Abstract
Automatic and reliable segmentation of hippocampus from MR brain images is of great importance in studies of neurological diseases, such as epilepsy and Alzheimer’s disease. In this paper, we proposed a novel metric learning method to fuse segmentation labels in multi-atlas based image segmentation. Different from current label fusion methods that typically adopt a predefined distance metric model to compute a similarity measure between image patches of atlas images and the image to be segmented, we learn a distance metric model from the atlases to keep image patches of the same structure close to each other while those of different structures are separated. The learned distance metric model is then used to compute the similarity measure between image patches in the label fusion. The proposed method has been validated for segmenting hippocampus based on the EADC-ADNI dataset with manually labelled hippocampus of 100 subjects. The experiment results demonstrated that our method achieved statistically significant improvement in segmentation accuracy, compared with state-of-the-art multi-atlas image segmentation methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Hippocampus is an important subcortical structure whose function is associated with learning and memory (den Heijer et al. 2012). Volumetric analysis of the hippocampus based on magnetic resonance imaging (MRI) has been widely adopted in studies of neurological diseases, such as epilepsy (Akhondi-Asl et al. 2011) and Alzheimer’s disease (Wolz et al. 2014). However, manual segmentation of the hippocampus from MRI brain images is time consuming (Carmichael et al. 2005) and suffers from high intra-operator and inter-operator variability (Chupin et al. 2007). Therefore, automatic and reliable segmentation of the hippocampus from MR brain images has been a hot research topic in medical image analysis.
In the last decade, multi-atlas based image segmentation (MAIS) methods have been developed and widely adopted in studies of the hippocampus segmentation (Warfield et al. 2004, Heckemann et al. 2006, Artaechevarria et al. 2009, Dill et al. 2015, Iglesias and Sabuncu 2015). A typical MAIS method consists of three steps: atlas image selection, atlas image registration, and segmentation label fusion. In the atlas image selection step, a subset of atlas images is selected for a given target image based on a pre-defined measurement of anatomical similarity, usually according to image intensities, e.g., sum of squared differences, correlation, or mutual information (Aljabar et al. 2009, Xie and Ruan 2014, Yan et al. 2015). In the atlas image registration step, the spatial correspondence between each atlas image and the target image is determined and the atlas images and their corresponding label maps are aligned to the target image (Lötjönen et al. 2010, Doshi et al. 2016). Finally, in the segmentation label fusion step, the warped label maps are fused to get a consensus label map for the target image (Warfield et al. 2004, Artaechevarria et al. 2009, Coupé et al. 2011, Hao et al. 2014).
Although a variety of atlas image selection strategies and different image registration techniques can be adopted in an MAIS method, the existing MAIS methods are typically characterized by their label fusion strategies. Among the existing label fusion strategies, weighted voting label fusion methods have attracted considerable attention. Assuming that the image registration from atlas images to the target image is reliable, traditional weighted voting label fusion strategies combine the corresponding labels based on predefined weighting models (Rohlfing et al. 2004, Heckemann et al. 2006, Artaechevarria et al. 2009, Sabuncu et al. 2010). The simplest method might be the majority voting which assigns a constant weight value for all atlases (Rohlfing et al. 2004, Heckemann et al. 2006). Better segmentation performance can be obtained with more sophisticated voting strategies, such as local weighted voting with inverse similarity metric (Artaechevarria et al. 2009) and local weighted voting with Gauss similarity metric (Sabuncu et al. 2010). It has been shown that local weighted voting strategies outperform global methods in segmenting high-contrast structures, but global techniques are less sensitive to noise when contrast between neighboring structures is low (Artaechevarria et al. 2009). Some of the weighted voting label fusion methods can be seen as special cases of a probabilistic generative model (Sabuncu et al. 2010).
Due to inter-subject anatomical variability, the registered atlas images are not always aligned with the target image perfectly. The image registration errors may hamper the label fusion if it is based on local image similarity measures with an assumption that voxel to voxel correspondence exists between atlas images and the target image. Such a problem can be effectively overcome by nonlocal patch based weighted voting methods (Coupé et al. 2011, Rousseau et al. 2011). In the nonlocal patch based weighted voting methods, all voxels in a searching region are selected and patches centered at these voxels are extracted as image patches in each warped atlas image. Voting weights are then computed according to the intensity similarities between the atlas image patches and the target image patch.
