Abstract
Autism spectrum disorder (ASD) is characterized by repetitive stereotypic behaviors, restricted interests, social withdrawal, and communication deficits. Aggression and insensitivity to pain are largely unexplained in these cases. We analyzed nine mRNA expressions of the candidate genes related to aggression and insensitivity to pain in the peripheral blood of patients with ASD. Whole blood samples were obtained from 40 autistic patients (33 boys, 7 girls) and 50 age- and sex-matched controls (37 boys and 13 girls) to isolate RNA. Gene expression was assessed by quantitative Real-Time PCR (qRT-PCR) in the Erciyes University Genome and Stem Cell Center (GENKOK). All of the gene expressions except CRHR1 and SLC6A4 were found to be statistically different between the ASD patients and controls. Gene expression also differed according to gender. Alterations in the mRNA expression patterns of the HTR1E, OPRL1, OPRM1, TACR1, PRKG1, SCN9A and DRD4 genes provide further evidence for a relevant effect of the respective candidate genes on the pathophysiology of ASD. Future studies may determine the sensitivity of these candidate markers in larger samples including further neuropsychiatric diagnosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Autism spectrum disorder (ASD) is manifested by common social interaction and communication abnormalities and a severe degree of limited interest and excessive repetitive behaviors (Sener et al. 2016). Approximately 50% of patients have intellectual disability, and comorbidity with neurodevelopmental and psychiatric disorders is also frequent (Woodbury-Smith and Scherer 2018). The prevalence of ASD is approximately 1/68, according to the results of recent studies (Rose et al. 2015). It is well established that ASD is more prevalent in males than in females (Waye and Cheng 2018). With the availability of new sequencing methodologies, major advances have been made in our understanding of the genetic background of autism (Woodbury-Smith and Scherer 2018). Autistic children may excessively react to stimulations from one or several senses (taste, touch, hearing, sight, etc.) or may be unresponsive. The prevalence of insensitivity to pain in autism is approximately 25–40% (Moore 2015). Children with ASD have an abnormal sensitivity to painful stimuli, and hypersensitive responses to pain have been described in individuals with ASD (Riquelme et al. 2018). Both experimental and case studies imply that individuals with ASD can show insensitivity to pain (Allely 2013; Ross-Russell and Sloan 2005; Mieres et al. 2011; Elwin et al. 2012; Tordjman et al. 2009). It has been considered that the stereotypic behaviors and self-mutilating behavior often observed in individuals with ASD are also related to the insensitivity to pain (Smith and Matson 2010; Furniss and Biswas 2012). Insensitivity to pain which is a part of an unusual response is the subject of our research, but the studies regarding the perception of pain and its forms of expression in individuals with ASD are limited. However, this relationship has not yet been fully understood (MaClean et al. 2010). Aggression is a complex social behavior that involves a similarly complex neurochemical background (Katsouni et al. 2009). The available data indicated that aggression is a common problem reported in individuals with ASD and intellectual disability (ID) (Farmer and Aman 2010; Benson and Brooks 2008; Holden and Gitlesen 2006). Although it is hypothesized that ASD is related to aggression, very few systematic studies have been conducted thus far (Farmer and Aman 2010).
