Abstract
The advances in neuroimaging methods reveal that resting-state functional fMRI (rs-fMRI) connectivity measures can be potential diagnostic biomarkers for autism spectrum disorder (ASD). Recent data sharing projects help us replicating the robustness of these biomarkers in different acquisition conditions or preprocessing steps across larger numbers of individuals or sites. It is necessary to validate the previous results by using data from multiple sites by diminishing the site variations. We investigated partial least square regression (PLS), a domain adaptive method to adjust the effects of multicenter acquisition. A sparse Multivariate Pattern Analysis (MVVPA) framework in a leave one site out cross validation (LOSOCV) setting has been proposed to discriminate ASD from healthy controls using data from six sites in the Autism Brain Imaging Data Exchange (ABIDE). Classification features were obtained using 42 bilateral Brodmann areas without presupposing any prior hypothesis. Our results showed that using PLS, SVM showed poorer accuracies with highest accuracy achieved (62%) than without PLS but not significantly. The regions occurred in two or more informative connections are Dorsolateral Prefrontal Cortex, Somatosensory Association Cortex, Primary Auditory Cortex, Inferior Temporal Gyrus and Temporopolar area. These interrupted regions are involved in executive function, speech, visual perception, sense and language which are associated with ASD. Our findings may support early clinical diagnosis or risk determination by identifying neurobiological markers to distinguish between ASD and healthy controls.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Autism spectrum disorder (ASD) is characterized by social interactions and communication impairments as well as restricted and stereotyped behavior. This neurodevelopmental disorder develops in early childhood and results in developmental differences in brain anatomy, functioning and connectivity that affect behavior across the lifetime. The diagnosis is typically done by behavioral observations and clinical interviews at this stage. Due to the complex nature and increasing number of ASD patients, attractions have been drawn by both the scientific community and society.
Disrupted network connectivity between distant brain regions has been reported among individuals with ASD (Wiggins et al. 2011; Cherkassky et al. 2006; Anderson et al. 2011a; Keehn et al. 2013; Lynch et al. 2013; Tyszka et al. 2013; DiMartino et al. 2011; Gotts et al. 2012; Von Hagen et al. 2013). These reports showed both increased and decreased connectivities in brain regions including default mode network, social brain regions, attentional regions, visual search regions and corticostriatal connections. However, knowing the specificity of diagnosis criteria (American Psychiatric Association 2013), there is hope that some (possibly complex) patterns of brain features may be unique to the disorder and it is worth to continue the research.
Given the fact that functional connectivity literature in ASD is complex and often inconsistent (Müller et al. 2011; Nair et al. 2014), machine learning (ML) techniques may provide valuable tool to discover the aberrant connectivity pattern that characterizes ASD. Several studies applied multivariate classification to functional connectivity MRI (fcMRI) (Van Dijk et al. 2010) for diagnostic classification, i.e. to characterize ASD using features that are predictive of a diagnosis on the level of individuals. A leave-one-out classifier (Anderson et al. 2011a, b) was performed using a large fcMRI connectivity matrix and achieved an overall classification accuracy of 79%. However the accuracy was lower in a separate small replication sample. Uddin et al. (2013) used a logistic regression classifier with features identified by ICA and achieved accuracies about 60–70% for all but one component identified as salience network, for which accuracy reached 77%.
Due to the advent of collective publicly dataset ABIDE (Di Martino et al. 2014), previous studies started investigating the potential diagnostic accuracy of ML algorithms with fMRI data gathered across different scanners, with different field strengths and different acquisition schemes. Nielsen et al. (2013) reported an overall accuracy of 60% for whole brain classification using a large dataset obtained from ABIDE dataset. In another study, Chen et al. (2015) showed that support vector machine classifier performed modestly (with accuracy <70%) whereas random forest classifier achieved 91% accuracy using Power atlas (Power et al. 2014). Plitt et al. (2015) implemented several machine learning tools and reported that individuals can be classified as having ASD with accuracy 76.67% using Destrieux atlas from their rs-fMRI scans alone. In a recent study (Chen et al. 2016) a MVPA was utilized to classify ASDs from HCs based on frequency-specific whole brain functional connectivity using a cross-site evaluation. These studies used aggregated data from all sites and evaluated using either LOOCV or 10-fold CV scheme.
