Abstract
Gliomas are the most common malignant tumors in the central nervous system. The tumor microenvironment (TME) plays a crucial role in tumor proliferation, invasion, angiogenesis, and immune escape. However, little is known about TME in gliomas. The purpose of this study was to explore the biomarkers associated with TME in glioblastoma (GBM) to predict immunotherapy effectiveness and prognosis in patients. Based on RNA-seq transcriptome data and clinical features of 1222 samples (113 normal samples and 1109 tumor samples) in The Cancer Genome Atlas (TCGA) database, the ImmuneScore, StromalScore, and ESTIMATEScore were calculated by ESTIMATE algorithm. The differentially expressed genes (DEGs) and differentially mutated genes (DMGs) were determined in the TCGA GBM cohort. Furthermore, gene set enrichment analysis (GSEA) was used to investigate the enrichment pathways of INSRR genes with abnormal expression. The proportion of tumor-infiltrating immune cells (TIICs) was evaluated by CIBERSORT. Frequent mutations of TP53, EGFR, and PTEN occurred in high and low immune scores. The cross-analysis of DEGs and DMGs revealed that INSRR was an immune-related biomarker in the TCGA GBM cohort. According to GSEA, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway with INSRR abnormal expression were IgA-produced intestinal immune network and Alzheimer’s disease, oxidative phosphorylation, and Parkinson’s disease, respectively. Additionally, INSRR expression was correlated with dendritic cells activated, dendritic cells resting, T cells CD8, and T cell gamma delta. INSRR is associated with the immune microenvironment in GBM and is used as a biomarker to predict immune invasion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gliomas are the most prevalent malignant tumors in the central nervous system [1]. According to the WHO classification, this kind of tumor can be divided into four grades, from I to IV [2], of which glioblastoma (GBM) is the most malignant type (WHO grade IV). Despite the application of active treatment strategies such as radiotherapy, temozolomide-based chemotherapy, and excision, GBM still has a high recurrence and death rate due to its strong plasticity and heterogeneity [3]. Therefore, exploring valuable biomarkers and therapeutic targets is essential for predicting the prognosis of GBM.
The tumor microenvironment (TME) plays an essential role in the formation and development of tumors [4]. Tumor-related immune cells can serve as tumor-antagonizing or tumor-promoting factors [5]. Tumor cells can release some immunosuppressive molecules to inhibit other immune cells, allowing them to elude the surveillance of the immune system and proliferate in the body [6]. Immunotherapies regulating the TME and immune checkpoints have shown good efficacy in cancer treatment over the past few years [7, 8]. Although multiple immune-related biomarkers have been found to be associated with the prognosis of tumors [9,10,11], there is still a lack of immune-related biomarkers that can effectively predict outcomes in GBM patients.
TME comprises tumor cells, stromal cells, immune cells, extracellular matrix (ECM), signaling molecules, and cytokines [12]. The immune cells in TME can affect tumor growth [13]. The infiltration level of stromal and immune cells in tumor samples can be predicted by the ESTIMATE algorithm, which calculates the ImmuneScore and StromalScore based on unique gene signatures [14]. Subsequent studies have suggested that using the TME-related ImmuneScore and the ratio of tumor-infiltrating immune cells (TIICs) in TME improves the prediction accuracy of the TNM cancer staging system [15, 16]. TME-related ImmuneScore is considered a valuable indicator of cancer recurrence, metastasis, and prognosis in patients with colorectal cancer [17]. Furthermore, the effectiveness of ImmuneScore in the classification of colorectal cancer is also widely recognized [18].
Based on the ESTIMATE algorithm, the present study comprehensively analyzed gene expression, somatic mutations, and immune cell infiltration in the TME in the TCGA GBM cohort. Differentially expressed genes (DEGs) and differentially mutated genes (DMGs) were identified according to the ImmuneScore, and the insulin receptor-related receptor (INSRR) was determined as an immune-related biomarker. The current study aimed to provide a new valuable target for the immunotherapy of GBM.
Materials and methods
Data source and preprocessing
We downloaded RNA-seq data (normalized FPKM expression level data), tumor mutation burden, and corresponding clinical data from the TCGA website (https://cancergenome.nih.gov/) in February 2022. R4.2.1 was used for data standardization, processing, and analysis. RNA-seq values for each sample were integrated into a matrix file using Perl (https://www.perl.org/).