This study was based on the commonly accepted terms of ASD, such as “pain” or “insensitivity to pain” and “aggression” to select genes. Our previous preliminary study supported this hypothesis in ASD (Sener et al. 2017). OPRL1 and OPRM1 (opiate receptors that are associated with pain management) and TACR1 genes are related to pain (Provasi 2015; Steinhoff et al. 2014; Becker et al. 2014). SCN9A also plays a role in pain mechanisms, especially in the development of inflammatory pain (Meijer et al. 2014; Spratt et al. 2015). PRKG1 has a major role in nociception (Zhao et al. 2013). Serotonin (5-HT) as a neurotransmitter plays a major role in neural plasticity as a neurotransmitter (Kepser and Homberg 2015). It has been determined in recent years that 5-HT acts as a neurotrophic factor and not only influences brain functioning but also influences brain development (Miceli et al. 2013). Some studies have found that autistic children have elevated whole blood serotonin levels (hyperserotonemia), which is considered to be the most commonly observed findings and well-replicated change in ASD patients (Coutinho et al. 2007). HTR1E (5-hydroxytryptamine (serotonin) receptor 1 E) is a gene that was studied in the groups with attention deficit hyperactivity disorder (ADHD), and it was found to be correlated with low serotonergic activity and aggressive behavior in the ADHD group (Turecki et al. 2003; Oades et al. 2008). There is a relationship between autism and serotonin (5-hydroxytryptamine; 5-HT); this relationship has been previously shown by several studies (Longo et al. 2009; Koishi et al. 2006). The SLC6A4 gene (SERT or 5-HTT) modulates serotoninergic neurotransmission by the active reuptake of 5-HT from the synaptic cleft into presynaptic nerves (Huang and Santangelo 2008). This gene is thought to have contributed to the clinical picture of ASD (Ro et al. 2013; Valencia et al. 2012). The DRD4 gene encodes the D4 subtype of the dopamine receptor and the dopaminergic system is implicated in certain human behaviors, such as aggression (Gizer et al. 2009). In addition, the polymorphisms of these genes have been investigated in anxiety disorders and ASD (Gadow et al. 2010). The mRNA expression of the DRD4 gene was studied in the ADHD and ASD groups, and further studies including other neuropsychiatric diseases have been suggested to reveal DRD4 as a biomarker (Taurines et al. 2011). The CRHR1 gene encodes a G protein coupled receptor that binds neuropeptides of the corticotropin releasing hormone family and regulates the hypothalamic-pituitary-adrenal (HPA) pathway. Diseases associated with CRHR1 include depressive and anxiety disorders (Bortoluzzi et al. 2015; Ressler et al. 2010).
With this study, starting from the clinical symptoms, such as aggression and insensitivity to pain, the genes related to aggression and pain insensitivity were investigated. It is predicted that these genes may be highly associated with autism. The aim of our study was to explain the differences in candidate gene expression in whole blood samples of ASD patients. It is not easy to obtain postmortem brain tissue; this study was performed on the whole blood samples. We also aimed to reveal the relationship between genes and the clinical aspects of autism and assess genes as potential disease markers. In our previous study, we investigated OPRL1, TACR1, and HTR1E gene expression in a small ASD group and we have indicated that these genes have a decreased expression in patients. To our knowledge, the expression of these genes in children with ASD has not been previously investigated in the literature.
Methods
Study Design and Patient Collection
The study was approved by the ethical committee of the Faculty of Medicine, Erciyes University (2014/26). Written informed consent forms were received from the parents. The study conformed to the code of ethics stated in the Declaration of Helsinki. The clinical evaluation of the ASD patients in the present study was based on the clinical history from clinical examination, and neuropsychiatric assessment. Among the individuals diagnosed with ASD, 40 children with monitored ASD or with the first complaints of speech, social, and communication delays were selected after an evaluation according to the DSM-V criteria in the outpatient clinic of Child Psychiatry and Child Neurology, Erciyes University Medical Faculty. Patients who enrolled in the study were not taking any medication when blood samples were collected and according to their examination, that there was no clinical infection or endocrine findings. Exclusion criteria included a history of other developmental disorders, psychiatric diseases or genetic disorders, such as Fragile X Syndrome, Rett Syndrome, cerebral palsy, chronic seizures, and other congenital diseases. Taking into account the patient’s age and sex, 50 healthy individuals between the ages of two and six without chronic medical, psychiatric and genetic conditions and not taking psychiatric medication were used as controls. All of the control subjects were without any psychiatric condition as confirmed by a child psychiatrist according to their examinations and parental interviews. None of the controls had delays in speech or psychomotor development as per parental interview. Additionally, the patients and control group were selected from the same location and ethnic origin in this work. Information about the demographic and clinical characteristics of patients was collected using a sociodemographic form and scale which were prepared by the researchers.
Study Measurements
In addition, the degree of the disease severity was assessed by using the Childhood Autism Rating Scale (CARS). The CARS score is widely used for investigating the severity of ASD in clinical and general populations (Geier et al. 2013). In CARS, scores of 38–60 points constitute an autistic group showing severe symptoms, scores of 30–38 points constitute an autistic group showing mild to moderate symptoms, while scores of 15–29 constitute a non-autistic group. In addition to CARS, the Ankara Developmental Screening Inventory (ADSI) was also used in the patients. The ADSI is a developmental screening instrument that assesses development in children between 0 and 6 years of age in 154 items and five categories (communication-cognitive, fine motor, gross motor, social self-care skills and total skills), based on information obtained from parent interviews. The intellectual profile of the patients was determined with the Wecshler Intelligence Scale for the Children-Revised (WISC-R) scale.
