Abstract
Main Purpose
This study aimed to determine any association of KRAS and BRAF mutations in colorectal cancer with clinicopathological features and overall survival (OS) of Southeast Iranian colorectal cancer (CRC) patients.
Methods
Overall, KRAS and BRAF status were assessed in 100 Iranian CRC subjects. A hundred consecutive stages I–IV CRC patients, who underwent surgical tumor resection from February 2012 to August 2015, were prospectively attained from three centers and were enrolled in the research. Direct sequencing and real-time PCR methods were used to the detection of KRAS and BRAF mutations, respectively. Logistic regression models were used to detect associations of KRAS and BRAF mutations with clinical/clinicopathological features. Kaplan–Meier model was used to estimate overall survival.
Results
In total, KRAS and BRAF mutations were detected in 29 (29%) and 7 (7%) of 100 CRC patients, respectively. BRAF mutations that all comprised V600E and KRAS mutations were found in codon 12, 13, and 61 (72.4%, 20.7 and 6.9%), respectively. In a multivariate analysis, older age (≥ 60) was significantly associated with higher KRAS mutations rate and high BRAF mutation rate was significantly associated with older age (≥ 60) and poorly differentiated tumors. KRAS and BRAF mutant vs. wild type of KRAS and BRAF, 5-year OS was 62.1% vs. 71.8% (p value > 0.05) and 57.1% vs. 67.7% (p value > 0.05), respectively.
Conclusion
Mutations were found in both KRAS and BRAF genes in Iranian colorectal cancers patients and were associated with clinical/clinicopathologic features. Our data emphasizes the importance of these molecular features in Iranian CRC patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the most prevalent cancers and accounts for over 8% of all deaths annually worldwide [1]. In the last few years, incidence and mortality of CRC is increasing rapidly in Iran [2]. CRC is a multifactorial disease and both genetic and environmental factors play an important role in development and susceptibility of the cancer. Multiple alternative genetic pathways involve in CRC tumorigenesis. KRAS and BRAF encode a downstream protein that belongs to the RAS-RAF-MEK-ERK signaling pathway. Hyperactivation of this signaling pathway plays a significant role in the proliferation, differentiation, invasiveness, and metastasis of tumor cells. Oncogenic mutations (especially KRAS and BRAF mutations) lead to the persistent activation of this pathway and accelerate the pathogenesis of CRC [3].
Various studies have reported that about 30–45% of CRC tumors harbor a KRAS mutation [4]. Somatic mutations in KRAS are frequently observed in patients with resistance to antiepidermal growth factor receptor (anti-EGFR) therapy and associated with poor prognosis in metastatic or recurrent CRC [5]. Similarly, several recent reports have suggested that the presence of BRAF mutations in about 10% of CRC tumors can also affect the response to anti-EGFR therapy [6]. BRAF mutations are known as an indicator of poor prognosis and negative predictive biomarkers of anti-EGFR therapy in advanced CRC [7]. The frequency of these somatic mutations varies considerably among different populations. Ethnicity, lifestyle and geographical factors seem to affect the frequency and prognosis of mutation [8,9,10]. Today, genotyping of KRAS and BRAF mutations is routinely undertaken as it is an important biomarker used to predict the poor efficacy of anti-EGFR therapy in patients with metastatic colorectal cancer (mCRC).
To date, most studies about association of KRAS and BRAF mutations with survival and clinicopathological features were from developed countries and only limited studies have been reported about prognostic value of KRAS and BRAF mutations in Iranian patients with CRC [11,12,13]. Therefore, it is needed to assess the prognostic value of these mutations and its relationship with clinical/clinicopathological features. The results from previous studies did not reach a consensus and very few studies were performed to show the prognostic value of KRAS and BRAF mutations on overall survival in Iranian CRC patients. Consequently, the present study aimed to identify the frequency of KRAS and BRAF gene mutations in Iranian CRC patients, and investigate the prognostic value of KRAS and BRAF mutations and their associations with clinicopathological features.
Methods
Patient Samples
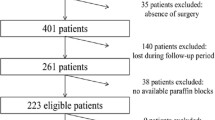
A hundred formalin-fixed, paraffin-embedded (FFPE) tumor blocks from patients diagnosed with colorectal cancer from February 2012 to August 2015 at the three different hospitals (Afzalipour, bahonar and mehregan hospitals) throughout Kerman province (southeast of Iran) were retrieved. Tumor sections from each FFPE tissue sample were stained with hematoxylin and eosin (H&E) and reviewed by two experienced pathologists (S.D and N.K.K) independently to estimate the percentage of tumor cells. Then, from different FFPE tissue blocks of each patient, the rich-tumor areas (more than 50% tumor cells) with lowest necrosis, hemorrhage, normal colonic cells, stromal cells, and blood-derived leukocytes were selected for genomic DNA extraction. The population study included patients with initial diagnosis of CRC and no patients had accepted adjuvant treatment at the time of sampling. Demographic, clinical, and clinicopathological data were obtained by reviewing the medical records to extract the following information which include age of diagnosis, sex, smoking status, alcohol intake, family history, tumor location (right, left or rectum), differentiation grade (well, moderate or poor), TNM stage (I, II, III, or IV), lymph node metastasis, and distant metastasis. Adjuvant chemotherapy was recommended to CRC patients according to Iranian Ministry of Health guidelines and international guidelines for diagnosis and treatment of CRC. According to the RAS driver variants status, patients were offered targeted agents as an adjunct to systemic chemotherapy. However, due to insurance and economic issues, no patients received anti-EGFR and/or anti-VEGF therapy during the study period. This study was performed under the license from Ethics Committee of Kerman University of Medical Sciences (Approval No. IR.KMU.REC.1397.209), and due to the retrospective nature of the study and unavailability of many patients, informed consent was rejected.
