Abstract
Autism spectrum disorder (ASD) is notoriously difficult to diagnose despite having a high prevalence. Existing studies have shifted toward using neuroimaging data to enhance the clinical applicability and the effectiveness of the diagnostic results. However, the time and financial resources required to scan neuroimages restrict the scale of the datasets and further weaken the generalization ability of the statistical results. Furthermore, multi-site datasets collected by multiple worldwide institutions make it difficult to apply machine learning methods due to their heterogeneity. We propose a deep learning approach combined with the F-score feature selection method for ASD diagnosis using a functional magnetic resonance imaging (fMRI) dataset. The proposed method is evaluated on the worldwide fMRI dataset, known as ABIDE (Autism Brain Imaging Data Exchange). The fMRI functional connectivity features selected using our method can achieve an average accuracy of 64.53% on intra-site datasets and an accuracy of 70.9% on the whole ABIDE dataset. Moreover, based on the selected features, the network topology analysis showed a significant decrease in the path length and the cluster coefficient in ASD, indicating a loss of small-world architecture to a random network. The altered brain network may provide insight into the underlying pathology of ASD, and the functional connectivity features selected by our method may serve as biomarkers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Autism spectrum disorder (ASD), a lifelong neurodevelopmental disorder, is characterized by a lack of social interaction and emotional intelligence, as well as repetitive, aberrant, stigmatized and fixated behavior [1]. Early diagnosis is crucial to control and treat this disorder [2]. However, due to the overlapping nature of its symptoms, the current psychiatric diagnostic procedure, lacking biological evidence, is largely based on narrative interactions between individuals and clinical professionals [3], which not only are prone to generate a high variance during the diagnosis [4] but also require a long period of time to detect abnormalities [5].
Noninvasive brain imaging techniques, such as functional magnetic resonance imaging (fMRI) and electroencephalography (EEG), have been extensively applied to explore the functional characteristics and properties of the brain [6]. Quantitative analysis of brain imaging data can reveal subtle variations in neural patterns or networks which can help in diagnosing brain disorders such as Alzheimer’s disease or ADHD [7,8,9,10,11,12].
Recently, a worldwide open-source multi-site neuroimaging database, Autism Brain Imaging Data Exchange (ABIDE), made it possible to detect ASD using machine learning methods applied on fMRI data. The ABIDE dataset is a collaborative effort involving 1,112 structural, resting-state fMRI datasets and phenotypic information obtained from 17 sites, of which 539 datasets are from ASD patients and 573 are from typical controls [13]. Many efforts such as using convolutional neural networks (CNNs), recurrent neural networks (RNNs) and autoencoders (AEs) have been made to identify ASD with the ABIDE dataset [14,15,16].
The ABIDE dataset presents a potential for the extraction of functional biomarkers for ASD classification and gains a lot of attention, but the ASD classification accuracy on this dataset is infancy, which may be caused by its own characteristics:
Insufficient samples
Since scanning neuroimages is time-consuming and costly, the average number of samples in each site is less than 100. For fMRI neuroimaging, functional connectivity (FC) is one of the most common analysis methods used to investigate the functionally integrated relationships between spatially separated brain regions. According to different brain atlases, there are at least tens of thousands of functional connectivity features for each sample. However, in machine learning problems that involve learning rules in a high-dimensional feature space, an enormous amount of data is typically required to ensure that there are enough samples with each combination of values. Training models on such small, high-dimensional MRI datasets is not quite easy. Not enough samples with thousands of features in each site may cause over-fitting and non-generalizability of the model [8], which may lead to a worse performance in ASD classification.
Heterogeneity
Heterogeneity in etiology, phenotype and outcome are hallmarks of ASD [17], which makes it unlikely for the sites to cover a wide spectrum of autism, as ASD patients in different regions may have different characteristics. On the other hand, with different fMRI acquisition devices or scan parameters, such a multi-site, agglomerative dataset is also heterogeneous. Training a model with only samples from multi-sites may also make it difficult for the network to capture the complex patterns in the functional connectivity, thereby reducing the classification accuracy.
The multi-site dataset has a data heterogeneity problem, while the single intra-site dataset is not efficient for training classification models. In this paper, we propose a deep learning approach with the F-score feature selection strategy for ASD diagnosis using fMRI functional connectivity data. With the F-score method applied on each site, the number of raw fMRI functional connectivity features are significantly reduced by 75% on average, which benefits the subsequent autoencoder training. The fMRI functional connectivity features selected by our method can achieve an average accuracy of 64.53% on intra-site datasets and an accuracy of 70.9% on the whole ABIDE dataset. Furthermore, based on the selected functional connectivity features, we also investigate the underlying pathology of ASD by using network topology analysis. The resulting decrease in the path length and the cluster coefficient in ASD indicates a loss of small-world architecture to a random network, which may provide insight into ASD diagnosis.