Gene Expression Studies with Quantitative Real-Time PCR
All of the genetic studies were performed in the Erciyes University Genome and Stem Cell Center (GENKOK). Two milliliter blood samples were taken before the patients’ treatment. Peripheral blood samples were collected in the morning between 09:00 and 11:00. After collection, the total blood was immediately used for RNA isolation. Total RNA was extracted from the whole blood samples with TRIzol Reagent (Roche, Germany). The quantity (absorbance at 260 nm) and quality (ratio of absorbances at 260 nm and 280 nm) of RNA were evaluated with a BioSpec-Nano Spectrophotometer. All total RNA concentrations were > 40 ng/μl. Following isolation, RNA samples were stored at −80 °C prior to expression studies. First-strand complementary DNA was synthesized from the total RNA with the Transcriptor High Fidelity cDNA Synthesis Kit (Roche, Germany). Gene expression was assessed by quantitative Real-Time PCR (qRT-PCR) with RotorGene (Qiagene, Germany). Cycling conditions were 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. All of the the samples were studied in duplicate. ACTB (beta-actine) and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) were used as the housekeeping genes. The changes in gene expression between the case and control groups were determined by the 2∆∆Ct method of relative quantification. Target gene copy numbers were normalized using both housekeeping genes (Bustin et al. 2009).
Statistical Analysis
Histogram and q-q plots were examined; Shapiro-Wilk’s tests were used to assess data normality. The Levene test was used to check the variance homogeneity. To compare the differences among groups, either a one-way analysis of variance (ANOVA) or Kruskal-Wallis H test was used for continuous variables; chi-square analysis was used for categorical variables. We applied the Bonferroni correction for the correlation of CARS scores and gene expression patterns. All interpretations of the correlation coefficients were classified according to Evans JD (Evans 1996). Analyses were conducted using R 3.2.0 (www.r-poject.org) software. A p value less than 5% was considered as statistically significant.
Results
We recruited 40 patients with the diagnosis of ASD and 50 healthy controls. In the study group, there were 33 (82.5%) males and 7 (17.5%) females. There were 37 (74%) male controls and 13 (26%) female controls. The mean age was 3.64 ± 1.17 in the study group and 3.64 ± 1.19 in the control group. There was no significant difference between the groups for gender or age (p = 0.335, p = 0.988). When the patients were evaluated according to their mental condition, 29 (72.5%) of the ASD patients were diagnosed with ID. There were ten cases with ADHD (25%) and ten cases with no additional features. However, 15 ASD cases (37.5%) had aggression, and 5 (12.5%) had insensitivity to pain. The clinical and demographic features of the groups are presented in Table 1.
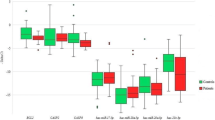
When the expressions of the OPRM1, PRKG1, SCN9A, DRD4, HTR1E, OPRL1 and TACR1 genes studied in the ASD patient group were compared to the control group, the results were found to be statistically significant (p = 0.014, p = 0.007, p < 0.001, p = 0.002, p = 0.010, p < 0.001 and p = 0.029, respectively); the expressions of the OPRM1, PRKG1 and HTR1E genes were low in the ASD cases, while the SCN9A, DRD4, OPRL1 and TACR1 gene expressions were high when compared to the control group (Fig. 1). No significant differences were recorded in the CRHR1 and SLC6A4 gene expressions between the ASD patients and controls (p = 0.372, p = 0.955).
Next, we compared the ASD group by dividing the patients into five subgroups based on clinical findings including the presence of ID, ADHD, aggression, insensitivity to pain and the ADSI scores. Comparisons of the DRD4 and OPRL1 gene expressions between the ID positive and negative groups, the OPRM1 and DRD4 gene expressions between the ADHD positive and negative groups, and the HTR1E gene expression between the aggression positive and negative groups were statistically significant (Table 2).