Formalin-Fixed Paraffin-Embedded Tissue DNA Extraction
For Genomic DNA extraction, 5–10-μm-thick sections were cut from FFPE tumor tissue blocks for each case and collected in 1.5 ml tubes. Paraffin was removed by using two washes with 1 ml of absolute xylol and a wash in 1 ml absolute ethanol. After each wash, the sections were vortexed and centrifuged at 13,000 × rpm for 10 min, and the supernatant was then removed. DNA was extracted from FFPE specimens using the QIAamp DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. The concentration and purity of all DNA samples were measured using NanoDrop ND-2000c Spectrophotometer (Thermo Scientific, USA). The mean concentration of DNA was 220.4 ng/μl, range from 40.2 to 358.6, and the ratio of A260/A280 was from 1.6 to 2.1. DNA was finally eluted in 50 μl of ATE buffer and stored at − 20 °C until use.
Detection of BRAFV600E Mutation
Detection of BRAFV600E mutation was performed using the therascreen BRAF RGQ PCR Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol. This kit utilizes a combination of ARMS (Amplification Refractory Mutation System) and Scorpion probes technology for mutation-specific amplification and detection of PCR products, respectively. This method allows detection of four different mutation including V600E, V600K, V600D, and V600R. Amplification, detection, and data analysis were performed on a Corbett Rotor-Gene 6000 Real-time PCR instrument (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions.
Detection of KRAS Mutations
The mutational analysis of KRAS (exon 2 and 3) was performed using PCR products and bidirectional sequencing from DNA samples. The primers used to evaluate exon 2 [14] and 3 [15] of KRAS were as previously described. PCR was performed on a Bio-Rad T100 Thermal Cycler in a total volume of 30 μl, containing 40–100 ng of template DNA, 15 μl 2× Taq DNA Polymerase Master Mix RED (Ampliqon, Denmark), and 10 mM of each primer. Amplification was carried out with the following condition: 95 °C for 10 min (first cycle); 35 cycle of 95 °C for 15 s, 56 °C for 15 s, 72 °C for 15 s; and final extension cycle at 72 °C for 10 min. PCR products were purified with a PCR product purification kit (Roche, Germany), according to the manufacturer’s instructions. Forward and reverse strands sequenced by the BigDye Terminator v3.1 kit (Applied Biosystems, Foster City, USA) on an ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, USA) and the sequence data were analyzed using Sequencing Analysis software v5.0 (Applied Biosystems, Foster City, USA). Visual evaluation of each chromatogram was done by two independent investigators and all abnormal or ambiguous sequences were confirmed by re-sequencing. Sequences were compared by the BLAST tool (www.ncbi.nim.nih.gov/BLAST/).
Statistical Analysis
The continuous data were presented in mean ± SD, analyzed by independent student t test. Normality of data was analyzed by Kolmogorov–Smirnov and Shapiro–Wilk test. Categorical variable data analysis was conducted using the χ2 or Fisher’s exact test. The χ2 test or Fisher’s exact test was used to compare the proportion of mutations among patients with different clinicopathological data. Logistic regression models were used to analyze the association based on the estimation of the odds ratios (ORs) and 95% confidence intervals (CIs). Overall survival (OS) was defined since the date of diagnosis up to the date of death or last of follow-up visit. The overall survival was plotted and analyzed by Kaplan–Meier (log-rank test). All statistical analyses were conducted by using SPSS 22.0 statistical package (SPSS Inc., Chicago, IL, USA). All p values were two-sided. The statistical significance was considered if the p value < 0.05.
Results
Characteristics of CRC Patients
In this study, we retrieved 100 FFPE tissue blocks from 3 different centers of Kerman province. Table 1 summarizes demographic data and clinicopathological characteristics of CRC patients. Briefly, the prevalence of CRC was 64% in males and 36% in females. The average patient age was 59.60 ± 15.24 years (range from 19 to 85 years), and patients who aged younger than 60 years and older than 60 years respectively represented in 46% and 54% of the patients. The proportion of familial history of CRC was 15%. The tumor size of the patients ranged approximately from 2 to 10 cm (median size was 5.85 ± 3.4). The tumors were located at the right side of the colon (29%), including cecum and transverse colon; left side of the colon (30%), including sigmoid colon and splenic flexure; and rectum (41%). Regarding the tumor differentiation, 8% of tumors were well differentiated, 78% were moderately differentiated and 14% were poorly differentiated. In total, the percentage of patients in stages I to IV were 11, 17, 59, and 13, respectively. The liver metastasis (10%) had the most frequently metastatic site, followed by nonregional lymph node and vessels (each 7%). There was no clinical information regarding previous chemotherapy.
Distribution of KRAS and BRAFV600E Mutations in CRC Patients
The prevalence and distribution of KRAS and BRAF mutations in the CRC patients is showed in Table 2. KRAS exon 2 and 3 amplification was done using polymerase chain reaction and detected in the presence of 293 bp and 289 bp fragments on 2% agarose gel electrophoresis, respectively. An example of representative electropherogram of KRAS mutant (exon 2 and 3) is shown in Fig. 1. KRAS mutation was identified in 29 (29%) of all the patient samples. Among 29 KRAS mutants, 27 (27%) had mutations in exon 2 and 2 (2%) in exon 3. Within KRAS exon 2, 21 (77.8%) of the mutations were identified in codon 12, and 6 (22.2%) were in codon 13. The most frequently observed mutation (13 of 27) was a 35G>A transition in codon 12 (G12D), followed by 38G>A (G13D) and 35G>T (G12V) (6 of 27 each). Also, 2 mutations were detected in codon 61 of exon 3 (183A>C, Q61H). Of the 100 tumor samples, 7 samples (7%) harbored a mutation in codon 600 of the BRAF gene (1799T>A, V600E). No other recurrent forms of BRAFV600 mutations (including V600K, V600D, and V600R) were identified in current study. Figure 2a–d shown the distributions of all three tumor subgroups (KRAS-mutant, BRAF-mutant, and negative) with respect to tumor sites, tumor differentiation, TNM stage, and distant metastasis. Additionally, Fig. 2e shown the distribution of KRAS and BRAF mutations by specific mutation in all included patients.