Estimation of immuneScore, stromalScore, and ESTIMATEscore
The ESTIMATE R package and the Expression Data (Estimate) algorithm were used to estimate the ratios of immune and matrix components in the TME in glioma samples. These ratios were presented as ImmuneScore, StromalScore, and ESTIMATEScore. A higher score indicated a larger proportion in the TME.
Correlation of immuneScores, stromalScores, and ESTIMATEscores with survival
Samples were divided into two groups according to the median of ImmuneScores, StromalScores, and ESTIMATEScores, respectively. Overall survival (OS) was the primary outcome. The survminer R package was adopted to perform a survival analysis, and a Kaplan–Meier survival curve was drawn accordingly. The log-rank test was used for comparison between subgroups. A P < 0.05 indicated a statistically significant difference.
Identification of DEGs
Samples were divided into the high and low ImmuneScore groups. Limma R package was used to identify DEGs, and the screening conditions were |log2(FC)|> 1 and p-adjust < 0.05. Phetmap R package was used to draw a heat map of DEGs. The ClusterProfile and Richlot packages were used for the DEG enrichment analysis based on the Gene Ontology (GO) and the Kyoto encyclopedia of genes and genomes (KEGG) databases. The enrichment analysis involved biological process (BP), molecular function (MF), and cell component (CC). The enrichment items with P < 0.05 had statistical significance. A bubble diagram was developed using the Ggplot2 R package.
Identification of DMGs
Samples were divided into the high and low ImmuneScore groups. The limma R package was used to identify the tumor mutation burden in GBM samples from the TCGA database. The Maftools package was used to identify DMGs, and the screening condition was set to P < 0.05. Forest plots and oncoplots were drawn using the Forestplot R package.
Gene set enrichment analysis
Gene Set Enrichment Analysis (GSEA) was conducted to determine the biological pathways associated with high and low expression of CD2. We downloaded two gene set GMT documents (c2.cp.kegg.v7.5.1.entrez.gmt and h.all.v7.5.1.entrez.gmt) from the Molecular Signatures Database (MsigDb) for reference. The number of gene conversion permutations was set to 1000, and a pathway with P < 0.05 was considered significantly enriched.
Survival analysis
PanCanSurvPlot (https://smuonco.shinyapps.io/PanCanSurvPlot/) was used to analyze the association of INSRR with OS and disease-specific survival (DSS) of GBM patients.
Differential analysis and correlation analysis of immune cell infiltration
The CIBERSORT algorithm was used to predict the abundance of 22 infiltrating immune cells in both glioma and normal samples, including B cells, dendritic cells, eosinophils, macrophages, mast cells, monocytes, neutrophils, natural killer (NK) cells, plasma cells, T cells, and their different subtypes. These immune cells were grouped into high and low INSRR expression groups to analyze the correlation between each immune cell and the INSRR gene. The Wilcoxon rank-sum test was used to compare the proportions of the 22 infiltrating immune cells in the high and low INSRR expression groups, and the Pearson correlation between INSRR expression and the proportion of immune cells was assessed. Violin and scatter plots were drawn using the Vioplot, Ggplot22, and Ggpubr R packages.
Results
The correlation between immune infiltration and the OS in patients with GBM
The analysis process is shown in Fig. 1. RNA-seq and clinical data of 169 GBM cases were downloaded from the TCGA database. To evaluate the association between immune infiltration scores and the prognosis in GBM patients, the samples were grouped into two groups based on the medians, respectively: high and low ImmuneScore groups, high and low StromalScore groups, as well as high and low ESTIMATEScore groups. Subsequently, Kaplam-Meier survival curves were developed. As shown in Fig. 2, different immune scores showed different OS in GBM samples. However, no correlation was found between the ImmuneScore and OS, the StromalScore and OS, and the ESTIMATE score and OS, indicating that the three scores were not correlated with the prognosis in patients with GBM.
