Abstract
Autism spectrum disorder (ASD) includes a number of severe neurodevelopmental disorders known by defects in social interaction, impaired verbal and non-verbal interactions, and stereotypic activities and limited interests. Dysregulation of apoptotic pathways have been demonstrated in brain tissues of affected individuals. In the present study, we evaluated expression levels of apoptosis-related genes and miRNAs in peripheral blood of ASD patients compared with healthy subjects. Transcript levels of BCL2, CASP8, and hsa-29c-3p were significantly lower in total ASD patients compared with total normal children (P values = 0.003, 0.002, and 0.01 respectively). When sex of study participants was considered in the analysis, the difference in transcript levels of these genes was significant only in male subjects. Peripheral expression of BCL2 and hsa-29c-3p had 100% sensitivity 92% specificity in ASD diagnosis. The diagnostic power of combination of transcript levels of these genes was estimated to be 78% based on the calculated AUC value. The present study provides evidences for dysregulation of apoptotic pathways in peripheral blood of ASD patients and suggests certain apoptosis-related genes as biomarkers in this regard.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Autism spectrum disorder (ASD) comprises a group of profound neurodevelopmental disorders described by destruction of social interaction, impaired verbal and non-verbal interactions, and stereotypic activities and limited interests (Nazeer and Ghaziuddin 2012). Although the molecular underlying mechanisms of autism have not been clarified yet, postmortem and animal studies have demonstrated abnormal regulation of apoptotic pathways in autism (Wei et al. 2014). Apoptosis is regarded as a critical element of typical development of the nervous system. Dimensional, chronological, or quantitative dysregulation of this process would result in an anomalous quantity of neurons and deficient neural development (Wei et al. 2014). The mitochondrial, death receptor, and inflammatory pathways have been recognized as the main apoptotic pathways in the nervous system (Jarskog et al. 2005). Neuroimaging studies and assessment of postmortem brain have shown several anomalies in ASD patients including diminished Purkinje cell numbers in the cerebellar hemispheres and vermis (Bauman and Kemper 1985), absence of granular cells (Bauman and Kemper 2005), atypical widening of brain (Amaral et al. 2008), decreased corpus callosum size (Egaas et al. 1995), and enlarged amygdala size (Schumann et al. 2004) all of which can be at least partially attributed to dysregulation of apoptotic pathways. However, discoveries in the field of autism neuropathology are being limited by inaccessibility of brain tissue for analysis (Petinou and Minaidou 2017). Blood tissue is regarded as an alternative in this regard due to its availability especially during early phases of disease and relatively similar expression pattern with brain (Sullivan et al. 2006). In the current study, we selected three apoptosis-related mRNA coding genes (BCL2, CASP2, and CASP8) and four microRNAs (miRNAs) (hsa-miR-17-5p, hsa-miR-20a-5p, hsa-miR-29a-3p, and hsa-29c-3p) to assess their expression in the peripheral blood of ASD patients compared with normally developed children to find whether their expression pattern could be recruited as a diagnostic marker for ASD. Former studies have shown interaction between peripheral and central inflammation in the pathogenesis of ASD. Among the mentioned assessed molecules were those linked with the apoptosis (Cristiano et al. 2018). Moreover, as miRNAs can be transmitted from peripheral tissues into the bloodstream (Creemers et al. 2012), it is convincible to assume that alterations in miRNA levels in the peripheral blood mirror their concentrations in the central nervous system (Vaccaro et al. 2018). This speculation can be also applied to apoptosis-associated miRNAs. BCL2 is a principal up-stream protein in the intrinsic apoptotic pathway with anti-apoptotic effects (Cristiano et al. 2018). CASP2 cooperates with c-Myc to cytochrome c release into the cytosol leading to activation of the intrinsic apoptosis pathway (Vaccaro et al. 2018). CASP8 as another apoptotic initiator is a cysteine protease with a crucial function in the extrinsic apoptotic signaling pathway through death receptors (Creemers et al. 2012). miR-20a participates in the regulation of the extrinsic pathway through downregulation of Fas expression (Huang et al. 2012). Decreased expression of miR-29a is associated with overexpression of a chief anti-apoptotic BCL-2 family member (Desjobert et al. 2011). hsa-miR-29a-3p and hsa-29c-3p are possibly involved in extrinsic pathway (Venkatadri et al. 2016).