Correlation of KRAS and BRAF Gene Mutations with Clinical and Clinicopathological Characteristics
We analyzed the correlation between KRAS or BRAF mutations and the clinicopathological characteristics of CRC samples. A summary of the relationships between KRAS or BRAF mutations and different clinicopathological characteristics is provided in Table 3. Statistical analysis of the various characteristics variables showed a significant correlation between KRAS mutations and the older age (≥ 60 years) (OR 1.045; 95% CI 1.018–1.093,p = 0.044), but there were no statistically significant correlation between KRAS mutations, and sex (OR 0.934; 95% CI 0.328–2.659,p = 0.898), smoking (OR 2.863; 95% CI 0.92–8.85,p = 0.410), alcohol intake (OR 2.225; 95% CI 0.447–11.075, p = 0.329), family history (OR 0.924; 95% CI 0.233–3.662, p = 0.910), tumor location (p = 0.759), tumor size (OR 1.119; 95% CI 0.589–2.093, p = 0.729), tumor differentiation (p = 0.728), TNM stage (p = 0.733), lymph node metastasis (p = 0.543), and distant metastasis (OR 0.929; 95% CI 0.289–2.982, p = 0.901). Although, no correlation was observed between specific mutations in codon 12 and 13 and different clinicopathological parameters (data not shown).
The BRAFV600E mutant tumors in clinical samples were associated with older age (≥ 60 years) (OR 2.947; 95% CI 0.283–8.761, p = 0.031) and poor differentiation (OR 3.162; 95% CI 1.131–14.437, p = 0.017). There were no statistically significant correlation between BRAFV600E mutation and sex (OR 1.129; 95% CI 0.468–2.724, p = 0.778), smoking (OR 0.507; 95% CI 0.582–4.437, p = 0.533), alcohol intake (OR 0.935; 95% CI 0.087–0.987, p = 0.488), family history (OR 1.731; 95% CI 0.273–10.998, p = 0.533), tumor location (p = 0.671), tumor size (OR 1.195; 95% CI 0.638–2.240, p = 0.578), TNM stage (p = 0.688), lymph node metastasis (p = 0.943), and distant metastasis (OR 1.457; 95% CI 0.612–3.454, p = 0.390).
Overall Survival Analysis
Median of survival time for KRAS mutant was 42.6 months and for KRAS wild-type patients was 45.9 months (overall = 44.9 months). The Kaplan–Meier survival curves (5-year OS) based on KRAS and BRAF status has been shown in Figs. 3 and 4, respectively. The analysis showed no difference of OS between KRAS mutant patients and KRAS wild-type patients in 5-year (62.1% vs. 71.8%; log rank p = 0.543) OS. Similarly, patients with BRAF mutations have no significant association with overall survival rate. Median of survival time in patients with BRAF mutation was 39.7 and in BRAF wild-type patients was 44.5 months (overall = 44.3 months). As shown in Fig. 3, the 5-year OS rate in BRAF-mutant and BRAF wild-type patients was 57.1% and 67.7%, respectively (log rank p = 0.673).
Discussion
In this study, we evaluated KRAS and BRAFV600E mutations frequencies in 100 CRC FFPE tissue samples from three different hospitals located in Kerman, Iran. The present study utilized direct sequencing and real-time PCR methods to analyze the mutational status of KRAS and BRAFV600E in Iranian CRC patients, respectively. According to our findings, the prevalence of KRAS and BRAFV600E mutations were 29% and 7%, respectively. The remaining 64% of patients had no mutation in any type of genes analyzed.
KRAS is the most studied gene of RAS-RAF-MAPK pathway in CRC. It triggers the downstream cascades including the PIK3-AKT pathway, which may affect the cell proliferation, differentiation, and invasion [16]. Mutations in KRAS negatively predict the response to anti-EGFR therapies in patients with metastatic CRC. According to previous studies, the prevalence of KRAS mutation in CRC patients varies from 12.9 to 66.1% (mostly 30–45%) across the globe [17]. To our findings, the frequency of Iranian CRC patients with KRAS mutant tumors was 29%, which was similar to published reports from Asia and Europe [18,19,20]. It means that, if KRAS mutant profiling is applied to select candidates for anti-EGFR treatment, the number of Iranian patients that would be excluded is similar to that of other populations. In CRC patients, most of the KRAS mutations occur in codon G12 or G13 (about 90%), and the G12D (35G>A) is the most common mutation which results in an amino acid substitution (from glycine to aspartic acid) in KRAS codon 12 [21, 22]. Our data were consistent with these reports. In comparison with Iranian studies, these data is similar to other published data from various regions of Iran [11, 23,24,25,26].
Several studies have reported the associations of KRAS mutation with different clinical and/or clinicopathological features, including female gender [27,28,29,30,31], age at diagnosis (> 50 years) [28], location of tumor (right side) [18, 27, 31], tumor differentiation (well/moderately differentiated) [18, 31,32,33], TNM stage [30], and microsatellite-stable phenotype [33] in Caucasian and Asian CRC populations, while others did not report any association [34, 35]. In the current study, our findings showed the association of KRAS mutation with older age (≥ 60 years), which is consistent with a recent report of Iranian patients [12]. This condition can be due to increased genetic alterations of tumors with age. However, this finding was contrary to the findings of a study conducted by Nazemalhosseini-Mojarad et al. in Iranian CRC patients [11]. Other clinicopathologic features did not correlate significantly with the occurrence of KRAS mutations.