Many approaches have been proposed to obtain weighting coefficients for improving segmentation accuracy and robustness of the nonlocal patch based weighted voting methods, for example reconstruction based methods (Liao et al. 2013, Wu et al. 2014) and joint label fusion (JLF) method (Wang et al. 2013). Reconstruction based methods computed the reconstruction coefficients of the target patch from a patch library by sparse representation (Liao et al. 2013) or local independent projection (Wu et al. 2014), and then used them to combine atlas labels to label the target voxel. Since different atlases may produce similar label errors (Wang et al. 2013), the JLF method minimized the total expectation of labeling error by explicitly modeling pair-wise dependency between atlases as a joint probability of two atlases that make similar segmentation errors.
The existing MAIS methods typically measure the similarity of image patches based on Euclidean distance metric. However, Euclidean distance metric is not necessarily optimal for the label fusion since they do not characterize any statistical distributions of image intensities in the patches. The statistical distributions of image intensities could be estimated from the atlas images and their associated segmentation labels, but might vary at different image locations. It has been reported that patches with similar intensity values may have different segmentation labels, which will lead to segmentation errors in MAIS methods (Bai et al. 2015). To overcome this problem, we present a kernel classification method for metric learning such that image patches of the same structure keep close to each other and those of different structures are separated. With the obtained metric, we develop an optimal nonlocal weighted voting label fusion method. We have validated the proposed method for segmenting the hippocampus from MRI brain images, and compared our method with state-of-the-art MAIS techniques, including majority voting method (MV) (Rohlfing et al. 2004, Heckemann et al. 2006), local weighted voting with Inverse similarity metric (LW-INV) (Artaechevarria et al. 2009), local weighted voting with Gauss similarity metric (LW-GU) (Sabuncu et al. 2010), nonlocal patch based weighted voting with Gauss similarity metric (NLW-GU) (Coupé et al. 2011, Rousseau et al. 2011), local label learning (LLL) (Hao et al. 2014), and the JLF method (Wang et al. 2013). The experimental results have demonstrated that our method could achieve better segmentation performance than the state-of-the-art MAIS methods.
Materials and Methods
Image Dataset
The proposed algorithm was validated for segmenting the hippocampus based on the first release of EADC-ADNI dataset, consisting MRI scans and their corresponding hippocampus labels of 100 subjects (www.hippocampal-protocol.net). These images were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI, RRID:SCR_003007) database (adni.loni.usc.edu/), and the subjects are from 3 diagnosis groups, including normal controls (NC), mild cognitive impairment (MCI), and patients with Alzheimer’s disease (AD).
The Principal Investigator of the ADNI is Michael W. Weiner, MD, VA Medical Center and University of California-San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research, approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years, and 200 people with early AD to be followed for 2 years. For up-to-date information, see www.adni-info.org.
Each of the MRI brain images was manually labeled according to a harmonized protocol (Boccardi et al. 2015). All images have been processed using a standard preprocessing protocol, including alignment along the line passing through the anterior and posterior commissures of the brain (AC-PC line) and bias field correction. And they have been warped into the MNI152 template space using linear image registration with affine transformation. We randomly select 40 subjects as training set, and other 60 subjects as testing set. Clinical scores and demographic information of these subjects are summarized in Table 1.
Metric Learning for Multi-Atlas Based Image Segmentation
Given a target image I, and N atlases \( {\overset{\sim }{A}}_i=\left({\overset{\sim }{I}}_i,{\overset{\sim }{L}}_i\right),i=1,2,\dots, N \), where \( {\overset{\sim }{I}}_i \) is the i-th image and \( {\overset{\sim }{L}}_i \) is its segmentation label with value 1 indicating foreground and 0 indicating background, the multi-atlas segmentation method registers each atlas image \( {\overset{\sim }{I}}_i \) to the target image and propagates the corresponding segmentation \( {\overset{\sim }{L}}_i \) to the target space, resulting N warped atlases A i = (I i , L i ) , i = 1 , 2 , … , N. Then, it infers the label of each voxel of the target image from the warped atlases. Figure 1 shows a flowchart for segmenting an image with a typical multi-atlas image segmentation method.
Identification of a Bounding Box of Hippocampus
Since all images were aligned to the MNI152 template using linear image registration with affine transformation and resampled to have voxel size of 1x1x1mm3, a bounding box can be identified for both the left and right hippocampus to cover the hippocampus of unseen target image. In particular, we scan all the atlases to find the minimum and maximum x, y, z positions of the hippocampus and add 7 voxels in each direction to cover the hippocampus of unseen testing images.