Identification of DEGs based on immunescore
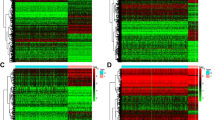
Based on transcriptome data from 169 GBM cases in the TCGA database, we compared the gene expression in the high ImmuneScore and low ImmuneScore groups to investigate the correlation between gene expression and ImmuneScore. There were 919 DEGs between the high and low ImmuneScore groups, with 754 up-regulated genes and 165 down-regulated genes. The main DEGs between the high and low ImmuneScore groups are presented in Fig. 3a. According to the GO enrichment analysis, DEGs were primarily associated with the inflammatory microenvironment and immune function, such as leukocyte migration and adhesion, cytokine production, and T cell activation. The GO enrichment analysis of DEGs is shown in Fig. 3b. The migration of leukocyte, myeloid leukocyte, granulocyte and neutrophil, cytokine production, and T cell activation are the most common terms for biological processes. The external side of plasma membrane, secretory granule membrane, and cytoplasmic vesicle lumen are the most common terms for cellular components. The receptor ligand activity, signaling receptor activator activity, cytokine receptor binding, and immune receptor activity are the most common terms for molecular functions. Similarly, the KEGG enrichment analysis showed a high enrichment of inflammation-associated processes, such as cytokine–cytokine receptor interaction, chemokine signaling pathway, NOD-like receptor signaling pathway, NF-κB pathway, and Toll-like receptor signaling (Fig. 3c). As a result, these inflammation-related DEGs indicate that immune factors play an important role in the TME in patients with GBM.
Identification of DMGs based on immunescore
To explore the correlation between gene mutations and immune components in TME, we compared somatic mutation data in the high and low ImmuneScore groups to identify DMGs. According to the DEGs mutation rate analysis, TP53, EGFR, and PTEN showed frequent mutations in both high and low ImmuneScore groups. The DEGs mutation rate in the high ImmuneScore group is presented in Fig. 4a, and the DEGs mutation rate in the low ImmuneScore group is presented in Fig. 4b. Additionally, forest map (Fig. 4c) and oncoplot (Fig. 4d) show the common DMGs between high ImmuneScore and low ImmuneScore groups. As shown in Fig. 4c and d, the two cohorts shared 16 DMGs. VENN analysis showed that INSRR overlapped in the cross-over analysis of DEGs and DMGs. Shared factors in DEGs and DMGs are presented in Fig. 4e.
GSEA in high and low expression of INSRR groups
GSEA was performed for both high and low expression of INSRR groups. According to the GSEA, the high expression of INSRR was mainly associated with two KEGG pathways, including asthma and the intestinal immune network for IgA production (P < 0.05). Important KEGG pathways associated with high expression of INSRR are shown in Fig. 5a. Meanwhile, the low expression of INSRR was primarily correlated with six KEGG pathways, including Alzheimer’s disease, GAP junction, olfactory transduction, oxidative phosphorylation, Parkinson’s disease, and Ribosome (P < 0.05). Important KEGG pathways associated with low expression of INSRR are shown in Fig. 5b. Furthermore, low-expression INSRR was enriched in three Hallmark gene sets, including E2F targets, MYC targets V1, and oxidative phosphorylation (P < 0.05). Important Hallmark pathways associated with low expression of INSRR are presented in Fig. 5c.
INSSR was related to the survival in GBM patients
PanCanSurvPlot (https://smuonco.shinyapps.io/PanCanSurvPlot/) was used to perform survival analysis on high and low expression of INSRR groups, and the results revealed that GBM patients in the high expression group had longer OS and DSS than those in the low-expression group (Fig. 6a and b).