The rest of the paper is organized as follows: before introducing the pipeline of our method, related studies will be briefly outlined in Sect. 2. A detailed description of the proposed method will be discussed in Sect. 3. Results of the classification accuracy on intra-site and on the whole dataset will be presented in Sect. 4. Hyperparameters and the brain network topology patterns in ASD will be discussed in Sect. 5. Finally, a conclusion will be drawn in Sect. 6.
Related Work
ASD Diagnosis based on Neuroimaging
Previous ASD studies based on neuroimaging have examined anatomical and functional abnormalities associated with ASD. Monk et al. [18] found that poorer social functioning in ASD subjects was correlated with connectivity between the posterior cingulate cortex and the superior frontal gyrus. Assaf et al. [19] discovered that adolescent ASD patients showed a decrease in the functional connectivity between the precuneus and the medial prefrontal cortex/anterior cingulate cortex, default mode network (DMN) core areas and other default mode sub-network areas. These findings indicated that ASD altered the intrinsic connectivity within the default network, and that the connectivity between these structures is associated with specific ASD symptoms.
It has been shown that ASD disrupts the functional connectivity between multiple brain regions, which affects global brain networks. Thus, it is possible to classify ASD subjects and control subjects by exploring the neural patterns of the functional connectivity [3, 16, 20,21,22]. For example, [3] developed a classifier which achieved a high accuracy of 85% for a Japanese discovery cohort and demonstrated a remarkable degree of generalization (75% accuracy) for two independent validation cohorts in the USA and in Japan. Parisot et al. [21] introduced the novel concept of graph convolutional networks (GCNs) for brain analysis in populations, combining functional connectivity features and demographic attributes leading to an accuracy of 69.5% for the ABIDE dataset. Aghdam et al. [20] used a deep belief network (DBN) to exploit the latent or abstract high-level features inside rs-fMRI (resting-state functional magnetic resonance imaging) and sMRI (structural magnetic resonance imaging) data from a subsample comprised of 185 individuals (116 ASD and 69 TC) and achieved an accuracy of 65.56%.
Classification of the ABIDE Dataset
In the field of ASD diagnosis, the Autism Brain Imaging Data Exchange (ABIDE) initiative has gained a lot of attention, as it offers a great potential for the extraction of functional biomarkers for ASD classification. However, it also makes the ASD classification task itself much more challenging due to its multi-site and multi-protocol aspects, which bring up significant issues related to patient heterogeneity, statistical noise and experimental differences in the rs-fMRI data [23]. Many studies and methods have been developed based on this dataset [1, 16, 23, 24]. Nielsen et al. [25] was the first one to use the ABIDE dataset to classify ASD, achieving an accuracy of 60%.
Machine learning techniques such as support vector machines (SVMs) and random forests have been explored in multiple studies [26, 27]. Fredo et al. [26] used conditional random forests to reduce the FC matrix and build random forests with 143 features to classify the ASD and control groups, achieving an accuracy of 65% on the whole dataset.
Recently, using neural networks and deep learning methods such as autoencoders (AEs), deep neural networks (DNNs), long short-term memory (LSTM) and convolutional neural networks (CNNs) has also become very popular for diagnosing ASD [16, 28, 29]. Brown et al. [28] obtained a 68.7% classification accuracy on 1,013 subjects using an element-wise layer for DNNs.
Most recently, [7] proposed a deep learning approach which achieved a maximum accuracy of 82% (only on the OHSU site) for classifying 26 subjects. In their method, half of the pairwise Pearson’s correlation coefficients, comprising of the 1/4 largest and 1/4 smallest values, were considered as features. Data augmentation was applied to generate synthetic samples using linear interpolation. Two stacked denoising autoencoders were first pre-trained in order to extract lower-dimensional data. After that, the autoencoder’s weights were applied to a single-layer perceptron classifier for the final classification. They evaluated their model on each site separately, and the average accuracy obtained was 63.8%.
In general, most studies related to classifying the ABIDE dataset using machine learning techniques either only considered a subset of the dataset or incorporate other information together with fMRI data. There are few studies that have only used fMRI data without any assumptions on the demographic information. To the best of our knowledge, ASD-DiagNet [7] is currently a state-of-the-art technique for ASD diagnosis and is therefore used as one of the baseline methods for evaluating the method proposed in this paper.
