezem, M. (2005). High-level acetaldehyde production in Lactococcus lactis by metabolic engineering. Applied and Environmental Microbiology, 71, 1109–1113.
Casadaban, M. J., & Cohen, S. N. (1980). Analysis of gene control signals by DNA-fusion and cloning in Escherichia coli. Journal of Molecular Biology, 138, 179–207.
Chaves, A. C. S. D., Fernandez, M., Lerayer, A. L. S., Mierau, I., Kleerebezem, M., & Hugenholtz, J. (2002). Metabolic engineering of acetaldehyde production by Streptococcus thermophilus. Applied and Environmental Microbiology, 68, 5656–5662.
Courtin, P., & Rul, F. (2004). Interactions between microorganisms in a simple ecosystem: yogurt bacteria as a study model. Lait, 84, 125–134.
de Felipe, F. L., Starrenburg, M. J. C., & Hugenholtz, J. (1997). The role of NADH-oxidation in acetoin and diacetyl production from glucose in Lactococcus lactis subsp. lactis MG1363. FEMS Microbiology Letters, 156, 15–19.
Derzelle, S., Bolotin, A., Mistou, M. Y., & Rul, F. (2005). Proteome analysis of Streptococcus thermophilus grown in milk reveals pyruvate formate lyase as the major upregulated protein. Applied and Environmental Microbiology, 71, 8597–8605.
Gezginc, Y., Topcal, F., Comertpay, S., & Akyol, I. (2015). Quantitative analysis of the lactic acid and acetaldehyde produced by Streptococcus thermophilus and Lactobacillus bulgaricus strains isolated from traditional Turkish yogurts using HPLC. Journal of Dairy Science, 98, 1426–1434.
Guo, X. W., Zhang, Y. H., Cao, C. H., Shen, T., Wu, M. Y., Chen, Y. F., et al. (2014). Enhanced production of 2,3-butanediol by overexpressing acetolactate synthase and acetoin reductase in Klebsiella pneumoniae. Biotechnology and Applied Biochemistry, 61, 707–715.
Hols, P., Hancy, F., Fontaine, L., Grossiord, B., Prozzi, D., Leblond-Bourget, N., et al. (2005). New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiology Reviews, 29, 435–463.
Jensen, N. B. S., Melchiorsen, C. R., Jokumsen, K. V., & Villadsen, J. (2001). Metabolic behavior of Lactococcus lactis MG1363 in microaerobic continuous cultivation at a low dilution rate. Applied and Environmental Microbiology, 67, 2677–2682.
Kang, X., Ling, N., Sun, G., Zhou, Q., Zhang, L., & Sheng, Q. (2012). Complete genome sequence of Streptococcus thermophilus strain MN-ZLW-002. Journal of Bacteriology, 194, 4428–4429.
Karakas-Sen, A., Ridout, M. J., & Narbad, A. (2012). Heterologous expression and purification of the dehydratase NisB involved in the biosynthesis of lantibiotic nisin. Annals of Microbiology, 62, 1099–1107.
Makarova, K., Slesarev, A., Wolf, Y., Sorokin, A., Mirkin, B., Koonin, E., et al. (2006). Comparative genomics of the lactic acid bacteria. Proceedings of the National Academy of Sciences of the United States of America, 103, 15611–15616.
McNulty, N. P., Yatsunenko, T., Hsiao, A., Faith, J. J., Muegge, B. D., Goodman, A. L., et al. (2011). The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science Translational Medicine, 3, 106.
Melchiorsen, C. R., Jensen, N. B., Christensen, B., Vaever Jokumsen, K., & Villadsen, J. (2001). Dynamics of pyruvate metabolism in Lactococcus lactis. Biotechnology and Bioengineering, 74, 271–279.
Melchiorsen, C. R., Jokumsen, K. V., Villadsen, J., Johnsen, M. G., Israelsen, H., & Arnau, J. (2000). Synthesis and posttranslational regulation of pyruvate formate-lyase in Lactococcus lactis. Journal of Bacteriology, 182, 4783–4788.