A positive and strong correlation was observed between the PRKG1 and OPRM1 genes, the PRKG1 and SLC6A4 genes, as well as the OPRL1 and DRD4 genes. Other positive or negative correlations are summarized in Table 3. The p value of the correlation coefficient between the CARS score and the HTR1E expression was detected as 0.054. This probability value is pretty close to the significance level, alpha = 0.05, and thus can be considered statistically significant.
We also compared the groups according to gender. When we analyzed the gender, we could not detect significant differences in gene expression in the female patients compared with the female controls. In contrast to this result, there were significant differences in the male patients and controls. OPRM1 and PRKG1 gene expression levels (0.014 and 0.024) were significantly lower and DRD4 and OPRL1 gene expression levels (0.001 and 0.004) were found to be increased in the males controls.
Discussion
In this study we assessed the clinical symptoms of ASD such as aggression and insensitivity to pain in relation to candidate gene expression in children with ASD. Except for CRHR1 and SLC6A4, we obtained significant differences between the patients and the control groups. In our preliminary study, we studied the gene expression of OPRL1, TACR1, and HTR1E in 22 patients with ASD. Positive and strong correlations were obtained between the three gene expressions in the ASD group. The expression levels of the three genes were found to be statistically significant between the groups (Sener et al. 2017). In this study, we supported our previous results regarding related gene expressions.
Recently, the knock-out mu opioid receptor gene (Oprm1−/−) was proposed as a monogenic mouse model of autism, based on severe deficits in social behavior and communication skills (Becker et al. 2014). The expression of these genes was statistically significant (p < 0.001); thus, our data support the relationship between insensitivity to pain and autism. In MECP2-TG mice, reducing Crh, or its receptor, Crhr1, suppresses anxiety-like behavior; in contrast, reducing Oprm1 improves abnormal social behavior. Reducing the levels of either of these two genes specifically corrected the respective phenotypes (Samaco et al. 2012). In our study, it was observed that the expression of CRHR1 and OPRM1 were decreased in the controls. This finding is consistent with the results of the above-mentioned study. However, the lower expression of the CRHR1 gene did not give a significant result between the groups. Therefore, we can think that the low expression of these genes makes a positive contribution to the autism phenotype. The decreased expression of the CRHR1 gene can be interpreted as a reduced CRH level or a lack of an adequate HPA axis response. When genders were considered, lower CRHR1 expression was detected in males than in females. Based on this finding, the CRHR2 gene may play a more active role in male patients. We suggest that a clearer finding should be obtained by further investigating CRH levels and CRHR2 gene expression.
In a study that conducted interviews with families, aggression in patients with ASD was found to be 65% independent of mental retardation (Totsika et al. 2011). Again, the method of interviewing the families of ASD patients was selected in another study. The parents of 67 children were interviewed, and it was reported that two-thirds of these children had severe temper tantrums and one-third had past aggression (Dominick et al. 2007). A broader study shows that in more than 50% of the ASD patients physical aggression was seen as a snapshot of clinical findings, while 70% stated that aggression against caregivers was present in their lives (Kanne and Mazurek 2011; Mazurek et al. 2013). Many studies in ASD have emphasized that there is no change in the level of aggression with age (Dominick et al. 2007; Lecavalier 2006). However, 15 ASD cases (37.5%) had aggression in our study group. The involvement of substance P (SP) and its potent tachykinin receptor (TACR1 or NK1) in the induction of both defensive rage and predatory attack appears to be a consistent finding. An overall understanding of the nature of the SP involvement in the induction of aggression has not been fully achieved yet (Katsouni et al. 2009). TACR1 expression, which was the first aggression-related gene, differed significantly among the groups in our study.
The first evidence for DRD4 receptor mRNA expression in ASD patients is an increase in peripheral blood lymphocytes compared to controls (Emanuele et al. 2010). This study was in accordance with our findings, and the expression of the DRD4 gene was also increased in our patient group. In a recent study, DRD4 mRNA expression in whole blood was significantly lower in ADHD and ASD children compared to healthy controls (Taurines et al. 2011).