Material and Methods
Study Participants
The current study was conducted on peripheral blood samples obtained from 50 ASD patients (38 males and 12 females, age ± SD (range) = 6 ± 1.46(4–9)) and 50 healthy subjects (37 males and 13 females, age ± SD (range) = 6.04 ± 1.74(4–13)). Patients were diagnosed according to the Diagnostic and Statistical Manual of Mental Disorders, 5th edition (DSM-V) (Association 2013). A team consisted of psychologists and psychiatrics confirmed the diagnosis of patients through structured developmental assessments such as Autism Diagnostic Observation Schedule and Autism Diagnostic Interview—Revised (Steiner et al. 2012). The results of developmental, speech, and motor testing were documented. Control subjects were chosen from normally developed children with no history of behavioral/psychiatric disorder or former psychiatric treatment in themselves or their first-degree relatives. The absence of behavioral/psychiatric disorder was verified by a psychologist. Informed consent forms were signed by the parents of study participants. The study protocol was approved by ethical committee of Hamadan University of Medical Sciences. This study was conducted in accordance with the Declaration of Helsinki.
Expression Study
Peripheral blood samples were collected from all study participants in EDTA tubes. All samples were gathered from study participants between 10 to 12 AM. Total blood cells were subjected to RNA extraction without enrichment of any kinds of blood cells. Hybrid-RTM blood RNA extraction Kit (GeneAll Biotechnology Co Ltd., South Korea) and the Applied Biosystems High-Capacity cDNA Reverse Transcription Kit were used for isolation of total RNA and cDNA synthesis. Approximately, 3 μg RNA per 0.25 ml of blood was obtained from each sample and 250 ng of RNA samples were used for cDNA production. The relative transcript levels of genes were compared between study groups using the Applied Biosystems TaqMan® Universal PCR Master Mix in rotor gene 6000 Corbett Real-Time PCR System. About 50–100 ng of cDNA was used in each reaction. Reactions were performed in duplicate. The HPRT1 gene was used as the reference gene. PCR program comprised a denaturation step at 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 60 s, and a final extension step in 72 °C for 5 min. The nucleotide sequences of primers and probes used for amplification of apoptosis-related genes are shown in Table 1.
Approximately, 3 μg RNA per 0.25 ml of blood was obtained from each sample and 250 ng of miRNA samples were used for cDNA production. TaqMan® MicroRNA Reverse Transcription Kit (Invitrogen) was used to convert miRNAs to cDNA. About 50–100 ng of cDNA was used in each reaction. Relative expressions of miRNAs were compared between cases and controls using TaqMan® MicroRNA Assays kit (Applied Biosystems). RNU6B gene was used as normalizer. Reactions were performed in duplicate. The mean CT values of this housekeeping gene were 17.79 and 17.87 in cases and controls respectively.
Statistical Analysis
SPSS version 18 (Chicago, IL, USA) was used for statistical analysis. Relative expression of genes in ASD patients compared with controls was calculated using Efficiency^-∆CT values. The significance of difference in mean values of gene expressions between patients and healthy subjects was assessed using independent T test. Spearman rank order correlation test was applied to assess the correlation between relative expression levels of genes. P values less than 0.05 were considered significant.