According to several earlier reports, there is no convincing evidence that KRAS mutations are independent prognostic biomarker for poor OS in CRC patients [5, 34, 36, 37]. Among these, Abubaker et al. [36] and Richman et al. [5] found that KRAS mutations are associated with a poorer overall survival. Additionally, a meta-analysis conducted by Qiu et al. [38] showed that OS was significantly shorter in KRAS mutant patients compared with that in KRAS wild-type patients. In contrast, a recent study on 353 Chinese CRC patients revealed that KRAS mutations were not associated with OS, but BRAF mutations were associated with poorer OS [34]. Conversely to many published reports, we identified no significant association between KRAS mutations and OS in Iranian patients with CRC and it can be due to our sample selection or low sample size. Similar to our findings, recent Iranian studies did not find any association between KRAS mutation and OS [11, 13].
BRAF is a downstream member of the RAS-RAF-MAPK signaling pathway and its mutation is the most commonly observed gene alteration after KRAS mutation in colorectal cancer. In the present study, the prevalence of BRAFV600E mutation was 7% (7/100), one of the highest prevalence reported in the Iranian CRC patients. Notably, most Iranian studies reported low frequency of BRAFV600E (mostly no BRAF mutation) in CRC patients [39,40,41,42]. In 2008, Brim et al. reported a very low frequency of BRAF mutation (2%) among Iranian CRC patients [42]. Additionally, Nazemalhosseini-Mojarad et al. [11] who aimed to explore the BRAFV600E mutation in 258 Iranian CRC patients also demonstrated that the prevalence of BRAFV600E mutation was 5.8%. According to best of our knowledge, this is the first Iranian study that used real-time PCR method in reporting the frequency of BRAF mutation among CRC patients, which can be a reason for the higher frequency of BRAF mutation. Although, a study conducted by Mohammadi-Asl et al. [43] showed that 46.25% (37/80) of the patients with colorectal cancer had BRAFV600E mutation. This high frequency of BRAFV600E mutation in CRC from Ahwaz city (southwest of Iran) could be due to different sample selections, different methodologies, different ethnicity of people, geographical and environmental features of this region, and common lifestyle. In addition, the frequency of BRAFV600E mutation is higher than several Asian populations (China, Japan, Korea, Taiwan, and Malaysia), but similar to that of western countries (France, UK, USA, Greece, and Italy [18, 20, 27, 33, 44,45,46,47,48,49,50,51,52]. According to published data from a distinct ancestral populations study, BRAF mutations occurring at a higher frequency in European patients with colorectal cancer with (17%) versus Asian (4%) [53]. BRAFV600E mutation frequency ranged from 0 to 22% within CRC patients from various geographical regions across the globe [17, 18, 40, 47, 54]. A recent meta-analysis conducted by Lowe et al. in 2019 [55] estimated that the global prevalence of BRAF mutation in CRC patients was 7.1%; however, two previous meta-analysis reported that it was 10.8% and 11.1% [56, 57]. These data indicate a slight reduction in the prevalence of BRAF mutation across the globe. The exact reason of this variation is still not clear, but the racial and/or environmental factors might contribute to the difference. Several studies have reported that BRAF mutation existed only in KRAS wild-type tumoral tissues, which is consistent with our results [17, 18, 32, 58]; however, few studies reported concurrent mutation of KRAS and BRAF mutation in CRC patients [27]. In this context, tumor heterogeneity may play a role [59].
We found that BRAF mutations occurred more frequently in older patients (age ≥ 60 years). BRAF mutations were also more common in poorly differentiated tumors, which is inconsistent with previous reports from different regions of Iran [11, 39]. Recent studies in Iranian populations did not find any correlations between BRAF mutations and clinical and/or clinicopathological features [11, 39]; however, most studies did not investigate the association of BRAF mutation with clinical and/or clinicopathological features [26, 40,41,42]. In contrast, a recent study reported association of BRAFV600E mutation with mucinous characteristics in Iranian CRC patients [11]. In several case series and meta-analysis studies, BRAF mutation was either strongly correlated with clinicopathological features such as older age, female gender, tumor location, poorly differentiated tumor, or at least showed a trend toward such correlations [27, 33, 46, 56, 57, 60].
Several studies widely reported that there is a correlation between BRAFV600E mutation and worse OS in CRC patients [34, 61]. Although, our findings showed that BRAFV600E mutation was not correlated with OS. Similarly, a previous research from Iran has reported that BRAF mutations are not associated with worse OS [11]. Anyway, this findings requires further confirmation in a larger Iranian CRC population and multicenter setting given the relatively low frequencies of BRAFV600E mutation (n = 7) in our study.
There were some limitations of this study. Part of our study limitations were small size and bias sample selection, incomplete information on clinicopathological data, observational retrospective nature of research, incomplete follow-up time such as recurrence and OS, and absence of epigenetic or MSI status of tumor tissues. Additionally, some hotspot mutations such as in exon 4 of the KRAS gene and in exon 11 and 15 of the BRAF gene were not screened. We collected tumoral tissues from three different hospitals in Kerman province (southeast of Iran) and evaluated the frequency of KRAS and BRAF mutations, associations with clinicopathological data, and correlations of these mutations with poor prognosis in Iranian CRC patients. These aspects make our findings more representative and prognostic for new CRC patients in Iran or southeast of Iran at least.