Methodology
In this section, the feature engineering approaches, such as the functional connectivity feature extraction method and the F-score feature selection method, will first be introduced in detail. Then, the classification model with its training and evaluation strategies will also be outlined.
Feature Extraction: Functional Connectivity Correlation Coefficient
Functional connectivity (FC) between brain regions is an important concept in fMRI analysis, and it has been shown to contain discriminatory patterns for fMRI classification [7]. The functional connectivity features are usually extracted from pairs of brain regions of interest (ROIs) based on the fMRI time series data by estimating the fluctuating coupling of the brain regions with respect to time, which is known as correlation. Among correlation measures, Pearson’s correlation coefficient is generally used for approximating the functional connectivity in fMRI data. It shows the linear relationship between the time series of two different ROIs. Given two time series, u and v, each of length T, the Pearson’s correlation coefficient can be computed using the following equation:
where \(\overline{u}\) and \(\overline{v}\) are the means of times series u and v, respectively. The Pearson’s correlation coefficient ranges from -1 to 1, where values close to −1 indicate that the time series are anti-correlated and values close to 1 indicate that the time series are positively correlated.
Computing all pairwise correlations generates a correlation matrix \(\mathbf {M}_{n \times n}\), where n is the number of regions. Due to the symmetric property of Pearson’s correlation, we only considered the top upper triangle part of the correlation matrix. The main diagonal of the matrix was also removed, since it represents a region correlating to itself. Later on, the remaining triangle was vectorized to retrieve a vector of features, with the purpose of using it for ASD classification. Thus, considering a brain atlas in which the brain is parceled into n regions, the number of original FC feature vectors is \(m=n \times (n-1)/2\).
Feature Selection: F-score
In order to reduce the number of functional connectivity features, we applied F-score [30] to preliminarily discriminate which features can characterize ASD. F-score is a simple technique used to measure the discrimination capability between two sets of real numbers. Given training feature vectors \(\mathbf {x}_j, j=1,...,m\), if the number of ASD and control subjects are a and c, respectively, then the F-score of the ith feature can be defined as:
where \(\overline{x}_i\), \(\overline{x}_i^a\), \(\overline{x}_i^c\) are the averages of the ith feature of the whole, ASD and control dataset, respectively; \(x_{j,i}^a\) is the ith feature of the jth ASD subject, and \(x_{j,i}^c\) is the ith feature of the jth control subject. The numerator indicates the discrimination between the ASD and control sets, while the denominator indicates the discrimination within each of the two sets. The larger the F-score is, the more likely it is that the feature is discriminative. Therefore, F-score can be used as a feature selection criterion.
In this part, the F-score feature selection method was only applied on the training dataset. After calculating the F-score value for each functional connectivity feature item, these values were sorted in descending order. Then, a threshold was set and the indices corresponding to the top-k largest values were picked up to form a feature selection mask. The mask was applied on both the FC vector training set and the test set to extract the FC feature vector.
Classification: Autoencoder and SLP
An autoencoder (AE) [31] is used to extract a lower-dimensional feature representation, which corresponds to the bottleneck layer in the network. An AE is a type of feed-forward neural network model, which first encodes its input x to a lower-dimensional representation:
where \(\tau\) is the tanh activation function, and \(W_{enc}\) and \(b_{enc}\) are the weight matrix and the bias for the encoder, respectively. Then, the decoder reconstructs the original input data:
where \(W_{dec}\) and \(b_{dec}\) are the weight matrix and the bias for the decoder, respectively.
For the classification task, a single-layer perceptron (SLP) was applied, which uses the bottleneck layer of the autoencoder \(h_{enc}\) as input and computes the probability of a sample belonging to the ASD patient class using a sigmoid activation function Sigmoid:
where \(W_{slp}\) and \(b_{slp}\) are the weight matrix and the bias for the SLP network, respectively.
The autoencoder can be trained to minimize its reconstruction error, which is computed as the mean squared error (MSE) between x and its reconstruction \(x^\prime\). The SLP network can be trained by minimizing the binary cross-entropy loss. Thus, the ASD classification problem can be described as:
where \(\mathcal {L}_{MSE}\) and \(\mathcal {L}_{BCE}\) are the loss of the autoencoder and the SLP, respectively. The hyperparameter \(\lambda\) is used to balance the importance of these two items.