The receiver operating characteristic (ROC) curve was plotted to assess the diagnostic power of genes transcript levels for categorizing disease status. In order to evaluate gene expression probability cut-off, the Youden index (j) was applied to maximize the difference between sensitivity (true-positive rate) and 1—specificity (false-positive rate). Sensitivity and specificity values were calculated based on the following equations:
The diagnostic power of transcript level of each gene was scored based on the area under curve (AUC) values using the following system: 0.90–1 = excellent (A), 0.80–0.90 = good (B), 0.70–0.80 = fair (C), 0.60–0.70 = poor (D), and 0.50–0.60 = fail (F).
Results
Relative Expression of Genes in ASD Patients and Controls
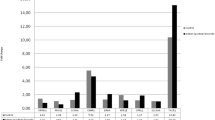
The level of HPRT1 and RNU6B were similar between ASD and controls. Transcript levels of BCL2, CASP8, and hsa-29c-3p were significantly lower in total ASD patients compared with total normal children (P values = 0.003, 0.002, and 0.01 respectively). When sex of study participants was considered in the analysis, the difference in transcript levels of these genes was significant only in male subjects. Figure 1 shows relative expression of genes in ASD patients and controls. Fold change values and the corresponding P values are shown in Table 2.
Correlation Between Relative Expressions of Genes and Age of Study Participants
No significant correlation was found between relative expression of genes and age of ASD cases. However, expressions of hsa-miR-17-5p and hsa-miR-20a-5p were inversely correlated with age in controls. Table 3 shows R and P values of partial correlations, controlling for sex, between levels of gene expression and age of study participants.
Pairwise Correlation Between Expression Levels of Genes
BCL2 expression was significantly correlated with expression of both CASP8 and CASP2 genes in both cases and controls. CASP2 and CASP8 expressions were correlated only in healthy subjects. hsa-miR-29a-3p expression was correlated with expression levels of other miRNAs only in healthy children. Table 4 shows pairwise correlation between expression levels of genes. The most significant pairwise correlation was the correlation between hsa-miR-20a-5p and hsa-miR-17-5p (Fig. 2).
ROC Curve Analysis
ROC curves were depicted to demonstrate the diagnostic ability of transcript levels of each gene as a binary classifier system when its discrimination threshold is altered. The true positive rate (sensitivity) was plotted against the false positive rate (1—specificity) at different threshold sites. The AUC was defined as the likelihood that transcript levels of a certain gene could rank a randomly chosen sample from a patient higher than a randomly chosen control sample. As transcript levels of BCL2, CASP8, and has-29c-3p were significantly different between ASD patients and healthy subjects, we evaluated the diagnostic power of these genes in differentiating disease status (Fig. 3 and Table 4). BCL2 had 100% sensitivity, while hsa-29c-3p had 92% specificity in this regard. The diagnostic power of combination of transcript levels of these genes was estimated to be 78% based on the calculated AUC value (Table 5).
Discussion
The role of apoptosis in the development of brain has been highlighted previously (Roth and D’Sa 2001). Several abnormal features of autistic brains can be attributed to dysregulation of apoptotic pathways. Recently, Dong et al. have shown the occurrence of endoplasmic reticulum (ER) stress and the consequent induction of apoptosis in distinct regions of autistic brains and suggest that cellular stress and apoptosis might participate in the pathogenesis of autism (Dong et al. 2018).
In the current case-control study, we demonstrated lower levels of BCL2 transcripts in the peripheral blood of ASD patient compared with healthy subjects. The role of BCL2 in the neuropsychiatric diseases has been highlighted previously. Jarskog et al. have assessed expression of apoptosis-related genes in middle temporal gyrus of postmortem tissues and found elevated Bax/Bcl-2 ratio in the schizophrenia patients compared with controls and proposed that cortical cells are susceptible to apoptosis in chronic schizophrenia (Jarskog et al. 2004). Fatemi et al. have evaluated expression of Bcl-2 in cerebellar cortex of autistic patients and normal controls and found significant decreased level of this protein in autistic cerebellum which is suggestive of its participation in certain structural and behavioral defects seen in autism (Fatemi et al. 2001). Moreover, Bcl-2 levels have been decreased in autistic superior frontal cortex of autistic patients compared to control tissues (Araghi-Niknam and Fatemi 2003). These results have been confirmed by other independent studies in both brain tissues (Sheikh et al. 2010a, b) and lymphoblast cell lines (Malik et al. 2011). Totally, these observations are consistent with the reported neuroprotective role for Bcl-2 which is exerted through inhibition of apoptosis and increasing cell survival (Sasaki et al. 2006).