Correlation between INSSR expression and TIICs
We also investigated INSRR-related immune cells. The CIBERSORT algorithm was adopted to assess the infiltration of 22 immune cells in GBM and analyze the relative proportion of tumor-infiltrating subtypes. Immune cell infiltration in GBM patients from the TCGA cohort is presented in Fig. 7a; the correlation between 21 TIICs in patients with GBM is depicted in Fig. 7b. Subsequently, the proportions of 22 immune cells were compared between the high and low INSRR expression groups. The proportions of 22 immune cells are described in Fig. 7c. Among the 22 immune cells, T cells CD8, T cells CD4 memory resting, and mast cells activated showed a significant difference in the infiltrating abundance between the high and low INSRR expression groups. Furthermore, four of the 22 immune cells showed a positive correlation with INSRR expression in GBM cases, including dendritic cells activated, dendritic cells resting, T cells CD8, and T cell gamma delta. The Pearson correlation between four important TIICs and INSRR expression is presented in Fig. 7d. The differential and correlation cross-over analysis identified one shared TIIC associated with INSRR expression, namely T cells CD8. The INSRR expression-related TIIC is shown in Fig. 7e. In addition, we analyzed the correlation between INSRR expression and common immune check point (ICPs) to assess GBM patients' response to INSRR expression-related immunotherapy. The results showed that the expression of PDCD1, CTLA4, LAG3, CD86, and HAVCR2 was significantly different between the high and low INSRR expression groups. The correlation between common ICPs and INSRR expression is shown in Fig. 7f.
Correlation between INSSR expression and TIICs. a Immune cell infiltration in GBM patients from TCGA cohort; b Correlation between 21 TIICs in patients with GBM; c Violin plot of the proportions of 22 immune cells in tumor tissue with low and high expression of INSSR; d Pearson correlation between four important TIICs and INSRR expression; e Venn diagram of INSRR expression-related TIICs; f Correlation between common ICPs and INSRR expression
Discussion
The cellular immune response plays a key role in the progression of cancers and is an influential factor in the prognosis of cancers [19, 20]. Therefore, immune-related genes are potential treatment targets and value in the prognosis of cancers. Currently, TME has become a research hotspot for its important value in treatment and prognosis [21, 22]. The TME in GBM also has attracted extensive attention worldwide. Based on the ESTIMATE and CIBERSORT algorithms and GBM somatic mutation data, this study found that INSRR, a core gene in TME, was associated with GBM, providing a novel target for the clinical diagnosis and treatment of GBM.
The ESTIMATE algorithm is a comprehensive method for estimating tumor purity based on gene expression and can help identify candidate TME-related biomarkers [23]. In the current study, it was used to calculate ImmuneScores, StromalScores, and ESTIMATEScores based on the TCGA transcriptome data. Then TME-related DEGs between the high and low ImmuneScore groups were determined. The biological functions of these DEGs are mainly the migration of leukocyte, granulocyte, myeloid leukocyte, and neutrophil, as well as cytokine production, and T cell activation. Further, these DEGs participate in a series of immune- and inflammation-related signaling pathways, such as NOD-like receptor signaling pathway, nuclear factor κB (NF-κB) pathway, and Toll-like receptor signaling pathway. The NOD-like receptor signaling pathway plays an oncogenic role in tumorigenesis by regulating innate and adaptive immunity, apoptosis, and cell differentiation in tumors [24]. The NF-κB pathway, as a major transcription factor produced by cytokine, recruits innate immune cells to participate in the regulation of the TME by creating an inflammatory environment [25]. The Toll-like receptor signaling pathway is a classic pattern recognition receptor pathway, and GBM tissues with high TLR2 levels have a similar mutualism pattern with MHC I molecules. Besides, the number and activity of infiltrating CD8 T cells are associated with TLR2 levels in GBM patients [26]. Therefore, understanding these biological functions and pathways is helpful for elucidating the underlying mechanisms of GBM.
A previous study revealed that cancers with more non-synonymous variants were correlated with favorable survival outcomes with cancer immunotherapy [4]. To investigate the correlation between gene mutations and immune components in TME, we identified DMGs between the high and low ImmuneScore groups. According to the DEG mutation rate analysis, TP53, EGFR, and PTEN showed frequent mutations in both high and low ImmuneScore groups. TP53 mutations may activate carcinogenic and inflammatory pathways, thereby accelerating the progression of GBM [27]. EGFR mutation is the most common mutation that presents 57% of genetic mutations in GBM [28]. PTEN deletion is the most prevalent genomic event in GBM, and its mutation is a crucial biomarker for GBM [29].