Conclusion
In this study, we identified KRAS and BRAFV600E mutations in 29 (29%) and 7 (7%) of the Iranian CRC patients, respectively. Distinctively, our study revealed a higher prevalence of BRAFV600E mutation in Iranian CRC patients. The prevalence of KRAS mutations was higher in older age CRC patients. Also, the BRAFV600E mutation was more common in older age patients, and patients who showed poor differentiation in clinical samples. This study adds to the evidence that KRAS and BRAF mutations in Iranian colorectal cancer patients occur at a similar status to that of other populations; however, prevalence of BRAF mutation is higher in this study than in previous Iranian studies. Our findings open the field to further studies investigating how these mutations can be variable in frequency in different populations. However, large-scale clinical studies are needed to confirm this finding in Iranian CRC patients.
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. https://doi.org/10.1002/ijc.29210.
Azadeh S, Moghimi-Dehkordi B, Fatemi SR, Pourhoseingholi MA, Ghiasi S, Zali MR. Colorectal cancer in Iran: an epidemiological study. Asian Pacific J Cancer Prev. 2008;9:123–6.
Eklöf V, Wikberg ML, Edin S, Dahlin AM, Jonsson BA, Öberg Å, Rutegård J, Palmqvist R. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br J Cancer. 2013;108:2153–63. https://doi.org/10.1038/bjc.2013.212.
Ren J, Li G, Ge J, Li X, Zhao Y. Is K-ras gene mutation a prognostic factor for colorectal Cancer. Dis Colon rectum. 2012;55:913–23. https://doi.org/10.1097/DCR.0b013e318251d8d9.
Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P. KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol. 2009;27:5931–7. https://doi.org/10.1200/JCO.2009.22.4295.
Rowland A, Dias MM, Wiese MD, Kichenadasse G, McKinnon RA, Karapetis CS, Sorich MJ. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer. 2015;112:1888–94. https://doi.org/10.1038/bjc.2015.173.
Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, Maggi C, Cabiddu M, Iacovelli R, Bossi I, Lonati V, Ghilardi M, De Braud F, Barni S. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer. 2015;51:587–94. https://doi.org/10.1016/j.ejca.2015.01.054.
Baldus SE, Schaefer K-L, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16:790–9. https://doi.org/10.1158/1078-0432.CCR-09-2446.
Jayasekara H, English DR, Haydon A, Hodge AM, Lynch BM, Rosty C, Williamson EJ, Clendenning M, Southey MC, Jenkins MA, Room R, Hopper JL, Milne RL, Buchanan DD, Giles GG, MacInnis RJ. Associations of alcohol intake, smoking, physical activity and obesity with survival following colorectal cancer diagnosis by stage, anatomic site and tumor molecular subtype. Int J Cancer. 2018;142:238–50. https://doi.org/10.1002/ijc.31049.
Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: the evolving field of molecular pathological epidemiology. J Natl Cancer Inst. 2010;102:365–7. https://doi.org/10.1093/jnci/djq031.
Nazemalhosseini-Mojarad E, Kishani Farahani R, Mehrizi M, Baghaei K, Yaghoob Taleghani M, Golmohammadi M, Peyravian N, Ashtari S, Pourhoseingholi MA, Asadzadeh Aghdaei H, Zali MR. Prognostic Value of BRAF and KRAS Mutation in Relation to Colorectal Cancer Survival in Iranian Patients: Correlated to Microsatellite Instability. In: Prognostic value of BRAF and KRAS mutation in relation to colorectal cancer survival in Iranian patients: correlated to microsatellite instability. Cancer: J. Gastrointest; 2019. https://doi.org/10.1007/s12029-019-00201-4.
Shahriari-Ahmadi A, Ansarinejad N, Fardad F, Abbaszadeh M, Sadeghi M. KRAS and NRAS testing in metastatic colorectal cancer in Central Iran (Tehran): a review on literature of the middle east. Indian J Med Paediatr Oncol. 2018;39:210. https://doi.org/10.4103/ijmpo.ijmpo_133_17.
Payandeh M, Shazad B, Sadeghi M, Shahbazi M. Correlation between RAS test results and prognosis of metastatic colorectal cancer patients: a report from Western Iran. Asian Pac J Cancer Prev. 2016;17:1729–32. https://doi.org/10.7314/apjcp.2016.17.4.1729.
S. Jancik, J. Drabek, J. Berkovcova, Y.Z. Xu, M. Stankova, J. Klein, V. Kolek, J. Skarda, T. Tichy, I. Grygarkova, D. Radzioch, M. Hajduch, A comparison of Direct sequencing, Pyrosequencing, High resolution melting analysis, TheraScreen DxS, and the K-ras StripAssay for detecting KRAS mutations in non small cell lung carcinomas, J. Exp. Clin. Cancer Res. 31 (2012). https://doi.org/10.1186/1756-9966-31-79.
Rechsteiner M, Von Teichman A, Rüschoff JH, Fankhauser N, Pestalozzi B, Schraml P, Weber A, Wild P, Zimmermann D, Moch H. KRAS, BRAF, and TP53 deep sequencing for colorectal carcinoma patient diagnostics. J Mol Diagnostics. 2013;15:299–311. https://doi.org/10.1016/j.jmoldx.2013.02.001.
Harari PM, Allen GW, Bonner JA. Biology of interactions: Antiepidermal growth factor receptor agents. J Clin Oncol. 2007;25:4057–65. https://doi.org/10.1200/JCO.2007.11.8984.
Bisht S, Ahmad F, Sawaimoon S, Bhatia S, Das BR. Molecular spectrum of KRAS, BRAF, and PIK3CA gene mutation: determination of frequency, distribution pattern in Indian colorectal carcinoma. Med Oncol. 2014;31:124. https://doi.org/10.1007/s12032-014-0124-3.