We also detected downregulation of CASP8 in peripheral blood of autistic patients compared with controls. Siniscalco et al. have previously assessed transcript levels of Caspases in peripheral blood mononuclear cells (PBMCs) of autistic patients and healthy subjects and demonstrated elevated mRNA levels for caspase-1, -2, -4, -5 in ASD children as compared to healthy subjects. However, they demonstrated no significant difference in transcript levels of caspase-8 between cases and controls (Siniscalco et al. 2012). The inconsistency between our results and Siniscalco et al. might be due to source of expression analysis (total blood vs. PBMCs) or their small sample size (15 cases and 10 controls). Alternatively, the ethnic-based differences including both environmental and genetic factors might be involved in alteration of genes expressions.
Although our study shows dysregulation of genes involved in apoptosis process in ASD patients, the consequence of this dysregulation should be assessed by functional studies. Moreover, assessment of protein levels of these genes could be beneficial in this regard.
In addition, we demonstrated lower levels of hsa-miR-29c-3p in peripheral blood of autistic patients compared with controls. Kichukova et al. have assessed the relative expression levels of 42 miRNAs in the serum of ASD patients compared to normal controls and found higher expression of 11 miRNAs while lower expression of 29 miRNAs (including hsa-miRNA-29c-5p) in ASD patients compared with controls (Kichukova et al. 2017). hsa-miR-29c-3p has been identified as a pro-apoptotic miRNA which targets both pro- and anti-apoptotic genes in breast cancer MCF-7 cell line (Venkatadri et al. 2016). On the other hand, this miRNA is expressed in in the brain and decreases beta secretase 1 (BACE1) protein as demonstrated by both in vitro and animal studies (Zong et al. 2011). Although evidences support the role of BACE1 in neuronal synapses, such function is dependent on synaptic developmental stages as its deletion has not changed synaptic proteins in the previously developed neuronal synapses (Kamikubo et al. 2017). So although the observed downregulation of hsa-miR-29c-3p in our study might be accompanied by elevation of BACE1, the contribution of such elevation in the remedy of autism is questionable as the development of neuronal synapses is expected to be accomplished at this diseased stage.
The lack of difference in expression of these genes in female subjects might be due to small number of subjects or sex-based difference in the underlying mechanism or phenotypes of ASD. Phenotypic differences have been noted between affected girls and boy. For instance, restricted interests and behaviors and stereotypes are less frequent in girls than boys. Such different patterns can be attributed to sexually dimorphic underlying causative mechanisms including those related to sex chromosomal gene dosage and sex hormone levels (Van Wijngaarden-Cremers et al. 2014).
We detected no significant correlation between relative expression of genes and age of ASD cases which means that their peripheral expression is independent of age. So the transcript levels of these genes might be regarded as age-independent markers of disease. The importance of such markers has been highlighted by a previous report which has suggested ineffectiveness of age as a character to illustrate neurodevelopment in ASD. Categorizing ASD patients by age might haze essential data about their developmental milestones and influence their clinical rehabilitations (Wu et al. 2018).
When evaluating the correlation between expression levels of these genes, we found absence of expected correlations in ASD patients in some cases. For instance, CASP2 and CASP8 expressions were correlated only in healthy subjects. hsa-miR-29a-3p expression was correlated with expression levels of other miRNAs only in healthy children. Such observations might imply disturbance of regulation of apoptotic pathways in ASD which should be assessed in future studies. A recent study has demonstrated correlations between expression level of hsa-miR-29a-3p and environment pollution. Such finding might indicate a possible correlation between ASD, gene expression, and environment (Espin-Perez et al. 2018).