Receptor tyrosine kinase is a key component of cell signaling pathways that participate in the regulation of cell interactions, cell proliferation and differentiation, cell migration, metabolism, and cell cycle control. Dysfunction of the receptor may lead to the development of cancers [30, 31]. To further identify the differentially mutated and expressed immune-related genes in the TME, a cross-analysis of DEGs and DMGs was performed. We identified an immune-related predictive biomarker, INSRR. INSRR is an insulin receptor that can be activated by basic extracellular mediators [32]. It does not recognize either insulin or any identified insulin-related peptides. INSRR mRNA is detected in the nervous system and peripheral tissues [33]. BDNF-activated INSRR receptor chimera promotes neuronal survival and PC12 cell differentiation [34]. Furthermore, INSRR activates the phosphorylation of the intracellular signaling protein IRS-1 (insulin receptor substrate-1) and Akt (protein kinase B) and contributes to cytoskeleton rearrangement [35]. However, few tumor researches focus on INSRR, so the association between INSRR and glioma remains unclear. In the current study, GSEA analysis showed that genes in the high INSRR expression group were mainly involved in the intestinal immune network for IgA production signaling pathway. The activation of this pathway was proven to promote the proliferation and migration of hepatocellular carcinoma cells [36]. Additionally, genes in the low INSRR expression group were mainly enriched in the oxidative phosphorylation pathway and E2F targets. Inhibiting oxidative phosphorylation can structurally relieve hypoxia and reactivate antitumor immune response [37]. Existing research has demonstrated that some oxidative phosphorylation inhibitors can improve the efficacy of radiotherapy combined with immunotherapy [38]. The E2F family is involved in tumorigenesis and progression of various tumors. The expression of E2F1-8 has been demonstrated to be elevated in brain cancer and central nervous system neoplasms. In GBM patients, an elevated expression of E2F3–6 is associated with poor prognosis and increased infiltration of CD8 + T cells, macrophages, neutrophils, and dendritic cells [39]. These findings suggest that INSRR may have a close association with immunity.
Immune infiltration destroys the immune microenvironment in GBM, promoting immune escape [40]. The current study adopted the CIBERSORT algorithm to select 22 immune cells from the TCGA GBM samples and investigated the correlation between INSRR expression and immunity-related factors. The results showed that INSRR expression was positively correlated with dendritic cells activated, dendritic cells resting, CD8 + T cell, and T cell gamma delta in GBM patients, indicating that GBM patients with high expression of INSRR may be immunocompromised. CD8 + T cells can differentiate into cytotoxic T cells to defend against pathogen invasion [41]. CD8 + T cell-mediated immunoediting inhibits genomic evolution and immune evasion in murine gliomas [42]. Therefore, it is speculated that INSRR may promote cancer by regulating tumor immunity, such as CD8 + T cell-mediated immunity. Furthermore, a relatively high expression of ICPs was observed in the high INSRR expression group, suggesting that INSRR has the potential to predict GBM’s response to immunotherapy. These results revealed that the biological behavior of INSRR may depend on its expression level and TIIC phenotype, which may provide new insights for targeted therapy and molecular biology. Nonetheless, the specific mechanism remains to be studied.
There are several limitations to our study. Firstly, the present study only included GBM samples from the TCGA database without comparison with other databases. Thus, there may be a selection bias in the transcripts. Secondly, the specific function and mechanism of INSRR in glioma cells were not evaluated. Additional in vivo and vitro studies are required to clinically verify its function. Lastly, the fundamental role of INSRR in some special gliomas, such as diffuse midline glioma, needs to be further discussed.
According to the comprehensive bioinformatics analysis, INSRR is identified as an indicator of remodeling TME, and it is closely correlated with the degree of infiltration of immune cells in GBM patients. The expression level of INSRR is associated with dendritic cells activated, dendritic cells resting, CD8 + T cell, and T cell gamma delta and may serve as a tumor promotor by regulating tumor immunity. In conclusion, INSRR exhibits immune-related activity in the TME in patients with GBM and may influence the biological behavior and phenotype of GBM.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Xu S, et al. Immunotherapy for glioma: current management and future application. Cancer Lett. 2020;476:1–12. https://doi.org/10.1016/j.canlet.2020.02.002.
Terashima K, Ogiwara H. [Pediatric Glioma]. No Shinkei Geka. Neurol Surg. 2021;49(3):640–6. https://doi.org/10.11477/mf.1436204438.