Zhang J, Zheng J, Yang Y, Lu J, Gao J, Lu T, Sun J, Jiang H, Zhu Y, Zheng Y, Liang Z, Liu T. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Sci Rep. 2015;5:18678. https://doi.org/10.1038/srep18678https://www.nature.com/articles/srep18678#supplementary-information.
Palomba G, Colombino M, Contu A, Massidda B, Baldino G, Pazzola A, Ionta M, Capelli F, Trova V, Sedda T, Sanna G, Tanda F, Budroni M, Palmieri G, Cossu A. Prevalence of KRAS, BRAF, and PIK3CA somatic mutations in patients with colorectal carcinoma may vary in the same population: clues from Sardinia. J Transl Med. 2012;10:178. https://doi.org/10.1186/1479-5876-10-178.
Aoyagi H, Iida S, Uetake H, Ishikawa T, Takagi Y, Kobayashi H, Higuchi T, Yasuno M, Enomoto M, Sugihara K. Effect of classification based on combination of mutation and methylation in colorectal cancer prognosis. Oncol Rep. 2011;25:789–94. https://doi.org/10.3892/or.2010.1118.
Yoon HH, Tougeron D, Shi Q, Alberts SR, Mahoney MR, Nelson GD, Nair SG, Thibodeau SN, Goldberg RM, Sargent DJ, Sinicrope FA. KRAS codon 12 and 13 mutations in relation to disease-free survival in BRAF-wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance). Clin Cancer Res. 2014;20:3033–43. https://doi.org/10.1158/1078-0432.CCR-13-3140.
Piton N, Lonchamp E, Nowak F, Sabourin JC, Christiane M, Marie-Pierre G, Philippe MJ, Pierre D. Real-life distribution of KRAS and NRAS mutations in metastatic colorectal carcinoma from French routine genotyping. Cancer Epidemiol Biomark Prev. 2015;24:1416–8. https://doi.org/10.1158/1055-9965.EPI-15-0059.
Vakil L, Najafipour R, Rakhshani N, Zamani F, Morakabati A, Javadi A. Investigation of FIH-1 and SOCS3 expression in KRAS mutant and wild-type patients with colorectal cancer. Tumor Biol. 2016;37:8841–8. https://doi.org/10.1007/s13277-015-4723-1.
Naghibalhossaini F, Hosseini HM, Mokarram P, Zamani M. High frequency of genes’ promoter methylation, but lack of BRAF V600E mutation among Iranian colorectal cancer patients. Pathol Oncol Res. 2011;17:819–25. https://doi.org/10.1007/s12253-011-9388-5.
Mohsen N, Ahmadreza S, Fatemeh H, Fatemeh H, Fariba ER. Frequency of K-RAS and N-RAS gene mutations in colorectal cancers in southeastern Iran. Asian Pac J Cancer Prev. 2016;17:4511–5. http://www.ncbi.nlm.nih.gov/pubmed/27880995
A. Koochak, N. Rakhshani, M.H. Karbalaie Niya, F.S. Tameshkel, M.R. Sohrabi, M.R. Babaee, H. Rezvani, B. Bahar, F. Imanzade, F. Zamani, M.R. Khonsari, H. Ajdarkosh, G. Hemmasi, Mutation analysis of KRAS and BRAF genes in metastatic colorectal cancer: a first large scale study from Iran., Asian Pac. J. Cancer Prev. 17 (2016) 603–8. http://www.ncbi.nlm.nih.gov/pubmed/26925650 ().
Z. Ye, W. Guan, T. Tang, F. Wang, … Y.Z.-A. at S., undefined 2019, Gene mutation profiling in chinese colorectal cancers patients and its association with clinicopathological characteristics and prognosis, Papers.Ssrn.Com. (n.d.). https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3405556 ().
Watanabe T, Yoshino T, Uetake H, Yamazaki K, Ishiguro M, Kurokawa T, Saijo N, Ohashi Y, Sugihara K. KRAS mutational status in Japanese patients with colorectal cancer: results from a Nationwide, multicenter, cross-sectional study. Jpn J Clin Oncol. 2013;43:706–12. https://doi.org/10.1093/jjco/hyt062.
Goetsch AL, Kimelman D, Woodruff TK. Disorders of the Sex Chromosomes and Sexual Development. In: Disorders of the sex chromosomes and sexual development, in: Fertil. Preserv. Restor. Patients with Complex Med. Cond: Springer International Publishing, Cham; 2017. p. 19–37. https://doi.org/10.1007/978-3-319-52316-3_3.
Li W, Qiu T, Zhi W, Shi S, Zou S, Ling Y, Shan L, Ying J, Lu N. Colorectal carcinomas with KRAS codon 12 mutation are associated with more advanced tumor stages. BMC Cancer. 2015;15:340. https://doi.org/10.1186/s12885-015-1345-3.
X. Fu, Y. Huang, X. Fan, Y. Deng, H. Liu, H. Zou, P. Wu, Z. Chen, J. Huang, J. Wang, H. Lin, S. Huang, X. Tan, P. Lan, L. Wang, J. Wang, Demographic trends and KRAS/BRAF V600E mutations in colorectal cancer patients of South China: a single-site report, Int. J. Cancer. (2019) ijc.31973. https://doi.org/10.1002/ijc.31973.
Shen Y, Wang J, Han X, Yang H, Wang S, Lin D, Shi Y. Effectors of epidermal growth factor receptor pathway: the genetic profiling of KRAS, BRAF, PIK3CA, NRAS mutations in colorectal cancer characteristics and personalized medicine. PLoS One. 2013;8:e81628. https://doi.org/10.1371/journal.pone.0081628.
Rimbert J, Tachon G, Junca A, Villalva C, Karayan-Tapon L, Tougeron D. Association between clinicopathological characteristics and RAS mutation in colorectal cancer. Mod Pathol. 2018;31:517–26. https://doi.org/10.1038/modpathol.2017.119.