Atlas Selection and Image Registration
For each target image, we select the top 20 most similar atlases based on normalized mutual information (NMI) between the target image and the atlas images within the bounding box (Hao et al. 2014). After the atlas selection, we register each atlas image to the target image using a nonlinear, cross-correlation-driven image registration algorithm, namely ANTs (Avants et al. 2008), with the following command: ANTS 3 -m CC [target.nii, source.nii, 1, 2] –i 100x100x10 –o output.nii -t SyN[0.25] –r Gauss[3,0]. The nonlinear registration was applied to the image blocks within the bounding box.
Initial Segmentation with Majority Voting
To reduce the computational cost, we adopt the majority voting based label fusion to obtain an initial segmentation result of the target image. For each voxel, the output of the majority voting label fusion is a probability value of the voxel belonging to the hippocampus. The segmentation result of voxels with 100 % certainty (probability value of 1 or 0) can be directly taken as the final segmentation result (Hao et al. 2014). Then, our method is applied to voxels with probability values greater than 0 and smaller than 1.
Training Patch Library Construction
To label a voxel of the target image, a set of voxel-wise training samples is identified from the warped atlases. Since the registered atlas images are not always aligned with the target image perfectly, we adopt the nonlocal patch based label fusion framework to construct a training library of image patches (Coupé et al. 2011, Rousseau et al. 2011). For labeling a target voxel, voxels in a cube-shaped searching neighborhood V with size (2r s + 1) × (2r s + 1) × (2r s + 1) of each atlas image are selected, and patches centered at these voxels are extracted and vectorized to form a patch library P = [p 1, p 2, … , p n ], where n = N ∙ (2r s + 1)3 is the number of selected patches. And the segmentation label of each image patch’s center voxel is used as the image patch’s label, l i , i = 1 , 2 , … , n. Thus, we construct a training dataset Δ = {(p i , l i )| i = 1, 2, … , n}, where p i is the i-th image patch in the patch library P and l i is the label of its center voxel.
Metric Learning
Learning a distance metric from training samples is an important machine learning topic. Many methods have been proposed to learn distance/similarity metrics (Xing et al. 2002). Among them, learning a Mahalanobis distance metric for k-nearest neighbor classification has been successfully applied to many computer vision problems (Guillaumin et al. 2009). In this study, we adopt a supervised metric learning method to learn a Mahalanobis distance metric from the training dataset of image patches (Wang et al. 2015).
Given any two samples (p i , l i ) and (p j , l j ) from the training dataset Δ, we obtain a doublet (p i , p j ) with a label h, where h = − 1 if l i = l j , and h = 1 otherwise. For each training sample p i , we find its m 1 nearest similar neighbors, denoted by \( \left\{{p}_{i,1}^s,\dots, {p}_{i,{m}_1}^s\right\} \), and its m 2 nearest dissimilar neighbors, denoted by \( \left\{{p}_{i,1}^d,\dots, {p}_{i,{m}_2}^d\right\} \), and construct (m 1 + m 2) doublets:
By collecting all possible doublets, we build a doublet set, denoted by \( \left\{{\boldsymbol{z}}_1,\dots, {\boldsymbol{z}}_{N_d}\right\} \), where z j = (p j , 1, p j , 2) , j = 1 , 2 , … , N d , and the label of z j is denoted by h j . Given the doublet set \( \left\{{\boldsymbol{z}}_1,\dots, {\boldsymbol{z}}_{N_d}\right\} \), we use a kernel method to learn a classifier
where z j is the j-th doublet, h j is its label, \( \boldsymbol{z}=\left({p}_{k_1},{p}_{k_2}\right) \) is a testing doublet, K(∙, ∙) is a degree-2 polynomial kernel, defined as.
Then, we have
where M = ∑ j h j α j (p j , 1 − p j , 2)(p j , 1 − p j , 2)T is the matrix to be learned in the Mahalanobis distance metric. Once M is obtained, the kernel decision function g(z) can be used to determine whether \( {p}_{k_1} \) and \( {p}_{k_2} \) are similar or dissimilar to each other.
To learn M in the Mahalanobis metric, we adopt a support vector machine (SVM) model:
where ‖∙‖ F is the Frobenius norm. The Lagrange dual problem of the above doublet-SVM model is
The optimization problem can be solved using SVM solvers. In the current study, we implemented the metric learning method based on LibSVM and metric learning codes (Chang and Lin 2011, Wang et al. 2015).
To ensure M to be positive semi-definite, we compute a singular value decomposition of M = UΛV, and preserve only the positive singular values in Λ to form another diagonal matrix Λ +. Then, we let M + = UΛ + V.