Due to large sample size, multi-site data provides the power necessary to identify the neuroanatomical differences between ASD and typical control. However, this aggregated data introduces an additional problem of heterogeneity. Previous studies (Nielsen et al. 2013; Castrillon et al. 2014) revealed that acquisition site has significant effects on image properties. One solution to alleviate the problem of between-site variation is to utilize the domain adaptation machine learning algorithms (Jiang 2008; Pan and Yang 2010). Domain adaptation is a new approach in machine learning that deals with the differences in data distribution between test and train domain. Recently, this strategy has been used in predicting symptom severity in ASD based on cortical thickness measures in ABIDE dataset (Moradi et al. 2016).
Most functional neuroimaging methods are based on voxel (volume element data); even though recent studies used regions of interest (ROI) data. Most often this ROI approach presupposes the prior hypothesis and the regions outside of this network are not fully explored leading to potentially undetected effects. In this study, we used 42 bilateral Brodmann areas to see the effect of functional connectivities in these regions in distinguishing the two groups.
Previous studies did not address whether certain feature selection methods are better than others in combination with certain learning methods, in terms of producing models with high prediction accuracy. Relatively little has been published on the combined impact of choices of feature selection method and learning method on the predictive performance in autism research. To address this issue we empirically evaluated two different feature selection algorithms (filter-based and Elastic Nets) together with SVM learner. We addressed classifier generalizability by including subjects from the ABIDE (Di Martino et al. 2014) preprocessed dataset (356 individuals) from scanners located at six sites. Based on this data set, we aimed to assess the accuracy of ML classifiers for the automated detection of ASD. First we utilized domain adaptation algorithm to address the problem of data distribution across different acquisition sites. We used partial least square regression (PLS) with site as response variable and functional connectivities as predictor variables to reduce between-sites variability. Then we used PLS approximation of predictor variables and ASD/Control status as input to SVM model. We hypothesize that by considering PLS approximations of predictor variables one can effectively reduce nuisance variation between the data from different sites. We performed feature selection within a classification framework using a cross-site evaluation strategy, where the data from one site used as test dataset, and the remaining sites data as training data to learn the model parameters.
Methods and Materials
ABIDE Preprocessed Dataset
Data were collected from ABIDE site which is an open-access multi-site image repositories consists of structural and rs-fMRI scans from ASD and TD individuals ((Di Martino et al. 2014). The included sites with sample size were New York University (NYU, 149), Stanford University (STANFORD, 40), Olin Center, Institute of Living at Hartford Hospital (OLIN, 35), Kennedy Krieger Institute (KKI, 55), San Diego State University (SDSU, 36), University of Pittsburgh School of Medicine (PITT, 57). Acquisition parameters, protocol information can be obtained at ABIDE site http://fcon_1000.projects.nitrc.org/indi/abide/. We used preprocessed data using Connectome Computation System (CCS) pipeline as described at ABIDE sites. Subject demographics for individuals satisfying inclusion criteria are shown in Table 1.
Region of Interest and Connectivity Matrix
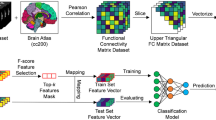
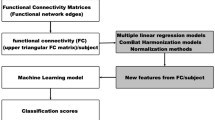
Connectivity maps were obtained utilizing CONN tool box (http://www.nitrc.org/projects/conn) running under Matlab 8.3 (2014a) (http://www.mathworks.com). Results were filtered to 0.01 to 0.1 Hz to limit spatial-temporal correlation to the spontaneous brain oscillation power; Brodmann’s areas ROIs provided by the same tool were utilized as seeding areas. Each Brodmann region was analyzed against all other Brodmann regions. Bivariate correlations were calculated between each pair of ROIs as reflections of connections. The list of 42 bilateral regions provided in CONN toolbox is given in supplementary materials. The rs-fMRI network was captured by an 84 × 84 symmetric matrix of nodes. We extracted the upper triangle elements of the functional connectivity matrix as classification features, i.e. the feature space for classification was spanned by the (84 × 83)/2 = 3486 dimensional feature vectors.
Significance Testing for Brain Connectivity and Site Effect
We performed a two sample t-test with Benjamin-Hochberg (Benjamini et al. 1995) correction using our data set to find any significant functional connectivity differences between healthy controls and ASD. A multivariate ANCOVA was used to see the effects of site for brain connectivities.