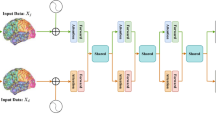
The whole pipeline of our method for ASD classification is shown in Fig. 1. With the help of a brain atlas, the time series for each brain region are extracted from the raw fMRI dataset. Then, pairwise Pearson correlation coefficients for each sample are calculated. After removing the lower triangular matrix and vectorizing, the FC vectors are extracted for further feature selection. Before applying the F-score method, the original FC vector dataset must be split into a training set and a test set. Then, the F-score method is applied on the training set to extract the feature mask, which can be used to extract feature vectors for both the training and test sets. With a proper training strategy, we can evaluate the quality of the selected features and the classification performance of the developed model.
Pipeline of the proposed method for ASD classification: (a) The original fMRI dataset is segmented by using a brain atlas, and pairwise Pearson’s correlations for each subject in the dataset are computed. The vectorized upper triangular FC matrix is used for further feature selection. (b) Firstly, the original FC vector dataset is split into a training set and a test set. Then, using the F-score feature selection method, the FC items in the training set with the top k largest F-score values are picked to form a feature mask, which is applied on the training set to select the discriminative FC features. After that, the mask is also applied to the test set to ensure that the indices and the dimensions of the features are consistent with the training set. Finally, the selected feature vector in the training set is used for neural network training, while the selected feature vector in the test set is used for model evaluation
Two phases are executed to train the model, which is shown in Fig. 2. For the joint training phase, the weights of the AE and the weights of SLP are trained simultaneously. In this phase, the inputs for SLP are the hidden outputs of AE model for each epoch. For the SLP optimizing phase, the weights of the AE in the are frozen, which means only SLP is trained during this phase. Other details in the training process, such as optimization method and its parameters, use the default settings [7].
The training model strategy consists of joint training phase and SLP optimizing phase. For the first phase, an AE and a SLP are trained together and both of their weights are optimized. Note that the inputs for the SLP are the outputs of the AE’s hidden layer. For the SLP optimizing phase, the weights of AE are frozen so that only SLP is modified during the following training epochs
Network Topology Analysis: Clustering Coefficient and Average Path Length
The graph theory can be used to check the brain network topology differences between normal and ASD. In general, graph can be represented by sets of nodes and edges between these nodes [32]. The functional connectivity matrix can be derived by calculating the correlation between paired brain regions, which can be easily transformed to a graph or network. In this case, the brain area is treated as the node and the value of correlation between brain regions is the edge, which is called weighted graph.
In this study, we constructed a weighted graph for each subject. For each weighted graph, the node set was a subset of all brain regions selected by F-score feature selection, and the edge weight between nodes was assigned as the correlation coefficient between the corresponding fMRI time series.
Topology features of weighted graph can be characterized by some network indexes [32]. In the network analysis field, the clustering coefficient and average path length are two most fundamental measures. These two measures depend on network structure but also connectivity values. In weighted graph, the clustering coefficient of a node indicates the proportion of its neighbors that are also connected with each other and quantifies the tendency to form local clusters. The clustering coefficient of node i in weighted graph is defined as
where w is the edge weight between paired nodes. And clustering coefficient of the whole network can be calculated by
The average path length can be computed by the harmonic mean of the shortest path between all pairs of nodes in the graph [32], which is defined as the path with the largest total weight. Average weighted path length of the whole network can be calculated by
where \(l_{ij}\) is the length of shortest path between nodes i and j.
Experiments and Results
Experimental Paradigm
Datasets
To evaluate the proposed approach, we performed experiments on ABIDE-I datasets provided by the ABIDE initiative, which is a consortium that provides previously collected rs-fMRI ASD and matched controls data for the purpose of data sharing in the scientific community. We included data from 505 ASD individuals and 530 typical controls (TC). The ABIDE datasets were collected at 17 different imaging sites and included rs-fMRI images, T1 structural brain images and phenotypic information for each patient. Parameters such as repetition time (TR), echo time (TE), number of voxels, number of volumes and openness or closeness of the eyes while scanning were different among sites. Table 1 shows the class information for each site.
ABIDE-I datasets provided the average time series extracted from ROIs based on different atlases. The atlas used in our experiments was generated by the spatially constrained spectral clustering algorithm (CC-200) [33]. The preprocessed rs-fMRI data were downloaded from the Preprocessed Connectomes Project. The fMRI data were slice-time corrected, motion corrected, and the voxel intensity was normalized. Nuisance signal removal was performed using 24 motion parameters and the global signal as the regressors. Functional data were band-pass filtered (0.01-0.1 Hz) and spatially registered using a nonlinear method to a template space (MNI152). The mean time series for each ROI was extracted for each subject.