F. Guo, H. Gong, H. Zhao, J. Chen, Y. Zhang, L. Zhang, X. Shi, A. Zhang, H. Jin, J. Zhang, Y. He, Mutation status and prognostic values of KRAS, NRAS, BRAF and PIK3CA in 353 Chinese colorectal cancer patients, Sci. Rep. 8 (2018). https://doi.org/10.1038/s41598-018-24306-1.
Akman T, Oztop I, Baskin Y, Unek I, Demir N, Ellidokuz H, Yilmaz A. The association of clinicopathological features and survival in colorectal cancer patients with KRAS mutation status. J Cancer Res Ther. 2016;12:96–102. https://doi.org/10.4103/0973-1482.148684.
Abubaker J, Bavi P, Al-Haqawi W, Sultana M, Al-Harbi S, Al-Sanea N, Abduljabbar A, Ashari LH, Alhomoud S, Al-Dayel F, Uddin S, Al-Kuraya KS. Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal carcinoma. J Pathol. 2009;219:435–45. https://doi.org/10.1002/path.2625.
Andreyev HJN, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85:692–6. https://doi.org/10.1054/bjoc.2001.1964.
Qiu LX, Mao C, Zhang J, Zhu XD, Liao RY, Xue K, Li J, Chen Q. Predictive and prognostic value of KRAS mutations in metastatic colorectal cancer patients treated with cetuximab: a meta-analysis of 22 studies. Eur J Cancer. 2010;46:2781–7. https://doi.org/10.1016/j.ejca.2010.05.022.
Dolatkhah R, Somi MH, Kermani IA, Bonyadi M, Sepehri B, Boostani K, Azadbakht S, Fotouhi N, Farassati F, Dastgiri S. Association between proto-oncogene mutations and clinicopathologic characteristics and overall survival in colorectal cancer in East Azerbaijan, Iran. Onco Targets Ther. 2016;9:7385–95. https://doi.org/10.2147/OTT.S116373.
Javadi F, Geramizadeh B, Mirzai M. BRAF gene mutation analysis in colorectal cancer in South of Iran. Ann Color Res. 2014;2:1–5.
Molaei M, Farahani RK, Maftouh M, Taleghani MY, Vahdatinia M, Khatami F, Nazemalhosseini-Mojarad E, Aghdae HA, Aboutorabi A, Zali MR. Lack of BRAFV600E mutation in stage I and II of colorectal cancer. Gastroenterol Hepatol from Bed to Bench. 2016;9:94–9.
Brim H, Mokarram P, Naghibalhossaini F, Saberi-Firoozi M, Al-Mandhari M, Al-Mawaly K, Al-Mjeni R, Al-Sayegh A. Impact of BRAF, MLH1 on the incidence of microsatellite instability high colorectal cancer in populations based study. Mol Cancer. 2008;7:1–11. https://doi.org/10.1186/1476-4598-7-68.
Asl JM, Almasi S, Tabatabaiefar MA. High frequency of BRAF proto-oncogene hot spot mutation V600E in cohort of colorectal cancer patients from Ahvaz City, Southwest Iran. Pakistan J Biol Sci PJBS. 2014;17:565–9. https://doi.org/10.3923/PJBS.2014.565.569.
Kwon MJ, Lee SE, Kang SY, La Choi Y. Frequency of KRAS, BRAF, and PIK3CA mutations in advanced colorectal cancers: comparison of peptide nucleic acid-mediated PCR clamping and direct sequencing in formalin-fixed, paraffin-embedded tissue. Pathol Res Pract. 2011;207:762–8. https://doi.org/10.1016/j.prp.2011.10.002.
Yip WK, Choo CW, Leong VC-S, Leong PP, Jabar MF, Seow HF. Molecular alterations of Ras-Raf-mitogen-activated protein kinase and phosphatidylinositol 3-kinase-Akt signaling pathways in colorectal cancers from a tertiary hospital at Kuala Lumpur, Malaysia. APMIS. 2013;121:954–66. https://doi.org/10.1111/apm.12152.
Kawazoe A, Shitara K, Fukuoka S, Kuboki Y, Bando H, Okamoto W, Kojima T, Fuse N, Yamanaka T, Doi T, Ohtsu A, Yoshino T. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer. 2015;15:258. https://doi.org/10.1186/s12885-015-1276-z.
Hsieh L-L, Er T-K, Chen C-C, Hsieh J-S, Chang J-G, Liu T-C. Characteristics and prevalence of KRAS, BRAF, and PIK3CA mutations in colorectal cancer by high-resolution melting analysis in Taiwanese population. Clin Chim Acta. 2012;413:1605–11. https://doi.org/10.1016/J.CCA.2012.04.029.
Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E, Freed E, Meyerhardt JA, Saridaki Z, Georgoulias V, Finkelstein D, Fuchs CS, Kulke MH, Shivdasani RA. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101:465–72. https://doi.org/10.1038/sj.bjc.6605164.
Janku F, Lee JJ, Tsimberidou AM, Hong DS, Naing A, Falchook GS, Fu S, Luthra R, Garrido-Laguna I, Kurzrock R. PIK3CA mutations frequently coexist with RAS and BRAF mutations in patients with advanced cancers. PLoS One. 2011;6:e22769. https://doi.org/10.1371/journal.pone.0022769.
Rosty C, Young JP, Walsh MD, Clendenning M, Sanderson K, Walters RJ, Parry S, Jenkins MA, Win AK, Southey MC, Hopper JL, Giles GG, Williamson EJ, English DR, Buchanan DD. PIK3CA activating mutation in colorectal carcinoma: associations with molecular features and survival. PLoS One. 2013;8:e65479. https://doi.org/10.1371/journal.pone.0065479.