Feature Selection Algorithms
Previous studies did not explore the impact of feature selection in ABIDE studies. We empirically evaluated two different feature selection algorithms such as filter-based (ttest) and embedded Elastic Nets (Tibshirani 1996, Zou and Hastie 2005) together with SVM learner and evaluated the impact of these methods on selecting the important connections. The popular LASSO regression method minimizes the Residual Sum of Squares (RSS), similar to Ordinary Least Squares (OLS) regression, but poses a constraint to the sum of the absolute values of the coefficients being less than a constant. This additional constraint is similar to that introduced in Ridge regression, where the constraint is to the sum of the squared values of the coefficients. This simple modification allows LASSO to perform also variable selection because the shrinkage of the coefficients is such that some coefficients can be shrunk exactly to zero. The LASSO computes model coefficient \( \widehat{\beta} \) by minimizing the following function R(β) + λ ||β||1, where R(β) is the mean square error on the training set and ||β||1 =\( \sum \limits_{i=1}^p\mid {\beta}_i\mid \). λ controls the degree of sparsity of the solution, i.e. the number of features selected.
Elastic Net (Zou and Hastie 2005) is similar to LASSO. It differs in that the l1 norm of β is replaced by a combination of l1 and l2 norms. In this case, we minimize R(β) + λ Pα(β), where \( \kern0.5em P\alpha \left(\beta \right)=\frac{\left(1-\alpha \right)}{2}{\left|\left|\beta \right|\right|}_2^2+\alpha\ {\left|\left|\beta \right|\right|}_1 \), for α strictly between 0 and 1, and a nonnegative λ. The λ parameter can be tuned in order to set the shrinkage level, and the higher the λ is, and the more coefficients are shrunk to 0. Elastic Net is the same as LASSO when α = 1. As α shrinks toward 0, Elastic Net approaches ridge regression. For other values of α, the penalty term P α (β) interpolates between the L1 norm of β and the squared L2 norm of β. The advantage of Elastic Net over LASSO is that the Elastic Net penalty completes automatic variable selection and continuous shrinkage simultaneously, and it can select from a group of correlated variables. It is especially useful for large p small n problems where the grouped variables situation is a particularly important concern (Hastie et al. 2001).
Both Elastic Net and Univariate t-test based feature subset ranking were implemented in MATLAB 2016a.
Partial Least Squares
Partial Least Squares (PLS) regression is based on linear transition from a large number of original predictors to a new variable space based on small number of orthogonal factors (latent variables). The advantage of PLS is that it finds components (latent variables) which explain the covariance between predictor and response variables. This method is particularly suitable with high dimensional and high correlated predictor variables. The general underlying model of multivariate PLS is X = TPT + E, Y = UQT + F, where X is an n x m matrix of predictors, Y is an n x p matrix of response variables; T and U are n x l matrices that are respectively projections of X and projections of Y. P and Q are m x l and p x l orthogonal loading matrices and matrices E and F are the error terms, which are assumed independent and identically distributed random normal variables. We denote the functional connectivities values by X, where n is the number of subjects and m is the number of connectivities (m = 3486). The response variable Y represents the site information and is denoted by Y = {y1, 1 .....,yn, p}, where p is the number of sites. yn, p is 1 if subject n belongs to site p, otherwise zero. We used the PLS approximation of the predictor matrix X as our feature set for predicting ASD. In this current work, we used SIMPLS method which calculates the PLS factors directly as linear combinations of the original variables. The rationale behind this method is to reduce the overall inter-site variance by using PLS approximation of the predictor matrix X. We hypothesize that using PLS approximation our classification framework will be robust.
Classification and Feature Selection Framework
Classification algorithms were implemented using MATLAB 2014a. We used Support Vector Machine (SVM) classifier (Vapnik 1995), which is a widely used method for binary classification in fMRI studies. We performed a cross-site validation approach to address the issue of site acquisition effect. More specifically, a leave-one-site-out CV (LOSOCV) was performed in such a way that the data from each site was in its own fold. In this way we are training the model using 5 sites and testing with the one site to avoid any double dripping. In each training fold for LOSOCV, we performed another 10-fold CV to determine the best α (elastic net) and k (ttest) parameters. Inside each 10-fold CV training fold another 10-fold CV was performed to determine the parameter C for SVM and lambda for Elastic Net. Once we found the connectivities using training subjects with best alpha and k parameters, we used these connections to train the SVM model and tested with the hold-out site for prediction.
We used PLS approximation for predictor variables and binary outcome of patient status ASD vs control as input to SVM algorithm. For elastic net features we selected non-zero coefficient of elastic net parameters as important features and ranked t-stat for t-test feature selection. The framework is given in Fig. 1. The implementation of PLS was done by PLSREGRESS functions in MATLAB software with a fixed number of components selected from a range of values {5, 10, 15, 20, 25, 30, 35} with highest percentage of variance explained by the model. We have chosen 30 components that explain most of the variance in the observed response variables.