Baseline Methods
We compared the proposed method with two baseline approaches: ASD-DiagNet [7] and stacked autoencoder (AE) [1]. Both methods are based on autoencoders.
ASD-DiagNet
The ASD-DiagNet [7] method involves combining an autoencoder and a SLP together. The autoencoder and SLP are trained jointly. Linear interpolation data augmentation methods are also introduced. For the feature selection method, ASD-DiagNet treated the 1/4 largest and 1/4 smallest Pearson’s correlation values as the features.
Stacked autoencoder
The stacked AE [1] trains a stacked denoising autoencoder model, which did not use any feature selection methods.
Evaluation Methods
The proposed model was evaluated using k-fold cross-validation. The original dataset was randomly partitioned into k equal sized subsamples. During the evaluation, a single subsample was retained as the validation data for testing the model, and the remaining \(k-1\) subsamples were used as training data. The process was then repeated k times, with each of the k subsamples used exactly once as the validation data. In the intra-site scenario, we executed the fivefold cross-validation with stratified sampling on each site separately. Besides, for the whole dataset evaluation, tenfold cross-validation was utilized on the whole samples. In this case, data in each fold were randomly selected from the whole dataset without stratified sampling.
Three metrics were employed to evaluate the classification performance of the different methods, including classification accuracy (ACC), sensitivity (SEN) and specificity (SPE). Accuracy measures the proportion of correctly classified subjects (actual ASD subjects classified as ASD and actual healthy subjects classified as healthy). Sensitivity represents the proportion of actual ASD subjects which are correctly classified as ASD, and specificity measures the proportion of actual healthy subjects which are classified as healthy.
In detail, the equations of accuracy, sensitivity and specificity are computed as follows:
TP, TN, FP, FN are the number of positive samples predicted as positive, negative samples predicted as negative, negative samples predicted as positive and positive samples predicted as negative, separately.
In order to examine the abnormal changes in the brain network topology of ASD subjects using features selected by our method, the weighted graph analysis method was adopted. In the weighted graph analysis, a graph of 200 nodes (i.e., 200 brain regions from the CC200 brain atlas) was constructed in which the edge weight between nodes was assigned as the functional connectivity between the corresponding brain regions. The topology features of the weighted graph can be characterized by some network indexes. The clustering coefficient [32] and the average shortest path length [32], which are the two most fundamental measures, were used to analyze the brain functional network.
Hyperparameters
For our proposed method, the two most important parameters are the threshold k, selecting the top-k most discriminative features during the feature selection, and the loss balance item \(\lambda\), balancing the AE and the SLP during the model training phase.
In the classification performance evaluation for each site step, the optimal parameters k and \(\lambda\) are not the same for each site, as shown in Table 2. This is because of the heterogeneity of the multi-site dataset. For evaluating the classification performance on the whole site, in this study the parameter k was set to 2000 and the parameter \(\lambda\) was set to 10, which are values that were chosen based on the discussion results of the two parameters outlined in Sect. 5.
Intra-Site Evaluation Results
In this step, fivefold cross-validation was performed on each site with our proposed method. For each site, the same epochs were set in both the joint training phase (300 epochs) and the SLP optimizing phase (100 epochs).
The number of features k and the loss balance item \(\lambda\) vary for each site (Table 2). The accuracy of each method is shown in Fig. 3 and Table 3.
Based on the results in intra-site situation, it can be observed that, in most sites, the proposed model achieves a better performance than other methods. In addition, for the USM, UCLA and OHSU sites, our proposed method can achieve an accuracy of over 74%. It should be noted that for the original ASD-DiagNet method, an augmentation method was imported to generate more training samples. In order to compare these three methods under the same dataset conditions, here for the ASD-DiagNet method only its accuracy was reported without using data augmentation.
Moreover, the stability of the accuracy among all sites was also investigated, as presented in Fig. 4. In this boxplot figure, the bar ranges from the first quartile (Q1) to the third quartile (Q3) of the percentage accuracy, the width between the upper and lower quartile represents the inter-quartile range (IQR), and the median value is indicated by a black line across the bar, while the outliers are represented as black diamonds. It shows that our method has more higher median value and smaller difference between upper and lower boundary, which indicates that our method is more stable among all sites.