Low serotonergic activity correlates with increased impulsive-aggressive behavior, while the opposite association may apply to cognitive impulsiveness (Oades et al. 2008). The SLC6A4 gene modulates serotoninergic neurotransmission by the active reuptake of serotonin from the synaptic cleft into the presynaptic nerves (Whyte et al. 2014). In our study, while the data were not significantly different, the low gene expression of SLC6A4 in the patient group implied that there is an abnormality of serotonin reuptake. HTR1E encodes one of the highly conserved serotonin receptor gene families (Shoval et al. 2014). The serotonin receptors are involved in several behavioral functions (e.g. nociception, aggression, and stress). This gene expression was significantly lower in the ASD patients compared to the controls. We can suggest that serotonin receptors have potential roles in the pathophysiology of these cases, as is consistent in the literature. In our study, the HTR1E gene expression pattern was significantly different in our groups. As a result of our findings mentioned above, TACR1, HTR1E and DRD4 expression patterns may be useful tools in the diagnostic procedures of ADHD and ASD in future studies.
Additionally in this study, a positive and strong correlation was observed between the PRKG1 and OPRM1, PRKG1 and SLC6A4, OPRL1 and DRD4 genes. These positively correlated genes may be thought to have contributed to the autism phenotype by interacting on the same or similar pathways.
Finally, our study revealed significant results with the clinical findings of ASD. This indicates a need for replication of this study both with mRNA and protein levels and for contributing influences to be explored and better understood. To the best of our knowledge, the expressions of the candidate genes that we tested have not yet been investigated in ASD patients displaying aggression and insensitivity to pain. Alterations in the mRNA expression patterns of the HTR1E, OPRL1, OPRM1, TACR1, PRKG1, SCN9A and DRD4 genes provide further evidence for a relevant effect of the respective candidate genes on the pathophysiology of ASD. Low expression of the OPRM1 and CRHR1 genes can make a positive contribution to the autism phenotype. We suggest that CRH levels and the CRHR2 gene may be further investigated in the same way. The presence of expression differences between genders also provides clues about the disease phenotype. Given their potential as markers, mRNA expression patterns may be useful tools in the diagnostic procedures of ASD. Future studies may use larger sample groups to determine the sensitivity and specificity of these putative markers in the mRNA and protein expression profiles of these genes, including other neuropsychiatric disorders.
References
Allely CS (2013) Pain sensitivity and observer perception of pain in individuals with autistic spectrum disorder. Sci World J 2013:916178
Becker JA, Clesse D, Spiegelhalter C et al (2014) Autistic-like syndrome in mu opioid receptor null mice is relieved by facilitated mGluR4 activity. Neuropsychopharmacology 39(9):2049–2060
Benson BA, Brooks WT (2008) Aggressive challenging behavior and intellectual disability. Curr Opin Psychiatry 21:454–458
Bortoluzzi A, Blaya C, da Rosa ED et al (2015) What can HPA axis-linked genes tell us about anxiety disorders in adolescents? Trends Psychiatry Psychother 37(4):232–237
Bustin SA, Benes V, Garson JA et al (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622
Coutinho AM, Sousa I, Martins M et al (2007) Evidence for epistasis between SLC6A4 and ITGB3 in autism etiology and in the determination of platelet serotonin levels. Hum Genet 121(2):243–256
Dominick KC, Davis NO, Lainhart J et al (2007) Atypical behaviors in children with autism and children with a history of language impairment. Res Dev Disabil 28:145–162
Elwin M, Ek L, Schroder A, Kjellin L (2012) Autobiographical accounts of sensing in Asperger syndrome and high-functioning autism. Arch Psychiatr Nurs 26:420–429
Emanuele E, Boso M, Cassola F et al (2010) Increased dopamine DRD4 receptor mRNA expression in lymphocytes of musicians and autistic individuals: bridging the music-autism connection. Neuro Endocrinol Lett 31(1):122–125