Consensus Features
Since we used a 10-fold CV strategy, the feature ranking was based on different training dataset in each cross validation (CV) fold. Therefore the feature (functional connections) contributions to classification were not evenly distributed. In this study we adopted the concept of consensus functional connectivity (Fair et al. 2012, Bhaumik et al., 2016), which is defined as the functional connectivity feature appearing in the final feature set of each CV iteration. We computed the percentages of occurrences of features that contributed to identification of depressed patients across all iterations of the cross validation. The functional connectivities which appeared in more than 70% of the 10-fold process are selected for each site and indicated the most discriminative features between those with ASD and TD subjects. We reported those connectivities (Table 5) which appeared in 3 or more sites in our results section.
Assessment of Classification Result
To evaluate the quality of the classification result, we will report three established measures, accuracy, sensitivity and specificity. The accuracy of a classifier is defined as the ratio of total number of correctly classified subjects and total subjects. The sensitivity and specificity evaluates the performance of a classifier to identify positive and negative instances, respectively.
Results
First we employed a two sample t-test with Benjamin-Hochberg (Benjamini et al. 1995) correction using our data set to find any significant (p < .05) functional connectivity differences between healthy controls and ASD. The significant connections were calculated using data from all sites but one site. We also reported significant connections combing data from all sites. The connections between Primary Auditory Cortex and bilateral Somatosensory Association Cortex have been seen as significant in the four data sets removing KKI, OLIN, SDSU and STANFORD respectively as well as combined sites (Table 2). Our MANCOVA analysis suggests that overall there is a site effect (F (5,366) = 4.97, P = .0002) on brain connectivities.
Classification Accuracy
Relatively moderate classification accuracies from holdout site have been achieved for different experimental strategies. First we found the connectivities using 10-fold CV with best alpha and k parameters and an internal 10-fold CV for SVM parameter C. We then used these connections to train the SVM model and tested with the hold-out site for prediction. The datasets from KKI (64%), OLIN (63%) and SDSU (64%) can achieve accuracy more than 60% using elastic net strategy. The sites PITT and STANFORD dataset could reach an accuracy of 61% and 70% respectively (Table 3) using t-test strategy. Prediction accuracies of all sites are always lowered in LASSO compared to t-test or Elastic Net except OLIN site. Previous result (Chen et al. 2016) also showed the similar classification accuracies for NYU (63%) and SDSU (60%) sites in cross-site evaluation.
While comparing overlapping confidence intervals between sites there is clearly significant differences in accuracy between STANFORD and other sites for ttest. Paired t-test showed no significant differences between ttest and Elastic Net for all sites.
The prediction accuracies are decreased when we applied PLS strategy to account for site variation (Table 4). Using t-test strategy, the accuracies were decreased in KKI (2%), PITT (19%) and STANFORD (8%) sites. On the other hand, the accuracies were decreased in KKI (10%), NYU (7%), OLIN (3%), PITT (9%) and SDSU (3%) sites using Elastic Net strategy. However no significant differences between SVM with PLS and without PLS for any feature selection algorithm.
Functional Networks Associated with Top Features
The most discriminative connections based on consensus functional connections are shown in Table 5 across all sites. These connections were selected more than 70% of training folds during classification. The connections which appeared in 3 or more sites are shown here. Several regions were noted to participate in two or more informative connections: Dorsolateral Prefrontal Cortex, Somatosensory Association Cortex, Primary Auditory Cortex, Inferior Temporal Gyrus and Temporopolar Area.
Discussions
To our knowledge, this is the first study to find the effect of partial least square regression (PLS) in conjunction with SVM algorithm using preprocessed Autism Brain Imaging Data Exchange (ABIDE) dataset, which is available online. Our goal was to overcome the issues of multi-site, multi-protocol variability by increasing sample sizes collected from these sites. We employed a cross-site evaluation strategy to generalize the classifier’s performance. We further evaluated different state-of-art feature selection strategies to find the important functional connectivities in classification of ASD versus TD subjects. Previous studies employed MVPA approach and achieved nearly 90% accuracy (Ecker et al. 2010, Uddin et al. 2011) when data set collected at a single center. These studies depend on specific scan parameters and are hard to utilize on other datasets. A multi-center study (Nielsen et al. 2013) achieved poorer accuracy (60%) than for single site results. Recent study showed (Chen et al. 2016) that using cross-site evaluation strategy the highest accuracy can be obtained for NYU and SDSU sites are 63% and 60% respectively. Our results showed similar accuracies for these two sites, 60% and 64% respectively. To overcome the problem of heterogeneity of sites we adopted PLS strategy and the accuracies obtained for these two sites are 53% and 61% respectively. Our results indicated that there is an effect of PLS strategy on classifier’s performance.