The average accuracy among each site using AE, ASD-DiagNet and the proposed method (AE+F-score). In this case, we calculated the average of the accuracy of each site. The IQR for our method is much smaller than other methods, which indicates our method is better in stability among these sites. Note that parameters k and \(\lambda\) are not the same for each site due to dataset heterogeneity
In Fig. 4, it can be seen that for our proposed method (AE+F-score), the median accuracy is higher than that of the other methods. Furthermore, the IQR is smaller than that of the other methods, which shows that our model is more stable compared to the baseline methods.
Whole Dataset Evaluation Results
Despite the heterogeneity of the multi-site dataset, the accuracy performance was still investigated with tenfold cross-validation for comparison purposes. In this case, the parameter k was set to 2000, while the parameter \(\lambda\) was set to 10, which are values based on the discussion results of the two parameters, as outlined in Sect. 5. Note that our proposed model achieves the highest accuracy (70.9%) out of the three methods.
The results presented in Table 4 show that the proposed method can achieve an accuracy of 70.9% on the whole dataset, which is the highest accuracy among the compared methods. Moreover, the whole dataset accuracy is even higher than the median accuracy on each site, which may indicate that a certain site does not cover all types of ASD. The sensitivity and specificity are also superior than those of the other methods, which shows that our method can deal with the heterogeneity present in multi-site datasets.
Discussion
In this section, the impact of two hyperparameters on the classification accuracy is first investigated; these two are the number of selected features k and the loss balance item \(\lambda\). Then, the weighted graph analysis is used to examine the abnormal changes in the brain network topology of ASD subjects with two indicators: the cluster coefficient and the average shortest path length.
The Effect of the Hyperparameters
In Fig. 5, it can be observed that the median accuracy can be maximized when the number of features k is set to 2000. Not enough features make the model unable to properly capture the characteristics of ASD, which in turn leads to a decrease in accuracy. However, an excessive number of features cause too much noise in the feature vectors, which in turn leads to an increase in errors.
When it comes to the second hyperparameter, it can be seen in Fig. 6 that the median accuracy is maximized when the value of the loss balance item \(\lambda\) is set to 10.
The Abnormal Network Topology Pattern in ASD Subjects
The weighted graph method is employed to analyze the brain function network. A graph of 200 nodes based on the 200 brain regions segmented by the CC200 brain atlas was constructed, and the functional connectivity was used as the edges between the nodes.
Note that for these edges, not all functional connectivities were used to construct the graph; instead, only those functional connectivities selected using the F-score feature selection method were used to build the graph network. There are 200*(200 − 1)/2 = 19900 functional connectivities in total. After the F-score feature selection, the top-2000 connectivities are kept as the reserved edges and the weights of the other edges are set as 0.
In the ABIDE dataset, there are significantly more male patients than female patients (Table 1), and some sites even does not contain any female patient. Therefore, we only selected the same number of male patients and ordinary people for each site. In terms of age, there was no significant difference between the ASD group and the control group for each site (Fig. 7).
There was no significant difference between ASD and control group in terms of age for each site. Error bars represent the standard error. The independent t test (Fisher LSD) is used to determine the significant difference between the two groups and marked with asterisks (\(*p\le 0.05, **p\le 0.01, ***p\le 0.001\)). The outliers are represented as black diamonds
We used an independent t test, with significant differences \(p\le 0.05, p\le 0.005, p\le 0.001\), to evaluate the differences between ASD and TD subjects in each site separately. Boxplots of the clustering coefficient and the path length are illustrated for the two groups in each site in Figs. 8 and 9, respectively. It can be observed that in those sites with significant differences, the ASD group showed a significant decrease in the clustering coefficient together with a decreasing trend in the average shortest path length. These findings suggest a disrupted segregation and an integration organization in their brain networks, which indicates that the brain network topology in ASD subjects went from a small-world network to a random network.
The cluster coefficients for the males in ASD (black) and TD (red) groups for each site. Error bars represent the standard error. Significant differences identified by an independent t test (\(*p\le 0.05, **p\le 0.01, ***p\le 0.001\)) between the two groups are marked by asterisks (Fisher LSD). The outliers are represented as black diamonds
The average shortest path lengths for males in the ASD (black) and TD (red) groups for each site. Error bars are the standard error. Significant differences with independent t test (\(*p\le 0.05, **p\le 0.01, ***p\le 0.001\)) between the two groups are marked by asterisks (Fisher LSD). The outliers are represented as black diamonds
Conclusion
Autism is a complex neurodevelopmental disorder that affects multiple cognitive domains and brain systems. In this study, the problem of correctly identifying and classifying subjects with ASD from healthy subjects was addressed. A deep learning approach combined with the F-score feature selection method was proposed for fMRI functional connectivity data analysis. Our model was evaluated on the ABIDE dataset, with experimental results suggesting a superiority of the proposed method in ASD diagnosis, not only for intra-site datasets but also for the whole dataset. Furthermore, we attempted to investigate the brain network topology in ASD using the features selected by our method and found that it departed from a small-world network to a random network, which may be a discriminative feature in patients with ASD. This finding also proves that the features selected by our method can characterize ASD and may serve as biomarkers for contributing to the diagnosis of the disorder.