Label Fusion with the Learned Metric
With the learned Mahalanobis distance metric M, we obtain a new metric space by introducing a norm ‖∙‖ M : \( {\left\Vert x\right\Vert}_M=\sqrt{x^T Mx} \). And the distance between two samples is defined by d(x, y) = ‖x − y‖ M .
Given a target image patch p x and training image patches p i , i = 1 , 2 , … , n, we compute their distances by
According to these distances, we select k nearest training samples \( \left\{\left({p}_{s_j},{l}_{s_j}\right)|j=1,2,\dots, k\right\} \) to form a nearest neighborhood set \( {\mathcal{N}}_k\left({p}_x\right) \), and assign their similarity weights to be one, others to be zero:
Then, we use \( \widehat{L}(x)=\frac{\sum_{i=1}^nw\left({p}_x,{p}_i\right){l}_i}{\sum_{i=1}^nw\left({p}_x,{p}_i\right)} \) to compute the target voxel’s label. Finally, the estimated label of \( \widehat{L}(x) \) is thresholded to obtain a binary segmentation label \( L(x)=\left\{\begin{array}{c}1,\kern0.75em \widehat{L}(x)>0.5\\ {}\ 0,\kern0.75em \widehat{L}(x)<0.5\end{array}\right.. \)
In the weighted voting label fusion, two strategies are available to achieve label fusion: single-point estimation strategy and multi-point strategy. In the single-point estimation strategy the label estimated from each image patch is applied to its center voxel. In the multi-point estimation strategy, the label estimated from each patch is applied to all voxels covered by the image patch itself (Rousseau et al. 2011, Wang et al. 2013, Sanroma et al. 2015). Since each voxel has multiple estimated labels from image patches that cover the voxel itself, majority voting of the multiple estimated labels can be adopted to compute a final segment label.
Experiments
We optimized the parameters of our method based on the training dataset, and then evaluated the segmentation performance based on the testing dataset. We adopted 9 segmentation evaluation measures to evaluate the image segmentation results (Jafari-Khouzani et al. 2011). By denoting A as the manual segmentation, B as the automated segmentation, and V(X) as the volume of segmentation result X, these evaluation measures are defined as:
HD95:similar to HD,except that 5% data points with the largest distance are removed before calculation,
In the above definition, ∂A denotes the boundary voxels of A, d(∙, ∙) is the Euclidian distance of two points, card{∙} is the cardinality of a set.
Optimization of Parameters
The proposed method has following parameters: patch radius r p , searching radius r s , regularization parameter C in SVM, numbers of the nearest similar and dissimilar neighbors m 1, m 2 for constructing doublets, and number of the nearest neighbors k for selecting the most similar samples for label fusion. According to (Wang et al. 2015), we set C = 1, m 1 = m 2 = 1. We also fixed the searching radius r s = 1 (within a searching neighborhood of 3 × 3 × 3), since a nonlinear image registration algorithm was used to warp atlas images to the target image.
The other two parameters r p and k were determined empirically in {1, 2, 3} and {3, 9, 27}, respectively, based on the training set with 40 leave-one-out cross-validation experiments. Figure 2 shows average segmentation accuracy measured by the Dice index across 40 leave-one-out cross-validation experiments with different combinations of parameters, indicating that the optimal segmentation performance could be obtained with r p = 1 and k = 9.
Comparison with Existing MAIS Methods
The proposed method, referred to as nonlocal patch based weighted voting with metric learning (NLW-ML) hereafter, was compared with 6 state-of-the-art MAIS methods, including MV (Rohlfing et al. 2004, Heckemann et al. 2006), LW-INV (Artaechevarria et al. 2009), LW-GU (Sabuncu et al. 2010), NLW-GU (Coupé et al. 2011, Rousseau et al. 2011), LLL (Hao et al. 2014), and JLF (Wang et al. 2013).
The parameters of all these methods were optimized based on the same training dataset with the same parameter selection strategy. For LW-GU, patch radius r p and σ x in the Gauss similarity metric need to be determined. With cross-validation, the optimal value of r p was 2 selected from {1, 2, 3}, and σ x was adaptively set as \( {\sigma}_x={\mathit{\min}}_{x_i}\left\{{\left\Vert P(x)-P\left({x}_i\right)\right\Vert}_2+\varepsilon \right\},i=1..N \), where ε is a small constant to ensure numerical stability with a value 1e-20. LW-INV has 2 parameters, namely patch radius r p and γ in the inverse function model. The optimal values were r p = 2 and γ = − 3, obtained from the range of {1, 2, 3} and {−0.5, −1, −2, −3} respectively. NLW-GU has 3 parameters, namely searching radius r s , patch radius r p , and σ x in the Gauss similarity metric model. Similar to the NLW-ML, the searching radius r s was set to be 1. Based on the same cross-validation strategy, the optimal value of r p was 1 selected from {1, 2, 3}, and σ x was adaptively set as \( {\sigma}_x={\mathit{\min}}_{x_{s,j}}\left\{{\left\Vert P(x)-P\left({x}_{s,j}\right)\right\Vert}_2+\varepsilon \right\},s=1..N,j\in V \), where ε is a small constant to ensure numerical stability with a value 1e-20.