Our results showed that several regions occurred in two or more informative connections are Dorsolateral Prefrontal Cortex, Somatosensory Association Cortex, Primary Auditory Cortex, Inferior Temporal Gyrus and Temporopolar Area. Autism spectrum disorder had been diagnosed by emphasizing the impairments seen and the wide range of their severity (Tyszka et al. 2013). Several basic processing deficits have been seen in autism including social cognition, both interpersonal social processes and self-referential thought (Lombardo et al. 2007, Uddin 2011), impaired global feature processing (Anderson et al. 2011a, b), impaired reward processing (Chevallier et al. 2012; Damiano et al. 2012; Lin et al. 2012), motivation (Chevallier et al. 2012), and sensorimotor impairment (Perry et al. 2007). The social and self-referential cognitive processes have been linked with a pair of cortical midline brain regions, the ventromedial prefrontal cortex (VMPFC) and posterior cingulate cortex (PCC), which serve as hubs of the default mode network (DMN) (Greicius et al. 2003; Raichle et al. 2001). A critical component of social communication is processing auditory information. People with autism spectrum disorders typically have problems processing this information. The auditory cortex is the region of the brain that is responsible for processing of auditory (sound) information. As language deficits are a core feature of ASD, the study of auditory processing is essential to considering the roots of ASD as well as to conceptualize rational interventions. Investigators argued that autism is better characterized as a disorder of higher cortical functions, and specifically of the dorsolateral prefrontal cortex (Minshew and Goldstein 1993, Ozonoff et al. 1991, Rogers and Pennington 1991). Studies (Bertone et al. 2005, Dakin and Frith 2005, Tommerdahl et al. 2008, Dinstein et al. 2012) found that Somatosensory ASD has long been associated with sensory abnormalities. The middle temporal gyrus and inferior temporal gyrus are involved in a number of cognitive processes, including semantic memory processing, language processes (middle temporal gyrus), visual perception (inferior temporal gyrus), and integrating information from different senses. Temporopolar area is located primarily in the most rostral portions of the superior temporal gyrus and the middle temporal gyrus and so responsible for language processes. The other regions are orbitofrontal cortex, which is responsible for cognitive processing; angular Gyrus, involved in a number of processes related to language, number processing and spatial cognition and attention; supramarginal gyrus, receives somatosensory, visual, and auditory inputs from the brain (Dubac 2014); Dorsal anterior cingulate cortex (dACC), is a brain region that serves cognition and motor control and responsible for abnormalities in the structural or functional connections of the ACC and its sub-regions contribute to ASD (Zhou et al. 2016). Our results suggest that the functional connections distinguished two groups are heavily related to speech and language. Inferior prefrontal, premotor cortex, supramarginal gyrus and auditory cortex together make a strong case for that. These results are expected based on existing literature.
Our study suggests that leave-one-site out cross validation can be a potential strategy to moderately classify ASD from healthy controls. Classification accuracies were comparable for two sites NYU and SDSU with a recent study (Chen et al. 2016) without PLS. However, adopting PLS as a site variation correction, the accuracies decreased but not significantly. Higher accuracies obtained without PLS indicate an effect of site variability. In future, we need to collect data for other sites from ABIDE dataset and evaluate these strategies. We would like to evaluate our proposed domain adaptation strategy for other classifiers and different preprocessing strategies used in ABIDE consortium.
Information Sharing Statement
We provide our entire source code written in Matlab at the link https://github.com/ashishpradhan1008/PredictingASDbyCrossSiteEval. This link contains the dataset and Matlab code used for this paper. A demo code has been provided to guide the users to run the code and reproduce the results. Please keep in mind that the source code has not been optimized for speed and RAM use.
References
American Psychiatric Association (2013) Diagnostic and statistical manual of mental dis- orders, fourth edn., Washington, DC: American Psychiatric Association.