References
Heinsfeld AS, Franco AR, Craddock RC, Buchweitz A, Meneguzzi F. Identification of autism spectrum disorder using deep learning and the ABIDE dataset. NeuroImage: Clinical. 2018;17:16–23. Publisher: Elsevier.
Maenner MJ, Shaw KA, Baio J. Prevalence of autism spectrum disorder among children aged 8 years–autism and developmental disabilities monitoring network, 11 sites, United States, 2016. MMWR Surveill Summ. 2020;69(4):1. Publisher: Centers for Disease Control and Prevention.
Yahata N, Morimoto J, Hashimoto R, Lisi G, Shibata K, Kawakubo Y, Kuwabara H, Kuroda M, Yamada T, Megumi F. A small number of abnormal brain connections predicts adult autism spectrum disorder. Nat Commun. 2016;7(1):1–12. Publisher: Nature Publishing Group.
Mandell DS, Ittenbach RF, Levy SE, Pinto-Martin JA. Disparities in diagnoses received prior to a diagnosis of autism spectrum disorder. J Autism Dev Disord. 2007;37(9):1795–1802. Publisher: Springer.
Sharma SR, Gonda X, Tarazi FI. Autism spectrum disorder: classification, diagnosis and therapy. Pharmacol Ther. 2018;190:91–104. Publisher: Elsevier.
Martin AR, Aleksanderek I, Cohen-Adad J, Tarmohamed Z, Tetreault L, Smith N, Cadotte DW, Crawley A, Ginsberg H, Mikulis DJ. Translating state-of-the-art spinal cord MRI techniques to clinical use: a systematic review of clinical studies utilizing DTI, MT, MWF, MRS, and fMRI. NeuroImage: Clinical. 2016;10:192–238. Publisher: Elsevier.
Eslami T, Mirjalili V, Fong A, Laird AR, Saeed F. ASD-DiagNet: a hybrid learning approach for detection of autism spectrum disorder using fMRI data. Front Neuroinform. 2019;13:70. Publisher: Frontiers.
Eslami T, Saeed F. Similarity based classification of ADHD using singular value decomposition. In: Proceedings of the 15th ACM International Conference on Computing Frontiers. 2018. p. 19–25.
Goceri E. Diagnosis of Alzheimer’s disease with Sobolev gradient-based optimization and 3D convolutional neural network. Int J Numer Methods Biomed Eng. 2019;35(7):e3225. Publisher: Wiley Online Library.
Peng X, Lin P, Zhang T, Wang J. Extreme learning machine-based classification of ADHD using brain structural MRI data. PloS one. 2013;8(11):e79476. Publisher: Public Library of Science San Francisco, USA.
Sewani H, Kashef R. An autoencoder-based deep learning classifier for efficient diagnosis of autism. Children. 2020;7(10):182. Publisher: Multidisciplinary Digital Publishing Institute.
Subah FZ, Deb K, Dhar PK, Koshiba T. A deep learning approach to predict autism spectrum disorder using multisite resting-state fMRI. Appl Sci. 2021;11(8):3636. Publisher: Multidisciplinary Digital Publishing Institute.
Di Martino A, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M, Bookheimer SY, Dapretto M. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol Psychiatry. 2014;19(6):659–667. Publisher: Nature Publishing Group.
Kunda M, Zhou S, Gong G, Lu H. Improving multi-site autism classification based on site-dependence minimisation and second-order functional connectivity. bioRxiv. 2020. Publisher: Cold Spring Harbor Laboratory.
Liu W, Li M, Yi L. Identifying children with autism spectrum disorder based on their face processing abnormality: A machine learning framework. Autism Res. 2016;9(8):888–898. Publisher: Wiley Online Library.
Sharif H, Khan RA. A novel framework for automatic detection of autism: A study on corpus callosum and intracranial brain volume. 2019. arXiv preprint arXiv:1903.11323.