The only difference between NLW-GU and LW-GU was the image patches that they used. Particularly, NLW-GU used nonlocal image patches, i.e., the searching radius r s > 0 was used to extract image patches. In contrast, LW-GU used local image patches, i.e., the searching radius r s = 0. Since both of the NLW-GU method and the proposed NLW-ML method use non-local image patches, the only difference between them is the distance metric for measuring similarity between image patches. In the experiment, we found that the multi-point estimate strategy was better than the single-point strategy in all of these label fusion methods. Thus, we only report the results obtained with the multi-point strategy.
Similar to NLW-ML and NLW-GU, the searching radius rs for LLL and JFL was set to be 1. Other parameters of these two methods were optimized based on the same training set with the same parameter optimization strategy as adopted by the proposed method. For the LLL method, the optimal patch radius rp and the optimal number of training samples K were rp = 3 and K = 300, selected from {1, 2, 3} and {300, 400, 500}, respectively. Sparse linear SVM classifiers with default parameter (C = 1) were built to fuse labels in the LLL method. The single-point label fusion strategy was used in the LLL method. For the JLF method, the optimal patch radius rp and the optimal parameter β in the pairwise joint label difference term were rp = 1 and β = 1, selected from {1, 2, 3} and {0.5, 1, 1.5, 2}, respectively.
Table 2 summarizes segmentation results of the testing images obtained by the segmentation methods under comparison, including MV, LW-INV, LW-GU, NLW-GU, LLL, JLF, and NLW-ML. For each segmentation evaluation measure, the best value is shown in bold. These results indicated that the proposed method achieved the best overall performance. Specifically, Wilcoxon signed rank tests indicated that the proposed method performed significantly better than MV, LW-INV, LW-GU, NLW-GU, LLL (p < 0.001) and JLF (p < 0.05) in terms of Dice and Jaccard index values of their segmentation results. The results also demonstrated that NLW-GU performed better than LW-GU, indicating that the non-local patch based methods had better performance than traditional methods that adopted only corresponding image patches for label fusion (Coupé et al. 2011, Rousseau et al. 2011).
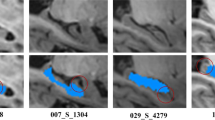
Figure 3 shows box plots of Dice and Jaccard index values of segmentation results obtained by different methods, indicating that our proposed method performed consistently better than other label fusion methods. The superior performance of our method was also confirmed by the visualization results, as shown in Fig. 4.
Hippocampal segmentation results obtained by different methods. One subject was randomly chosen from the dataset. The first row shows the segmentation results produced by different methods, the second row shows their corresponding surface rendering results, and the difference between results of manual and automatic segmentation methods was showed in the third row (red: manual segmentation results, green: automated segmentation results, blue: overlap between manual and automated segmentation results)
Discussion
The proposed method is a voting based label fusion method (Liao et al. 2013, Wu et al. 2014, Tong et al. 2015, Wu et al. 2015) with an integrated learning component (Hao et al. 2012b, Hao et al. 2014, Wang et al. 2014, Bai et al. 2015, Zhu et al. 2015). The voting based label fusion methods compute the voting weights by comparing the target image patch with each atlas image patches, and use them to combine atlas labels. In contrast, machine learning based methods utilize machine learning techniques to build a mapping between the segmentation label and the image appearance. The voting based methods typically assume that image patches with similar intensity information have the same segmentation label. Although this assumption is valid in most cases, a recent study has shown that similar image patches could bear different labels (Bai et al. 2015). The machine learning based methods overcome this limitation by learning a mapping function between the image patch and the label. The proposed method combines advantages of the existing methods by first adopting a classification method to learn the relationship between image patches and segmentation labels and then fusing the labels based on weights obtained with the learned distance metric.