We also assessed diagnostic power of transcript levels of genes in ASD and found that BCL2 had 100% sensitivity, while hsa-29c-3p had 92% specificity in this regard. The diagnostic power of combination of transcript levels of these genes was estimated to be 78% based on the calculated AUC value. Consequently, we proposed these genes as putative elements in a diagnostic panel for ASD. Based on the observed dysregulation of genes only in male subjects, such panel is only appropriate for male subjects at this step. However, lack of significant changes in female subgroup might be due to small number of subjects in this subgroup. Consequently, assessment of their diagnostic power in larger samples of females is needed to explore their significance in this regard. Future studies are also needed to find more suitable genes to be incorporated in this panel to increase the diagnostic power.
Finally, although we reported dysregulation of apoptosis-related genes in ASD, there may be an environmental component/interaction at play. There are many types of exposures that can cause apoptosis, e.g., fluoride (Jothiramajayam et al. 2014), microwave (Zuo et al. 2014), mercury (Shenker et al. 2000), high glucose (Peng et al. 2013), diesel exhaust (Yun et al. 2009), etc. These environmental factors might affect gene activity as well.
Our study has some limitations. We state lack of assessment of other pro-/anti-apoptotic genes such as other members of CASP family as a limitation of our study. Moreover, the small sample size of the study is another shortcoming of our study which should be overcome in future studies.
References
Amaral DG, Schumann CM, Nordahl CW (2008) Neuroanatomy of autism trends. Neuroscience 31:137–145. https://doi.org/10.1016/j.tins.2007.12.005
Araghi-Niknam M, Fatemi SH (2003) Levels of Bcl-2 and P53 are altered in superior frontal and cerebellar cortices of autistic subjects. Cell Mol Neurobiol 23:945–952
Association AP (2013) Diagnostic and statistical manual of mental disorders (DSM-5®). American Psychiatric Pub
Bauman M, Kemper TL (1985) Histoanatomic observations of the brain in early infantile. Autism Neurol 35:866–874. https://doi.org/10.1212/Wnl.35.6.866
Bauman ML, Kemper TL (2005) Neuroanatomic observations of the brain in autism: a review and future directions. Int J Dev Neurosci 23:183–187. https://doi.org/10.1016/j.ijdevneu.2004.09.006
Creemers EE, Tijsen AJ, Pinto YM (2012) Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res 110:483–495. https://doi.org/10.1161/circresaha.111.247452
Cristiano C, Lama A, Lembo F, Mollica MP, Calignano A, Mattace Raso G (2018) Interplay between peripheral and central inflammation in autism spectrum disorders: possible nutritional and therapeutic strategies. Front Physiol 9:184. https://doi.org/10.3389/fphys.2018.00184
Desjobert C et al (2011) MiR-29a down-regulation in ALK-positive anaplastic large cell lymphomas contributes to apoptosis blockade through MCL-1 overexpression. Blood 117:6627–6637. https://doi.org/10.1182/blood-2010-09-301994
Dong D, Zielke HR, Yeh D, Yang P (2018) Cellular stress and apoptosis contribute to the pathogenesis of autism spectrum disorder. Autism Res. https://doi.org/10.1002/aur.1966
Egaas B, Courchesne E, Saitoh O (1995) Reduced-size of corpus-callosum in autism arch. Neurol-Chicago 52:794–801. https://doi.org/10.1001/archneur.1995.00540320070014