Evans JD (1996) Straight forward statistics for the behavioral sciences. Brooks/Cole Publishing, Pacific Grove
Farmer CA, Aman MG (2010) Psychometric properties of the children's scale of hostility and aggression: reactive/proactive (c-sharp). Res Dev Disabil 31:270–280
Furniss F, Biswas AB (2012) Recent research on aetiology, development and phenomenology of self-injurious behaviour in people with intellectual disabilities: a systematic review and implications for treatment. J Intellect Disabil Res 56:453–475
Gadow KD, Devincent CJ, Olvet DM et al (2010) Association of DRD4 polymorphism with severity of oppositional defiant disorder, separation anxiety disorder and repetitive behaviors in children with autism spectrum disorder. Eur J Neurosci 32(6):1058–1065
Geier DA, Kern JK, Geier MR (2013) A comparison of the autism treatment evaluation checklist (ATEC) and the childhood autism rating scale (CARS) for the quantitative evaluation of autism. J Ment Health Res Intellect Disabil 6(4):255–267
Gizer IR, Ficks C, Waldman ID (2009) Candidate gene studies of ADHD: a meta-analytic review. Hum Genet 126(1):51–90
Holden B, Gitlesen JP (2006) A total population study of challenging behavior in the county of Hedmark, Norway: prevalence, and risk markers. Res Dev Dis 27:456–465
Huang CH, Santangelo SL (2008) Autism and serotonin transporter gene polymorphisms: a systematic review and meta-analysis. Am J Med Genet B Neuropsychiatr Genet 147B(6):903–913
Kanne SM, Mazurek MO (2011) Aggression in children and adolescents with ASD: prevalence and risk factors. J Autism Dev Disord 41:926–937
Katsouni E, Sakkas P, Zarros A et al (2009) The involvement of substance P in the induction of aggressive behavior. Peptides 30(8):1586–1591
Kepser LJ, Homberg JR (2015) The neurodevelopmental effects of serotonin: a behavioural perspective. Behav Brain Res 277:3–13
Koishi S, Yamamoto K, Matsumoto H et al (2006) Serotonin transporter gene promoter polymorphism and autism: a family-based genetic association study in Japanese population. Brain Dev 28(4):257–260
Lecavalier L (2006) Behavioral and emotional problems in young people with pervasive developmental disorders: relative prevalence, effects of subject characteristics, and empirical classification. J Autism Dev Disord 36:1101–1114
Longo D, Schüler-Faccini L, Brandalize AP et al (2009) Influence of the 5-HTTLPR polymorphism and environmental risk factors in a Brazilian sample of patients with autism spectrum disorders. Brain Res 1267:9–17
MacLean WE, Tervo RC, Hoch J et al (2010) Self-injury among a community cohort of young children at risk for intellectual and developmental disabilities. J Pediatr 157:979–983
Mazurek MO, Kanne SM, Wodka EL (2013) Physical aggression in children and adolescents with autism spectrum disorders. Res Autism Spectr Disord 7:455–465
Meijer IA, Vanasse M, Nizard S et al (2014) An atypical case of SCN9A mutation presenting with global motor delay and a severe pain disorder. Muscle Nerve 49(1):134–138
Miceli S, Negwer M, van Eijs F et al (2013) High serotonin levels during brain development alter the structural input-output connectivity of neural networks in the rat somatosensory layer IV. Front Cell Neurosci 7:88
Mieres AC, Smallwood V, Nicholson SK (2011) Retrospective case report: evaluation of pain in a child with pervasive developmental disorder. Pediatr Phys Ther 23:194–200
Moore DJ (2015) Acute pain experience in individuals with autism spectrum disorders: a review. Autism 19:387–399
Oades RD, Lasky-Su J, Christiansen H et al (2008) The influence of serotonin- and other genes on impulsive behavioral aggression and cognitive impulsivity in children with attention-deficit/hyperactivity disorder (adhd): findings from a family-based association test (fbat) analysis. Behav Brain Funct 4:48
Provasi D (2015) Computational structural biology of opioid receptors. Methods Mol Biol 1230:13–38
Ressler KJ, Bradley B, Mercer KB et al (2010) Polymorphisms in CRHR1 and the serotonin transporter loci: gene x gene x environment interactions on depressive symptoms. Am J Med Genet B Neuropsychiatr Genet 153B(3):812–824
Riquelme I, Hatem SM, Montoya P (2018) Reduction of pain sensitivity after somatosensory therapy in children with autism spectrum disorders. J Abnorm Child Psychol. https://doi.org/10.1007/s10802-017-0390-6