Anderson, J. S., Druzgal, T. J., Froehlich, A., Dubray, M. B., Lange, N., Alexander, A. L., Abildskov, T., Nielsen, J. A., Cariello, A. N., Cooperrider, J. R., et al. (2011a). Decreased interhemispheric functional connectivity in autism. Cereb Cortex, 21, 1134–1146.
Anderson, J. S., Nielsen, J. A., Froehlich, A. L., DuBray, M. B., Druzgal, T. J., Cariello, A. N., Cooperrider, J. R., Zielinski, B. A., Ravichandran, C., Fletcher, P. T., Alexander, A. L., Bigler, E. D., Lange, N., & Lainhart, J. E. (2011b). Functional connectivity magnetic resonance imaging classification of autism. Brain, 134(12), 3742–3754. https://doi.org/10.1093/brain/awr26322006979.
Benjamini Y, Hochberg Y. , (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing., J Roy Statist Soc Ser B (Methodological) 57:289-300.
Bertone, A., Mottron, L., Jelenic, P., & Faubert, J. (2005). Enhanced and diminished visuo-spatial information processing in autism depends on stimulus complexity. Brain, 128(Pt 10), 2430–2441.
Bhaumik, R., Jenkins, L. M., Gowins, J. R., Jacobs, R. H., Barba, A., Bhaumik, D. K., & Langenecker, S. A. (2016). Multivariate pattern analysis strategies in detection of remitted major depressive disorder using resting state functional connectivity. NeuroImage: Clinical. https://doi.org/10.1016/j.nicl.2016.02.018.
Castrillon, J. G., Ahmadi, A., Navab, N., & Richiardi, J. (2014). Learning with multi-site fmri graph data. In 2014 48th Asilomar conference on signals, systems and computers, IEEE (pp. 608–612).
Chen, C. P., Keown, C. L., Jahedi, A., Nair, A., Pflieger, M. E., Bailey, B. A., & Müller, R. (2015). Diagnostic classification of intrinsic functional connectivity highlights somatosensory, default mode, and visual regions in autism. NeuroImage: Clinical, 8, 238–245. https://doi.org/10.1016/j.nicl.2015.04.002.
Chen, H., Duan, X., Liu, F., Lu, F., Ma, X., Zhang, Y., Uddin, L. Q., & Chen, H. (2016). Multivariate classification of autism spectrum disorder using frequency-specific resting-state functional connectivity—A multi-center study. Progress in Neuro Psychopharmacology and Biological Psychiatry, 64, 1–9.
Cherkassky, V. L., Kana, R. K., Keller, T. A., & Just, M. A. (2006). Functional connectivity in a baseline resting-state network in autism. Neuroreport, 17, 1687–1690. https://doi.org/10.1097/01.wnr.0000239956.45448.4c.
Chevallier, C., Kohls, G., Troiani, V., Brodkin, E. S., & Schultz, R. T. (2012). The social motivation theory of autism. Trends in Cognitive Sciences, 16, 231–239.
Dakin, S., & Frith, U. (2005). Vagaries of visual perception in autism. Neuron, 48, 497–507.
Damiano, C. R., Aloi, J., Teadway, M., Bodfish, J. W., & Dichter, G. S. (2012). Adults with autism spectrum disorders exhibity decreased sensitivity to reward parameters when making effort-based decisions. Journal of Neurodevelopmental Disorders, 4, 13.
Di Martino, A., Yan, C. G., Li, Q., Denio, E., Castellanos, F. X., Alaerts, K., Anderson, J. S., Assaf, M., Bookheimer, S. Y., Dapretto, M., Deen, B., Delmonte, S., Dinstein, I., Ertl-Wagner, B., Fair, D. A., Gallagher, L., Kennedy, D. P., Keown, C. L., Keysers, C., Lainhart, J. E., Lord, C., Luna, B., Menon, V., Minshew, N. J., Monk, C. S., Mueller, S., Müller, R. A., Nebel, M. B., Nigg, J. T., O’Hearn, K., Pelphrey, K. A., Peltier, S. J., Rudie, J. D., Sunaert, S., Thioux, M., Tyszka, J. M., Uddin, L. Q., Verhoeven, J. S., Wenderoth, N., Wiggins, J. L., Mostofsky, S. H., & Milham, M. P. (2014). The autism brain imaging data exchange: towards a large- scale evaluation of the intrinsic brain architecture in autism. Molecular Psychiatry, 19(6), 659–667. https://doi.org/10.1038/mp.2013.7823774715.