Espin-Perez A et al (2018) Short-term transcriptome and microRNAs responses to exposure to different air pollutants in two population studies. Environ Pollut (barking, Essex : 1987) 242:182–190. https://doi.org/10.1016/j.envpol.2018.06.051
Fatemi SH, Stary JM, Halt AR, Realmuto GR (2001) Dysregulation of Reelin and Bcl-2 proteins in autistic cerebellum. J Autism Dev Disord 31:529–535
Huang G, Nishimoto K, Zhou Z, Hughes D, Kleinerman ES (2012) miR-20a encoded by the miR-17-92 cluster increases the metastatic potential of osteosarcoma cells by regulating Fas expression. Cancer Res 72:908–916. https://doi.org/10.1158/0008-5472.can-11-1460
Jarskog LF, Selinger ES, Lieberman JA, Gilmore JH (2004) Apoptotic proteins in the temporal cortex in schizophrenia: high Bax/Bcl-2 ratio without caspase-3 activation. Am J Psychiatry 161:109–115. https://doi.org/10.1176/appi.ajp.161.1.109
Jarskog LF, Glantz LA, Gilmore JH, Lieberman JA (2005) Apoptotic mechanisms in the pathophysiology of schizophrenia. Prog Neuro-Psychoph 29:846–858. https://doi.org/10.1016/j.pnpbp.2005.03.010
Jothiramajayam M, Sinha S, Ghosh M, Nag A, Jana A, Mukherjee A (2014) Sodium fluoride promotes apoptosis by generation of reactive oxygen species in human lymphocytes. J Toxicol Environ Health A 77:1269–1280. https://doi.org/10.1080/15287394.2014.928658
Kamikubo Y, Takasugi N, Niisato K, Hashimoto Y, Sakurai T (2017) Consecutive analysis of BACE1 function on developing and developed neuronal cells. J Alzheimer's Dis 56:641–653. https://doi.org/10.3233/jad-160806
Kichukova TM, Popov NT, Ivanov IS, Vachev TI (2017) Profiling of circulating serum microRNAs in children with autism spectrum disorder using stem-loop qRT-PCR Assay. Folia Med 59:43–52. https://doi.org/10.1515/folmed-2017-0009
Malik M, Sheikh AM, Wen G, Spivack W, Brown WT, Li X (2011) Expression of inflammatory cytokines, Bcl2 and cathepsin D are altered in lymphoblasts of autistic subjects. Immunobiology 216:80–85. https://doi.org/10.1016/j.imbio.2010.03.001
Nazeer A, Ghaziuddin M (2012) Autism Spectrum disorders: clinical features and diagnosis. Pediatr Clin N Am 59:19. https://doi.org/10.1016/j.pcl.2011.10.007
Peng CM, Ma JL, Gao X, Tian P, Li WZ, Zhang L (2013) High glucose induced oxidative stress and apoptosis in cardiac microvascular endothelial cells are regulated by FoxO3a. Plos One 8:e79739. https://doi.org/10.1371/journal.pone.0079739
Petinou K, Minaidou D (2017) Neurobiological bases of autism spectrum disorders and implications for early intervention: a brief overview. Folia Phoniatr Logop 69:38–42. https://doi.org/10.1159/000479181
Roth KA, D’Sa C (2001) Apoptosis and brain development. Ment Retard Dev Disabil Res Rev 7:261–266. https://doi.org/10.1002/mrdd.1036
Sasaki T et al (2006) Bcl2 enhances survival of newborn neurons in the normal and ischemic hippocampus. J Neurosci Res 84:1187–1196. https://doi.org/10.1002/jnr.21036
Schumann CM et al (2004) The amygdala is enlarged in children but not adolescents with autism; the hippocampus is enlarged at all ages. J Neurosci 24:6392–6401
Sheikh AM, Li X, Wen G, Tauqeer Z, Brown WT, Malik M (2010a) Cathepsin D and apoptosis related proteins are elevated in the brain of autistic subjects. Neuroscience 165:363–370. https://doi.org/10.1016/j.neuroscience.2009.10.035
Sheikh AM et al (2010b) BDNF-Akt-Bcl2 antiapoptotic signaling pathway is compromised in the brain of autistic subjects. J Neurosci Res 88:2641–2647. https://doi.org/10.1002/jnr.22416