The metric learning is essentially a preprocessing step in pattern recognition, aiming to learn from a given training dataset a distance metric, with which data samples can be more effectively classified (Weinberger and Saul 2009). In this study, we empirically demonstrated that the metric learning in conjunction with a k-NN classifier could lead to better performance for segmenting the hippocampus from MRI scans than state-of-the-art MAIS methods, including the LLL and JFL methods. We postulate that its promising performance might due to that the k-NN classifier could potentially capture nonlinear relationship that better model the image patches of background and hippocampus than linear models built by the other methods, such as the sparse linear SVM adopted in the LLL method. In fact, many metric learning methods have been demonstrated to achieve state-of-the-art performance on pattern recognition problems (Weinberger and Saul 2009).
In our method, we used nonlinear image registration to register image blocks of the hippocampus. Our results demonstrated that a small patch size was good enough to capture inter-subject anatomical differences. Since the metric learning could adaptively learn a distance metric for image patches from training data, our method is not sensitive to the patch size as the traditional patch based methods.
The computational burden for image registration is a major issue in the multi-atlas segmentation methods. To avoid the high computational cost of non-rigid image registration, non-local patch-based image labeling strategies were proposed so that linear image registration could be used to align the image to be segmented and the atlas images (Coupé et al. 2011). However, a non-local image patch searching procedure has to be adopted to identify similar image patches in the label fusion step, which often leads to higher computational cost than using non-rigid image registration in the atlas image registration (Rousseau et al. 2011). More recently, an optimized patch match strategy was proposed to improve the segmentation (Giraud et al. 2016). In the current study, we adopted an atlas selection strategy to reduce the computational cost associated with the nonlinear image registration (Aljabar et al. 2009, Hao et al. 2014). Particularly, the most informative atlases were first selected before the nonlinear image registration. Following (Hao et al. 2014), we selected 20 atlas images for segmenting each target image. The computational complexity of our label fusion method is similar to classification based methods (Hao et al. 2014, Bai et al. 2015). For a MATLAB based implementation of our algorithm, it took ~20 min to fuse labels for segmenting one side of the hippocampus on a personal computer with 4 cores of 3.4G HZ CPU.
It is straightforward to extend the metric learning method for multi-class classification problems in that the metric learning maximizes margin between differences of intra-class and inter-class samples. However, for most brain region segmentation problems with multiple regions to be segmented we could formulate the multi-class classification problem as multiple one-against-the-rest binary classification problems. Such a setting might be better to handle unbalanced training samples since we build local classifiers for different voxels of the brain instead of a global one for the whole brain voxels.
Our future work will integrate the supervised metric learning method and more sophisticated weighted voting label fusion methods, such as joint label fusion (Wang et al. 2013), in which label error is measured by the distance of patches with a predefined distance metric. Furthermore, our method can also be adopted in the shape constrained segmentation framework (Hao et al. 2012a). We will also combine our method with functional MRI image based hippocampus parcellation (Cheng and Fan 2014).
Conclusion
In the paper, we propose a novel nonlocal patch based weighted voting label fusion method with a learned distance metric for measuring similarity between image patches. The validation experimental results have demonstrated that the proposed method could achieve better segmentation performance than start-of-the-art MAIS methods, indicating that the learned distance metric for measuring similarity of image patches could improve the segmentation performance.
Information Sharing Statement
Software developed in this manuscript is available upon request from Dr. Fan or Dr. Zhu.
References
Akhondi-Asl, A., Jafari-Khouzani, K., Elisevich, K., & Soltanian-Zadeh, H. (2011). Hippocampal volumetry for lateralization of temporal lobe epilepsy: automated versus manual methods. NeuroImage, 54, S218–S226.
Aljabar, P., Heckemann, R., Hammers, A., Hajnal, J., & Rueckert, D. (2009). Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. NeuroImage, 46, 726–738.
Artaechevarria, X., Munoz-Barrutia, A., & Ortiz-de-Solorzano, C. (2009). Combination strategies in multi-atlas image segmentation: Application to brain MR data. IEEE Transactions on Medical Imaging, 28, 1266–1277.
Avants, B. B., Epstein, C. L., Grossman, M., & Gee, J. C. (2008). Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis, 12, 26–41.
Bai, W., Shi, W., Ledig, C., & Rueckert, D. (2015). Multi-atlas segmentation with augmented features for cardiac MR images. Medical Image Analysis, 19, 98–109.
Boccardi, M., Bocchetta, M., Morency, F. C., Collins, D. L., Nishikawa, M., Ganzola, R., Grothe, M. J., Wolf, D., Redolfi, A., & Pievani, M. (2015). Training labels for hippocampal segmentation based on the EADC-ADNI harmonized hippocampal protocol. Alzheimer's & Dementia, 11, 175–183.