Masi A, DeMayo MM, Glozier N, Guastella AJ. An overview of autism spectrum disorder, heterogeneity and treatment options. Neurosci Bull. 2017;33(2):183–193. Publisher: Springer.
Monk CS, Peltier SJ, Wiggins JL, Weng SJ, Carrasco M, Risi S, Lord C. Abnormalities of intrinsic functional connectivity in autism spectrum disorders. Neuroimage. 2009;47(2):764–772. Publisher: Elsevier.
Assaf M, Jagannathan K, Calhoun VD, Miller L, Stevens MC, Sahl R, O’Boyle JG, Schultz RT, Pearlson GD. Abnormal functional connectivity of default mode sub-networks in autism spectrum disorder patients. Neuroimage 2010;53(1):247–256. Publisher: Elsevier.
Aghdam MA, Sharifi A, Pedram MM. Combination of rs-fMRI and sMRI data to discriminate autism spectrum disorders in young children using deep belief network. J Digit Imaging. 2018;31(6):895–903. Publisher: Springer.
Parisot S, Ktena SI, Ferrante E, Lee M, Moreno RG, Glocker B, Rueckert D. Spectral graph convolutions for population-based disease prediction. In: International conference on medical image computing and computer-assisted intervention. Springer, 2017. p. 177–185.
Plitt M, Barnes KA, Martin A. Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage: Clinical. 2015;7:359–366. Publisher: Elsevier.
Abraham A, Milham MP, Di Martino A, Craddock RC, Samaras D, Thirion B, Varoquaux G. Deriving reproducible biomarkers from multi-site resting-state data: An autism-based example. NeuroImage. 2017;147:736–745. Publisher: Elsevier.
Sherkatghanad Z, Akhondzadeh M, Salari S, Zomorodi-Moghadam M, Abdar M, Acharya UR, Khosrowabadi R, Salari V. Automated detection of autism spectrum disorder using a convolutional neural network. Front Neurosci. 2020;13:1325. Publisher: Frontiers.
Nielsen JA, Zielinski BA, Fletcher PT, Alexander AL, Lange N, Bigler ED, Lainhart JE, Anderson JS. Multisite functional connectivity MRI classification of autism: ABIDE results. Front Hum Neurosci. 2013;7:599. Publisher: Frontiers.
Fredo AJ, Jahedi A, Reiter M, Müller RA. Diagnostic classification of autism using resting-state fMRI data and conditional random forest. Age (years). 2018;12(2):6–41.
Subbaraju V, Suresh MB, Sundaram S, Narasimhan S. Identifying differences in brain activities and an accurate detection of autism spectrum disorder using resting state functional-magnetic resonance imaging: A spatial filtering approach. Med Image Anal. 2017;35:375–389. Publisher: Elsevier.
Brown CJ, Kawahara J, Hamarneh, G. Connectome priors in deep neural networks to predict autism. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE; 2018. p. 110–113.
Guo X, Dominick KC, Minai AA, Li H, Erickson CA, Lu LJ. Diagnosing autism spectrum disorder from brain resting-state functional connectivity patterns using a deep neural network with a novel feature selection method. Front Neurosci. 2017;11:460. Publisher: Frontiers.
Chen YW, Lin CJ. Combining SVMs with various feature selection strategies. In: Feature extraction. Springer; 2006. p. 315–324.
Liou CY, Cheng WC, Liou JW, Liou DR. Autoencoder for words. Neurocomputing. 2014;139:84–96. Publisher: Elsevier.
Zeng K, Kang J, Ouyang G, Li J, Han J, Wang Y, Sokhadze EM, Casanova MF, Li X. Disrupted brain network in children with autism spectrum disorder. Sci Rep. 2017;7(1):1–12. Publisher: Nature Publishing Group.
Craddock RC, James GA, Holtzheimer III PE, Hu XP, Mayberg HS. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Hum Brain Mapp. 2012;33(8):1914–1928. Publisher: Wiley Online Library.
Funding
This work was supported by the Tianjin Natural Science Foundation for Distinguished Young Scholars (No. 18JCJQJC46100) and the Tianjin Science and Technology Plan Project (No. 18ZXJMTG00260).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
This article does not contain any studies with human participants performed by any of the authors.
Conflicts of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, J., Feng, F., Han, T. et al. Detection of Autism Spectrum Disorder using fMRI Functional Connectivity with Feature Selection and Deep Learning. Cogn Comput 15, 1106–1117 (2023). https://doi.org/10.1007/s12559-021-09981-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12559-021-09981-z