Ro M, Won S, Kang H et al (2013) Association of the FGA and SLC6A4 genes with autistic spectrum disorder in a Korean population. Neuropsychobiology 68(4):212–220
Rose S, Wynne R, Frye RE et al (2015) Increased susceptibility to ethylmercury-induced mitochondrial dysfunction in a subset of autism lymphoblastoid cell lines. J Toxicol 2015:573701
Ross-Russell M, Sloan P (2005) Autoextraction in a child with autistic spectrum disorder. Br Dent J 198:473–474
Samaco RC, Mandel-Brehm C, McGraw CM, Shaw CA, McGill BE, Zoghbi HY (2012) Crh and Oprm1 mediate anxiety-related behavior and social approach in a mouse model of MECP2 duplication syndrome. Nat Genet 44(2):206–211
Sener EF, Cıkılı Uytun M, Korkmaz Bayramov K et al (2016) The roles of CC2D1A and HTR1A gene expressions in autism spectrum disorders. Metab Brain Dis 31(3):613–619
Sener EF, Sahin M, Taheri S et al (2017) A preliminary study of the genes related with aggression and insensitivity to pain in the autism spectrum disorders. Psychiatry Clin Psychopharmacol 27(1):24–29
Shoval G, Bar-Shira O, Zalsman G et al (2014) Transitions in the transcriptome of the serotonergic and dopaminergic systems in the human brain during adolescence. Eur Neuropsychopharmacol 24(7):1123–1132
Smith KR, Matson JL (2010) Behavior problems: differences among intellectually disabled adults with co-morbid autism spectrum disorders and epilepsy. Res Dev Disabil 31:1062–1069
Spratt EG, Granholm AC, Carpenter LA et al (2015) Pilot study and review: physiological differences in BDNF, a potential biomarker in males and females with autistic disorder. Int Neuropsychiatr Dis J 3(1):19–26
Steinhoff MS, von Mentzer B, Geppetti P et al (2014) Tachykinins and their receptors: contributions to physiological control and the mechanisms of disease. Physiol Rev 94:265–301
Taurines R, Grünblatt E, Schecklmann M et al (2011) Altered mRNA expression of monoaminergic candidate genes in the blood of children with attention deficit hyperactivity disorder and autism spectrum disorder. World J Biol Psychiatry 12(Suppl 1):104–108
Tordjman S, Anderson GM, Botbol M et al (2009) Pain reactivity and plasma beta-endorphin in children and adolescents with autistic disorder. PLoS One 4(8):e5289
Totsika V, Hastings RP, Emerson E et al (2011) A population-based investigation of behavioural and emotional problems and maternal mental health: associations with autism spectrum disorder and intellectual disability. J Child Psychol Psychiatry 52:91–99
Turecki G, Sequeira A, Gingras Y, Séguin M, Lesage A, Tousignant M et al (2003) Suicide and serotonin: study of variation at seven serotonin receptor genes in suicide completers. Am J Med Genet Part B Neuropsychiat Genet 118B:36–40
Valencia AV, Páez AL, Sampedro ME et al (2012) Evidence for association and epistasis between the genetic markers SLC6A4 and HTR2A in autism etiology. Biomedica 32(4):585–601
Waye MMY, Cheng HY (2018) Genetics and epigenetics of autism. Psychiatry Clin Neurosci 72(4):228–244
Whyte A, Jessen T, Varney S, Carneiro AM (2014) Serotonin transporter and integrin beta 3 genes interact to modulate serotonin uptake in mouse brain. Neurochem Int 73:122–126
Woodbury-Smith M, Scherer SW (2018) Progress in the genetics of autism spectrum disorder. Dev Med Child Neurol 60(5):445–451
Zhao Z, Webb BT, Jia P, International Schizophrenia Consortium, Riley BP, Kendler KS, Fanous AH et al (2013) Association study of 167 candidate genes for schizophrenia selected by a multi-domain evidence-based prioritization algorithm and neurodevelopmental hypothesis. PLoS One 8(7):e67776
Acknowledgements
This study has been supported by a grant (Project Number: 114S742) from The Scientific & Technological Research Council (TUBITAK) of Turkey.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors have no conflicts of interest or other disclosures to report.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sener, E.F., Taheri, S., Sahin, M.C. et al. Altered Global mRNA Expressions of Pain and Aggression Related Genes in the Blood of Children with Autism Spectrum Disorders. J Mol Neurosci 67, 89–96 (2019). https://doi.org/10.1007/s12031-018-1213-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-018-1213-0