DiMartino, A., Kelly, C., Grzadzinski, R., Zuo, X. N., Mennes, M., Mairena, M. A., et al. (2011). Aberrant striatal functional con- nectivity in children with autism. Biological Psychiatry, 69, 847–856. https://doi.org/10.1016/j.biopsych.2010.10.029.
Dinstein, I., Heeger, D. J., Lorenzi, L., Minshew, N. J., Malach, R., & Behrmann, M. (2012). Unreliable evoked responses in autism. Neuron, 75, 981–991.
Dubac, B. (2014). The brain from top to bottom. McGill, 2002. Web.
Ecker, C., Marquand, A., Mourao-Miranda, J., Johnston, P., Daly, E. M., Brammer, M. J., et al. (2010). Describing the brain in autism in five dimensions—Magnetic resonance imaging-assisted diagnosis of autism spectrum disorder using a multi parameter classification approach. The Journal of Neuroscience, 30, 10612–10623. https://doi.org/10.1523/JNEUROSCI.5413-09.2010.
Fair D. A., Bathula D., Nikolas M. A., Nigg J. T., (2012) Distinct neuropsychological subgroups in typically developing youth inform heterogeneity in children with ADHD. Proceedings of the National Academy of Sciences 109 (17):6769-6774
Gotts, S. J., Simmons, W. K., Milbury, L. A., Wallace, G. L., Cox, R. W., & Martin, A. (2012). Fractionation of social brain circuits in autism spectrum disor- ders. Brain, 135, 2711–2725. https://doi.org/10.1093/brain/aws160.
Greicius, M. D., Krasnow, B., Reiss, A. L., & Menon, V. (2003). Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proceedings of the National Academy of Sciences of the United States of America, 100, 253–258.
Hastie, T., Tibshirani, R., & Friedman, J. (2001). New York: Springer-Verlag.
Jiang, J. (2008). A literature survey on domain adaptation of statistical classifiers. Technical report, Computer Science Department at University of Illinois at Urbana-Champaign. Available at URL <http://sifaka.cs.uiuc.edu/jiang4/domainadaptation/survey>.
Keehn, B., Shih, P., Brenner, L. A., Townsend, J., & Muller, R. A. (2013). Functional connectivity for an “island of sparing” in autism spectrum disorder: An fMRI study of visual search. Human Brain Mapping, 34, 2524–2537. https://doi.org/10.1002/hbm.22084.
Lin, A., Tsai, K., Rangel, A., & Adolphs, R. (2012). Reduced social preferences in autism: Evidence from charitable donations. Journal of Neurodevelopmental Disorders, 4, 8.
Lombardo, M. V., Barnes, J. L., Wheelwright, S. J., & Baron-Cohen, S. (2007). Self-referential cognition and empathy in autism. PLoS ONE, 2, e883.
Lynch, C. J., Uddin, L. Q., Supekar, K., Khouzam, A., Phillips, J., & Menon, V. (2013). Default mode network in childhood autism: Pos- teromedial cortex heterogeneity and relationship with social deficits. Biological Psychiatry, 74, 212–219. https://doi.org/10.1016/j.biopsych.2012.12.013.
Minshew, N. J., & Goldstein, G. (1993). Is autism an amnesic disorder? Evidence from the California verbal learning test. Neuropsychology, 7, 209–216.
Müller, R.-A., Shih, P., Keehn, B., Deyoe, J. R., Leyden, K. M., & Shukla, D. K. (2011). Underconnected, but how? A survey of functional connectivity MRI studies in autism spectrum disorders. Cerebral Cortex, 21(10), 2233–2243. https://doi.org/10.1093/cercor/bhq29621378114.
Nair, A., Keown, C. L., Datko, M., Shih, P., Keehn, B., & Müller, R. A. (2014). Impact of methodolog- ical variables on functional connectivity findings in autism spectrum disorders. Human Brain Mapping, 35(8), 4035–4048. https://doi.org/10.1002/hbm.2245624452854.
Nielsen, J. A., Zielinski, B. A., Fletcher, P. T., Alexander, A. L., Lange, N., Bigler, E. D., Lainhart, J. E., & Anderson, J. S. (2013). Multisite functional connectivity MRI classification of au- tism: ABIDE results. Frontiers in Human Neuroscience, 7, 599. https://doi.org/10.3389/fnhum.2013.0059924093016.
Ozonoff, S., Pennington, B., & Rogers, S. (1991). Executive function deficits in high-functioning autistic individuals: Relationship to theory-of-mind. Journal of Child Psychology and Psychiatry, 32, 1081–1105.