Saridaki Z, Tzardi M, Papadaki C, Sfakianaki M, Pega F, Kalikaki A, Tsakalaki E, Trypaki M, Messaritakis I, Stathopoulos E, Mavroudis D, Georgoulias V, Souglakos J. Impact of KRAS, BRAF, PIK3CA mutations, PTEN, AREG, EREG expression and skin rash in ≥2nd line Cetuximab-based therapy of colorectal cancer patients. PLoS One. 2011;6:e15980. https://doi.org/10.1371/journal.pone.0015980.
Simi L, Pratesi N, Vignoli M, Sestini R, Cianchi F, Valanzano R, Nobili S, Mini E, Pazzagli M, Orlando C. High-resolution melting analysis for rapid detection of KRAS, BRAF, and PIK3CA gene mutations in colorectal cancer. Am J Clin Pathol. 2008;130:247–53. https://doi.org/10.1309/LWDY1AXHXUULNVHQ.
Hanna MC, Go C, Roden C, Jones RT, Pochanard P, Javed AY, Javed A, Mondal C, Palescandolo E, Van Hummelen P, Hatton C, Bass AJ, Chun SM, Na DC, Kim T-I, Jang SJ, Osarogiagbon RU, Hahn WC, Meyerson M, Garraway LA, MacConaill LE. Colorectal cancers from distinct ancestral populations show variations in BRAF mutation frequency. PLoS One. 2013;8:e74950. https://doi.org/10.1371/journal.pone.0074950.
Sorbye H, Dragomir A, Sundström M, Pfeiffer P, Thunberg U, Bergfors M, Aasebø K, Eide GE, Ponten F, Qvortrup C, Glimelius B. High BRAF mutation frequency and marked survival differences in subgroups according to KRAS/BRAF mutation status and tumor tissue availability in a prospective population-based metastatic colorectal cancer cohort. PLoS One. 2015;10:e0131046. https://doi.org/10.1371/journal.pone.0131046.
Lowe K, Bylsma LC, Levin-Sparenberg ED, Sangaré L, Fryzek J, Alexander DD. Prevalence of KRAS , NRAS , and BRAF gene mutations in metastatic colorectal cancer patients: a systematic literature review and meta-analysis. J Clin Oncol. 2019;37:523–3. https://doi.org/10.1200/jco.2019.37.4_suppl.523.
Clancy C, Burke JP, Kalady MF, Coffey JC. BRAF mutation is associated with distinct clinicopathological characteristics in colorectal cancer: a systematic review and meta-analysis. Color Dis. 2013;15:e711–8. https://doi.org/10.1111/codi.12427.
Chen D, Huang J-F, Liu K, Zhang L-Q, Yang Z, Chuai Z-R, Wang Y-X, Shi D-C, Huang Q, Fu W-L. BRAFV600E mutation and its association with Clinicopathological features of colorectal cancer: a systematic review and meta-analysis. PLoS One. 2014;9:e90607. https://doi.org/10.1371/journal.pone.0090607.
Benvenuti S, Sartore-bianchi A, Di Nicolantonio F, Benvenuti S, Sartore-bianchi A, Di Nicolantonio F, Zanon C. Oncogenic activation of the RAS / RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies oncogenic activation of the RAS / RAF signaling pathway impairs epidermal growth. Fa Cancer Res. 2007;67:2643–8. https://doi.org/10.1158/0008-5472.CAN-06-4158.
Lee J-H, Ahn J, Park WS, Choe EK, Kim E, Shin R, Heo SC, Jung S, Kim K, Chai YJ, Chae H. Colorectal cancer prognosis is not associated with BRAF and KRAS mutations-a STROBE compliant study. J Clin Med. 2019;8:111. https://doi.org/10.3390/jcm8010111.
Eachkoti R, Farooq S, Syeed SI, Wani HA, Majid S, Pampori MR. Prevalence and prognostic relevance of BrafV600E mutation in colorectal carcinomas from Kashmir (North India) valley. Mutagenesis. 2018;33:225–30. https://doi.org/10.1093/mutage/gey008.
Phipps AI, Buchanan DD, Makar KW, Burnett-Hartman AN, Coghill AE, Passarelli MN, Baron JA, Ahnen DJ, Win AK, Potter JD, Newcomb PA. BRAF mutation status and survival after colorectal cancer diagnosis according to patient and tumor characteristics. Cancer Epidemiol Biomark Prev. 2012;21:1792–8. https://doi.org/10.1158/1055-9965.EPI-12-0674.
Acknowledgments
The authors thank Mohammad Ali Mohammadi (Ph.D. candidate in Biomedicine, Kerman University of Medical Sciences, Iran) for molecular testing guidance.
Funding
This study was supported by the research fund of Kerman University of Medical Sciences (project No. 97000217).
Author information
Authors and Affiliations
Contributions
A.Y conceived and designed the study. A. Y, A.S, Z.M.K and N.K.K performed the experiments and data analysis. A.Y and A.A interpreted the data and drafted the manuscript. M.S.F and Z.M.K helped with sample preparation, patient’s data collection and interpretation. A.Y and M.R.Z helped with statistical analysis. S.D. supervised the findings of this work, reviewed and revised the final manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
The Ethical Committee of the Kerman University of Medical Sciences approved this research (Approval No. IR.KMU.REC.1397.209).
Conflict of Interests
The authors declare that they have no conflict of interests.
Disclosure Statement
The authors have nothing to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yari, A., Samoudi, A., Afzali, A. et al. Mutation Status and Prognostic Value of KRAS and BRAF in Southeast Iranian Colorectal Cancer Patients: First Report from Southeast of Iran. J Gastrointest Canc 52, 557–568 (2021). https://doi.org/10.1007/s12029-020-00426-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-020-00426-8