Gusyatiner O, Hegi ME. Glioma epigenetics: from subclassification to novel treatment options. Semin Cancer Biol. 2018;51:50–8. https://doi.org/10.1016/j.semcancer.2017.11.010.
Li H, et al. Establishment of a novel ferroptosis-related lncRNA pair prognostic model in colon adenocarcinoma. Aging. 2021;13(19):23072–95. https://doi.org/10.18632/aging.203599.
Lei X, et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–33. https://doi.org/10.1016/j.canlet.2019.11.009.
Helmy KY, et al. Cancer immunotherapy: accomplishments to date and future promise. Ther Deliv. 2013;4(10):1307–20. https://doi.org/10.4155/tde.13.88.
Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557–66. https://doi.org/10.1158/0008-5472.Can-18-3962.
Gaggero S, et al. Cytokines orchestrating the natural killer-myeloid cell crosstalk in the tumor microenvironment: implications for natural killer cell-based cancer immunotherapy. Front Immunol. 2020;11:621225. https://doi.org/10.3389/fimmu.2020.621225.
Darvin P, et al. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med. 2018;50(12):1–11. https://doi.org/10.1038/s12276-018-0191-1.
Khan S, Gerber DE. Autoimmunity, checkpoint inhibitor therapy and immune-related adverse events: a review. Semin Cancer Biol. 2020;64:93–101. https://doi.org/10.1016/j.semcancer.2019.06.012.
Gibney GT, et al. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17(12):e542–51. https://doi.org/10.1016/s1470-2045(16)30406-5.
Chen R, et al. Dihydroartemisinin prevents progression and metastasis of head and neck squamous cell carcinoma by inhibiting polarization of macrophages in tumor microenvironment. Onco Targets Ther. 2020;13:3375–87. https://doi.org/10.2147/ott.S249046.
Hegde PS, Chen DS. Top 10 challenges in cancer immunotherapy. Immunity. 2020;52(1):17–35. https://doi.org/10.1016/j.immuni.2019.12.011.
Chen Y, et al. CD2 is a novel immune-related prognostic biomarker of invasive breast carcinoma that modulates the tumor microenvironment. Front Immunol. 2021;12:664845. https://doi.org/10.3389/fimmu.2021.664845.
Ma C, et al. AUNIP expression is correlated with immune infiltration and is a candidate diagnostic and prognostic biomarker for hepatocellular carcinoma and lung adenocarcinoma. Front Oncol. 2020;10:590006. https://doi.org/10.3389/fonc.2020.590006.
Wang J, et al. Low immune index correlates with favorable prognosis but with reduced benefit from chemotherapy in gallbladder cancer. Cancer Sci. 2020;111(1):219–28. https://doi.org/10.1111/cas.14239.
Mlecnik B, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44(3):698–711. https://doi.org/10.1016/j.immuni.2016.02.025.
Bruni D, et al. The immune contexture and immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer. 2020;20(11):662–80. https://doi.org/10.1038/s41568-020-0285-7.
Xia L, et al. The cancer metabolic reprogramming and immune response. Mol Cancer. 2021;20(1):28. https://doi.org/10.1186/s12943-021-01316-8.
Chen L, Musa AE. Boosting immune system against cancer by resveratrol. Phytother Res. 2021;35(10):5514–26. https://doi.org/10.1002/ptr.7189.
Abbott M, Ustoyev Y. Cancer and the immune system: the history and background of immunotherapy. Semin Oncol Nurs. 2019;35(5):150923. https://doi.org/10.1016/j.soncn.2019.08.002.
Muenst S, et al. The immune system and cancer evasion strategies: therapeutic concepts. J Intern Med. 2016;279(6):541–62. https://doi.org/10.1111/joim.12470.
Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. https://doi.org/10.1038/ncomms3612.
Liu P, et al. NOD-like receptor signaling in inflammation-associated cancers: from functions to targeted therapies. Phytomedicine. 2019;64:152925. https://doi.org/10.1016/j.phymed.2019.152925.
Yang M, et al. The application of nanoparticles in cancer immunotherapy: targeting tumor microenvironment. Bioact Mater. 2021;6(7):1973–87. https://doi.org/10.1016/j.bioactmat.2020.12.010.