Mollet, B., Constable, A., Delley, M., Knol, J., Marciset, O., & Pridmore, D. (1993). Molecular Genetics in Streptococcus thermophilus from transformation to gene-expression. Lait, 73, 175–180.
Ott, A., Germond, J. E., Baumgartner, M., & Chaintreau, A. (1999). Aroma comparisons of traditional and mild yogurts: Headspace gas chromatography quantification of volatiles and origin of alpha-diketones. Journal of Agricultural and Food Chemistry, 47, 2379–2385.
Ott, A., Germond, J. E., & Chaintreau, A. (2000). Origin of acetaldehyde during milk fermentation using C-13-labeled precursors. Journal of Agricultural Food Chemistry, 48, 1512–1517.
Pastink, M. I., Teusink, B., Hols, P., Visser, S., de Vos, W. M., & Hugenholtz, J. (2009). Genome-scale model of Streptococcus thermophilus LMG18311 for metabolic comparison of lactic acid bacteria. Applied and Environmental Microbiology, 75, 3627–3633.
Perez, P. F., Deantoni, G. L., & Anon, M. C. (1991). Formate production by Streptococcus thermophilus cultures. Journal of Dairy Science, 74, 2850–2854.
Sambrook, J., Fritsch, F., & Maniatis, T. (2001). Molecular cloning: a laboratory manual (2nd ed.). Cold Spring Harbor: Cold Spring Harbor Laboratory Press.
Sieuwerts, S., Molenaar, D., van Hijum, S. A. F. T., Beerthuyzen, M., Stevens, M. J. A., Janssen, P. W. M., et al. (2010). Mixed culture transcriptome analysis reveals the molecular basis of mixed culture growth in Streptococcus thermophilus and Lactobacillus bulgaricus. Applied and Environmental Microbiology, 76, 7775–7784.
Sun, Z., Chen, X., Wang, J., Zhao, W., Shao, Y., Wu, L., et al. (2011). Complete genome sequence of Streptococcus thermophilus strain ND03. Journal Bacteriology, 193, 793–794.
Treu, L., Vendramin, V., Bovo, B., Campanaro, S., Corich, V., & Giacomini, A. (2014). Genome Sequences of Streptococcus thermophilus strains MTH17CL396 and M17PTZA496 from Fontina, an Italian PDO cheese. Genome Announcements 2.
Treu, L., Vendramin, V., Bovo, B., Campanaro, S., Corich, V., & Giacomini, A. (2014). Whole-genome sequences of Streptococcus thermophilus strains TH1435 and TH1436, isolated from raw goat milk. Genome Announcements 2.
van den Bogaard, P. T. C., Hols, P., Kuipers, O. P., Kleerebezem, M., & de Vos, W. M. (2004). Sugar utilisation and conservation of the gal-lac gene cluster in Streptococcus thermophilus. Systematic and Applied Microbiology, 27, 10–17.
Vos, P., Vanasseldonk, M., Vanjeveren, F., Siezen, R., Simons, G., & Devos, W. M. (1989). A maturation protein is essential for production of active forms of lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. Journal of Bacteriology, 171, 2795–2802.
Wu, Q., Tun, H. M., Leung, F. C., & Shah, N. P. (2014). Genomic insights into high exopolysaccharide producing dairy starter bacterium Streptococcus thermophilus ASCC 1275. Scientific Reports, 4, 4974.
Zhang, M., Eddy, C., Deanda, K., Finkestein, M., & Picataggio, S. (1995). Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science, 267, 240–243.
Acknowledgments
This work was supported by the Scientific and Technological Research Council of Turkey (TÜBİTAK), Project No: 110 O 218.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Akyol, I., Ozcelik, F.G., Karakas-Sen, A. et al. Cloning and Overexpression of the als, pflA, and adhB Genes in Streptococcus thermophilus and Their Effects on Metabolite Formation. Mol Biotechnol 57, 923–930 (2015). https://doi.org/10.1007/s12033-015-9882-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-015-9882-1