Carmichael, O. T., Aizenstein, H. A., Davis, S. W., Becker, J. T., Thompson, P. M., Meltzer, C. C., & Liu, Y. (2005). Atlas-based hippocampus segmentation in Alzheimer’s disease and mild cognitive impairment. NeuroImage, 27, 979–990.
Chang, C. C., & Lin, C. J. (2011). LIBSVM: A library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST), 2(3), 27.
Cheng, H., & Fan, Y. (2014). Functional parcellation of the hippocampus by clustering resting state fMRI signals. In: 2014 I.E. 11th International Symposium on Biomedical Imaging (ISBI), pp 5–8.
Chupin, M., Mukuna-Bantumbakulu, A. R., Hasboun, D., Bardinet, E., Baillet, S., Kinkingnéhun, S., Lemieux, L., Dubois, B., & Garnero, L. (2007). Anatomically constrained region deformation for the automated segmentation of the hippocampus and the amygdala: method and validation on controls and patients with Alzheimer’s disease. NeuroImage, 34, 996–1019.
Coupé, P., Manjón, J. V., Fonov, V., Pruessner, J., Robles, M., & Collins, D. L. (2011). Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. NeuroImage, 54, 940–954.
den Heijer, T., van der Lijn, F., Vernooij, M. W., de Groot, M., Koudstaal, P., van der Lugt, A., Krestin, G. P., Hofman, A., Niessen, W. J., & Breteler, M. M. (2012). Structural and diffusion MRI measures of the hippocampus and memory performance. NeuroImage, 63, 1782–1789.
Dill, V., Franco, A. R., & Pinho, M. S. (2015). Automated methods for hippocampus segmentation: the evolution and a review of the state of the art. Neuroinformatics, 13, 133–150.
Doshi, J., Erus, G., Ou, Y., Resnick, S. M., Gur, R. C., Gur, R. E., Satterthwaite, T. D., Furth, S., & Davatzikos, C. (2016). MUSE: MUlti-atlas region Segmentation utilizing Ensembles of registration algorithms and parameters, and locally optimal atlas selection. NeuroImage, 127, 186–195.
Giraud, R., Ta, V.-T., Papadakis, N., Manjón, J. V., Collins, D. L., Coupé, P., & Initiative, A. D. N. (2016). An Optimized PatchMatch for multi-scale and multi-feature label fusion. NeuroImage, 124, 770–782.
Guillaumin M, Verbeek J, Schmid C (2009) Is that you? Metric learning approaches for face identification. In: Computer Vision, 2009 I.E. 12th International Conference on, pp 498–505: IEEE.
Hao, Y., Jiang, T., & Fan, Y. (2012a). Shape-constrained multi-atlas based segmentation with multichannel registration. SPIE Medical Imaging. International Society for Optics and Photonics, pp. 83143N-83143N-83148.
Hao, Y., Liu, J., Duan, Y., Zhang, X., Yu, C., Jiang, T., & Fan, Y. (2012b). Local label learning (L3) for multi-atlas based segmentation. SPIE Medical Imaging. International Society for Optics and Photonics, pp. 83142E-83142E-83148.
Hao, Y., Wang, T., Zhang, X., Duan, Y., Yu, C., Jiang, T., & Fan, Y. (2014). Local label learning (LLL) for subcortical structure segmentation: Application to hippocampus segmentation. Human Brain Mapping, 35, 2674–2697.
Heckemann, R. A., Hajnal, J. V., Aljabar, P., Rueckert, D., & Hammers, A. (2006). Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage, 33, 115–126.
Iglesias, J. E., & Sabuncu, M. R. (2015). Multi-atlas segmentation of biomedical images: A survey. Medical Image Analysis, 24, 205–219.
Jafari-Khouzani, K., Elisevich, K. V., Patel, S., & Soltanian-Zadeh, H. (2011). Dataset of magnetic resonance images of nonepileptic subjects and temporal lobe epilepsy patients for validation of hippocampal segmentation techniques. Neuroinformatics, 9, 335–346.
Liao, S., Gao, Y., Lian, J., & Shen, D. (2013). Sparse patch-based label propagation for accurate prostate localization in CT images. IEEE Transactions on Medical Imaging, 32, 419–434.
Lötjönen, J. M. P., Wolz, R., Koikkalainen, J. R., Thurfjell, L., Waldemar, G., Soininen, H., & Rueckert, D. (2010). Fast and robust multi-atlas segmentation of brain magnetic resonance images. NeuroImage, 49, 2352–2365.