Chang CY, et al. Glial TLR2-driven innate immune responses and CD8(+) T cell activation against brain tumor. Glia. 2019;67(6):1179–95. https://doi.org/10.1002/glia.23597.
Ham SW, et al. TP53 gain-of-function mutation promotes inflammation in glioblastoma. Cell Death Differ. 2019;26(3):409–25. https://doi.org/10.1038/s41418-018-0126-3.
Furnari FB, et al. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15(5):302–10. https://doi.org/10.1038/nrc3918.
Choi SW, et al. Mutation-specific non-canonical pathway of PTEN as a distinct therapeutic target for glioblastoma. Cell Death Dis. 2021;12(4):374. https://doi.org/10.1038/s41419-021-03657-0.
Trenker R, Jura N. Receptor tyrosine kinase activation: from the ligand perspective. Curr Opin Cell Biol. 2020;63:174–85. https://doi.org/10.1016/j.ceb.2020.01.016.
Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer. 2018;17(1):58. https://doi.org/10.1186/s12943-018-0782-4.
Deyev IE, et al. Insulin receptor-related receptor as an extracellular alkali sensor. Cell Metab. 2011;13(6):679–89. https://doi.org/10.1016/j.cmet.2011.03.022.
Dissen GA, et al. Expression of the insulin receptor-related receptor is induced by the preovulatory surge of luteinizing hormone in thecal-interstitial cells of the rat ovary. Endocrinology. 2006;147(1):155–65. https://doi.org/10.1210/en.2005-0386.
Kelly-Spratt KS, et al. BDNF activated TrkB/IRR receptor chimera promotes survival of sympathetic neurons through Ras and PI-3 kinase signaling. J Neurosci Res. 2002;69(2):151–9. https://doi.org/10.1002/jnr.10172.
Mozhaev AA, et al. Optimization of heterologous expression of insulin receptor-related receptor ectodomain. Dokl Biochem Biophys. 2019;485(1):101–3. https://doi.org/10.1134/s1607672919020017.
Yang Z, et al. Bufalin inhibits cell proliferation and migration of hepatocellular carcinoma cells via APOBEC3F induced intestinal immune network for IgA production signaling pathway. Biochem Biophys Res Commun. 2018;503(3):2124–31. https://doi.org/10.1016/j.bbrc.2018.07.169.
Chen D, et al. Combination treatment with radiotherapy and a novel oxidative phosphorylation inhibitor overcomes PD-1 resistance and enhances antitumor immunity. J Immunother Cancer. 2020. https://doi.org/10.1136/jitc-2019-000289.
Boreel DF, et al. Targeting oxidative phosphorylation to increase the efficacy of radio- and immune-combination therapy. Clin Cancer Res. 2021;27(11):2970–8. https://doi.org/10.1158/1078-0432.Ccr-20-3913.
Krajnik A, et al. Phosphoinositide signaling and mechanotransduction in cardiovascular biology and disease. Front Cell Dev Biol. 2020;8:595849. https://doi.org/10.3389/fcell.2020.595849.
Huang S, et al. Identification of immune cell infiltration and immune-related genes in the tumor microenvironment of glioblastomas. Front Immunol. 2020;11:585034. https://doi.org/10.3389/fimmu.2020.585034.
Nair S, et al. Zika virus oncolytic activity requires CD8+ T cells and is boosted by immune checkpoint blockade. JCI Insight. 2021. https://doi.org/10.1172/jci.insight.144619.
Kane JR, et al. CD8(+) T-cell-mediated immunoediting influences genomic evolution and immune evasion in murine gliomas. Clin Cancer Res. 2020;26(16):4390–401. https://doi.org/10.1158/1078-0432.Ccr-19-3104.
Funding
This work was supported by the National Natural Science Foundation of China (Nos. 81871023, 82171458, 81771322, 82171363, 82171321) and the Youth Nova Program of Shaanxi (No. 2021KJXX-19).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by HZ, MB, and DL. The first draft of the manuscript was written by HZ and all authors commented on the previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, H., Bao, M., Liao, D. et al. Identification of INSRR as an immune-related gene in the tumor microenvironment of glioblastoma by integrated bioinformatics analysis. Med Oncol 40, 161 (2023). https://doi.org/10.1007/s12032-023-02023-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-023-02023-8