Pan, S. J., & Yang, Q. (2010). A survey on transfer learning. IEEE Transactions on Knowledge and Data Engineering, 22(10), 1345–1359. https://doi.org/10.1109/TKDE.2009.191.
Perry, W., Minassian, A., Lopez, B., Maron, L., & Lincoln, A. (2007). Sensorimotor gating deficits in adults with autism. Biological Psychiatry, 61, 482–486.
Plitt, M., Barnes, K. A., & Martin, A. (2015). Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage: Clinical, 7, 359–366. https://doi.org/10.1016/j.nicl.2014.12.013.
Power J. D., Mitra A., Laumann T. O., Snyder A. Z., Schlaggar B. L., Petersen S. E., (2014) Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage 84:320–341
Raichle, M. E., MacLeod, A. M., Snyder, A. Z., Powers, W. J., Gusnard, D. A., & Shulman, G. L. (2001). A default mode of brain function. Proc Natl Acad Sci U S A, 98, 676–682.
Rogers, S., & Pennington, B. (1991). A theoretical approach to the deficits in infantile autism. Dev Psychopathol, 3, 137–162.
Tibshirani, R. (1996). Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (methodological), 58(1). Wiley: 267-88.
Tommerdahl, M., Tannan, V., Holden, J. K., & Baranek, G. T. (2008). Absence of stimulus-driven synchronization effects on sensory perception in autism: Evidence for local underconnectivity? Behavioral and Brain Functions, 4, 19.
Tyszka, J. M., Kennedy, D. P., Paul, L. K., & Adolphs, R. (2013). Largely typical patterns of resting-state functional connectivity in high- functioning adults with autism. Cerebral Cortex https://doi.org/10.1093/cercor/bht040.
Uddin, L. Q. (2011). The self in autism: An emerging view from neuroimaging. Neurocase, 17, 201–208.
Uddin L. Q., Menon V., Young C. B., Ryali S., Chen T., Khouzam A., Minshew N. J., Hardan A. Y., (2011). Multivariate searchlight classification of structural magnetic resonance imaging in children and adolescents with autism. Biological Psychiatry, 70(9): 833-841.
Uddin, L.Q., Supekar, K., Lynch, C.J., Khouzam, A., Phillips, J., Feinstein, C., Ryali, S., & Menon, V. (2013). Salience network-based classification and prediction of symptom severity in children with autism. J.A.M.A. Psychiatry 70, 869–879. 10.1001/ jamapsychiatry.2013.10423803651.
Van Dijk, K. R., Hedden, T., Venkataraman, A., Evans, K. C., Lazar, S. W., & Buckner, R. L. (2010). Intrinsic functional connectivity as a tool for human connectomics: Theory, proper- ties, and optimization. Journal of Neurophysiology, 103(1), 297–321. https://doi.org/10.1152/jn.00783.200919889849.
Vapnik, V. (1995). The natures of statistical learning theory. New York: Springer-Verlag.
Von dem Hagen, E. A., Stoyanova, R. S., Baron-Cohen, S., & Calder, A. J. (2013). Reduced func- tional connectivity within and between ‘social’ resting state networks in autism spec- trum conditions. Social Cognitive and Affective Neuroscience, 8, 694–701. https://doi.org/10.1093/scan/nss05322563003.
Wiggins, J. L., Peltier, S. J., Ashinoff, S., Weng, S. J., Carrasco, M., Welsh, R. C., et al. (2011). Using a self-organizing map algorithm to detect age-related changes in functional connectivity during rest in autism spectrum disorders. Brain Res, 1380, 187–197. https://doi.org/10.1016/j.brainres.2010.10.102.
Zhou, Y., Shi, L., Cui, X., Wang, S., & Lou, X. (2016). *Functional connectivity of the caudal anterior cingulate cortex is decreased in autism. PLoS One, 11(3), e0151879.
Zou, H., & Hastie, T. (2005). Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society, Series B, 67, 301–320.
Acknowledgments
The authors wish to acknowledge Dr. Heide Klumpp, Assistant Professor of Psychiatry at University of Illinois at Chicago and Dr. Cameron Craddock from Child Mind Institute in New York for their valuable comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 22 kb)
Rights and permissions
About this article
Cite this article
Bhaumik, R., Pradhan, A., Das, S. et al. Predicting Autism Spectrum Disorder Using Domain-Adaptive Cross-Site Evaluation. Neuroinform 16, 197–205 (2018). https://doi.org/10.1007/s12021-018-9366-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-018-9366-0