Rohlfing, T., Brandt, R., Menzel, R., & Maurer Jr., C. R. (2004). Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. NeuroImage, 21, 1428–1442.
Rousseau, F., Habas, P. A., & Studholme, C. (2011). A supervised patch-based approach for human brain labeling. IEEE Transactions on Medical Imaging, 30, 1852–1862.
Sabuncu, M. R., Yeo, B. T. T., Van Leemput, K., Fischl, B., & Golland, P. (2010). A generative model for image segmentation based on label fusion. IEEE Transactions on Medical Imaging, 29, 1714–1729.
Sanroma, G., Wu, G., Gao, Y., Thung, K.-H., Guo, Y., & Shen, D. (2015). A transversal approach for patch-based label fusion via matrix completion. Medical Image Analysis, 24, 135–148.
Tong, T., Wolz, R., Wang, Z., Gao, Q., Misawa, K., Fujiwara, M., Mori, K., Hajnal, J. V., & Rueckert, D. (2015). Discriminative dictionary learning for abdominal multi-organ segmentation. Medical Image Analysis, 23, 92–104.
Wang, H., Suh, J. W., Das, S. R., Pluta, J. B., Craige, C., & Yushkevich, P. A. (2013). Multi-atlas segmentation with joint label fusion. IEEE Transactions on Pattern Analysis and Machine Intelligence, 35, 611–623.
Wang, H., Cao, Y., & Syeda-Mahmood, T. (2014). Multi-atlas segmentation with learning-based label fusion. Machine learning in Medical Imaging, 256–263.
Wang, F., Zuo, W., Zhang, L., Meng, D., & Zhang, D. (2015). A kernel classification framework for metric learning. IEEE Transactions on Neural Networks and Learning Systems, 26, 1950–1962.
Warfield, S. K., Zou, K. H., & Wells, W. M. (2004). Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Transactions on Medical Imaging, 23, 903–921.
Weinberger, K. Q., & Saul, L. K. (2009). Distance metric learning for large margin nearest neighbor classification. Journal of Machine Learning Research, 10, 207–244.
Wolz, R., Schwarz, A. J., Yu, P., Cole, P. E., Rueckert, D., Jack, C. R., Raunig, D., Hill, D., & Initiative AsDN (2014). Robustness of automated hippocampal volumetry across magnetic resonance field strengths and repeat images. Alzheimer's & Dementia, 10, 430–438 e432.
Wu, Y., Liu, G., Huang, M., Guo, J., Jiang, J., Yang, W., Chen, W., & Feng, Q. (2014). Prostate segmentation based on variant scale patch and local independent projection. IEEE Transactions on Medical Imaging, 33, 1290–1303.
Wu, G., Kim, M., Sanroma, G., Wang, Q., Munsell, B. C., Shen, D., & Initiative, A. D. N. (2015). Hierarchical multi-atlas label fusion with multi-scale feature representation and label-specific patch partition. NeuroImage, 106, 34–46.
Xie, Q., & Ruan, D. (2014). Low-complexity atlas-based prostate segmentation by combining global, regional, and local metrics. Medical Physics, 41, 041909.
Xing, E. P., Jordan, M. I., Russell, S., & Ng, A. Y. (2002). Distance metric learning with application to clustering with side-information. In: Advances in Neural Information Processing Systems, pp 505–512.
Yan, P.-g., Cao, Y., Yuan, Y., Turkbey, B., & Choyke, P. L. (2015). Label Image Constrained Multiatlas Selection. IEEE transactions on Cybernetics, 45, 1158–1168.
Zhu, H., Cheng, H., & Fan, Y. (2015). Random local binary pattern based label learning for multi-atlas segmentation. SPIE Medical Imaging. International Society for Optics and Photonics, pp. 94131B-94131B-94138.
Acknowledgments
This work was supported in part by National Key Basic Research and Development Program (No. 2015CB856404), National Natural Science Foundation of China (No. 81271514, 61473296, 61602307), and NIH grants EB022573, CA189523, and AG014971.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Alzheimer’s Disease Neuroimaging Initiative (ADNI) is a Group/Institutional Author.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Rights and permissions
About this article
Cite this article
Zhu, H., Cheng, H., Yang, X. et al. Metric Learning for Multi-atlas based Segmentation of Hippocampus. Neuroinform 15, 41–50 (2017). https://doi.org/10.1007/s12021-016-9312-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-016-9312-y