Shenker BJ, Guo TL, Shapiro IM (2000) Mercury-induced apoptosis in human lymphoid cells: evidence that the apoptotic pathway is mercurial species dependent. Environ Res 84:89–99. https://doi.org/10.1006/enrs.2000.4078
Siniscalco D, Sapone A, Giordano C, Cirillo A, de Novellis V, de Magistris L, Rossi F, Fasano A, Maione S, Antonucci N (2012) The expression of caspases is enhanced in peripheral blood mononuclear cells of autism spectrum disorder patients. J Autism Dev Disord 42:1403–1410. https://doi.org/10.1007/s10803-011-1373-z
Steiner AM, Goldsmith TR, Snow AV, Chawarska K (2012) Practitioner's guide to assessment of autism spectrum disorders in infants and toddlers. J Autism Dev Disord 42:1183–1196. https://doi.org/10.1007/s10803-011-1376-9
Sullivan PF, Fan C, Perou CM (2006) Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet 141B:261–268. https://doi.org/10.1002/ajmg.b.30272
Vaccaro TDS, Sorrentino JM, Salvador S, Veit T, Souza DO, de Almeida RF (2018) Alterations in the MicroRNA of the blood of autism Spectrum disorder patients: effects on epigenetic regulation and potential biomarkers. Behav Sci (Basel, Switzerland) 8. https://doi.org/10.3390/bs8080075
Van Wijngaarden-Cremers PJ, van Eeten E, Groen WB, Van Deurzen PA, Oosterling IJ, Van der Gaag RJ (2014) Gender and age differences in the core triad of impairments in autism spectrum disorders: a systematic review and meta-analysis. J Autism Dev Disord 44:627–635. https://doi.org/10.1007/s10803-013-1913-9
Venkatadri R, Muni T, Iyer AKV, Yakisich JS, Azad N (2016) Role of apoptosis-related miRNAs in resveratrol-induced breast cancer cell death Cell Death Dis 7 E2104 https://doi.org/10.1038/Cddis.2016.6
Wei H, Alberts I, Li X (2014) The apoptotic perspective of autism. Int J Dev Neurosci 36:13–18. https://doi.org/10.1016/j.ijdevneu.2014.04.004
Wu D, Jose JV, Nurnberger JI, Torres EB (2018) A biomarker characterizing neurodevelopment with applications in autism. Sci rep-Uk 8:614. https://doi.org/10.1038/s41598-017-18902-w
Yun YP, Lee JY, Ahn EK, Lee KH, Yoon HK, Lim Y (2009) Diesel exhaust particles induce apoptosis via p53 and Mdm2 in J774A.1 macrophage cell line. Toxicology In Vitro 23:21–28. https://doi.org/10.1016/j.tiv.2008.09.018
Zong Y et al (2011) miR-29c regulates BACE1 protein expression. Brain Res 1395:108–115. https://doi.org/10.1016/j.brainres.2011.04.035
Zuo HY et al (2014) Neural cell apoptosis induced by microwave exposure through mitochondria-dependent caspase-3 pathway. Int J Med Sci 11:426–435. https://doi.org/10.7150/ijms.6540
Acknowledgements
The current study was supported by a grant from Hamadan University of Medical Sciences.
Funding
This study was financially supported by Hamadan University of Medical Sciences (Grant Number 961177275).
Author information
Authors and Affiliations
Contributions
SGF wrote the manuscript. VKO analyzed the data. MT and AK supervised the study. MME and AN performed the laboratory tests. All authors contributed to and have approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Eftekharian, M.M., Komaki, A., Oskooie, V.K. et al. Assessment of Apoptosis Pathway in Peripheral Blood of Autistic Patients. J Mol Neurosci 69, 588–596 (2019). https://doi.org/10.1007/s12031-019-01387-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-019-01387-9